Abstract
Genome-wide RNAi screening is a powerful, yet relatively immature technology that allows investigation into the role of individual genes in a process of choice. Most RNAi screens identify a large number of genes with a continuous gradient in the assessed phenotype. Screeners must then decide whether to examine just those genes with the most robust phenotype or to examine the full gradient of genes that cause an effect and how to identify the candidate genes to be validated. We have used RNAi in Drosophila cells to examine viability in a 384-well plate format and compare two screens, untreated control and treatment. We compare multiple normalization methods, which take advantage of different features within the data, including quantile normalization, background subtraction, scaling, cellHTS2 1, and interquartile range measurement. Considering the false-positive potential that arises from RNAi technology, a robust validation method was designed for the purpose of gene selection for future investigations. In a retrospective analysis, we describe the use of validation data to evaluate each normalization method. While no normalization method worked ideally, we found that a combination of two methods, background subtraction followed by quantile normalization and cellHTS2, at different thresholds, captures the most dependable and diverse candidate genes. Thresholds are suggested depending on whether a few candidate genes are desired or a more extensive systems level analysis is sought. In summary, our normalization approaches and experimental design to perform validation experiments are likely to apply to those high-throughput screening systems attempting to identify genes for systems level analysis.
Keywords: RNAi, high-throughput screen, normalization, validation
Introduction
Genome-wide RNA interference (RNAi)-based screening is a powerful technology to assign phenotype to a gene and is facilitating investigations into complex network interactions in a variety of organisms 5. Its compatibility with high-throughput technology 4 has revolutionized the quest for identification of novel genes in signal transduction pathways, human diseases, and drug targets 2,5,7. Publications from analysis of these types of screens are constantly being added to the literature, with relatively few publications addressing experimental design and data analysis. Although publications have addressed general protocols and logical approaches for gene selection 8 or introduced novel methods of data analysis and criteria for assessment of quality 1, there is a lack of data in the literature to compare and contrast data analysis methods and a lack of design of rigorous validation methods. Because RNAi-based screening is labor-intensive technology involving complex experimental setup, there is a deficiency of appropriate standards, and the current analytical tools are relatively limited, we address some of the deficiencies related to appropriate experimental design for Drosophila RNAi genomic screening, examine the available analytical methods, and develop a rigorous validation scheme.
Methods
Tissue Culture
Kc167 cells were grown in Schneider medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum (FBS) (65 °C, 10 minutes), 100 U/mL penicillin, and 100 μg/mL streptomycin, at 24 °C in a humidified chamber.
Genome-wide viability screen following MMS exposure
Methods were adapted from2, with 1.2 × 104 Kc167 cells suspended in 10 μL of serum-free Schneider media (Invitrogen) added to each well of 63 different 384-well plates. Each well contained, on average, 0.05 μg (0.01–0.43 μg) of a double-stranded (ds) RNA, with 22,915 individual dsRNA represented in the whole library (original Drosophila RNAi Screening Center (DRSC) library). One hour at 24 °C allowed phagocytic uptake of the dsRNA, then 20 μL of serum containing medium (15% heat-inactivated FBS) was added and the plates incubated for a further 72 h. Medium was exchanged for 30 μL of fresh serum containing medium (10% heat-inactivated FBS), with or without 0.004% (w/v) methyl methanesulfonate (MMS) (Sigma-Aldrich, St. Louis, MO). Following an additional 72 h incubation, medium was removed and the number of viable cells assessed using pre-diluted Celltitre-Glo (Promega, Madison, WI) per manufacturers instructions and an Analyst GT Reader (Molecular Devices, Sunnyvale, CA). Screens were performed in duplicate, with the replicate screen initiated on a different day.
Data analysis of library screen
It was assumed that the majority of the dsRNA on any one plate did not have any significant effect on viability, thus the median value of each plate was used for all normalization methods. Five different normalization methods were applied to the data, with two additional normalizations conducted by combining different methods.
Background subtraction and scaling
Background luminescence for each plate was calculated based on the median value of the plate and the value obtained for the well containing dsRNA targeting Thread (Th; CG12284), an anti-apoptotic gene. This positive control is present in well B02 of each library plate provided by the DRSC and consistently results in a loss of cell viability. For background subtraction, the luminescence value of the well containing Th was subtracted from the value of each other well in the plate. If after normalization the value was negative, the normalized value was set to zero.
Data was scaled as follows. For untreated plates the median value (muntrt) of each plate was assumed to be equivalent to 100% viability plus background (0% viability) and Thread (Thuntrt) to be equivalent to 40% viability plus background. 100% viability (100%untrt) was then calculated for each plate by:
| Eq. (1) |
0% viability (0%untrt) was calculated for each plate by:
| Eq. (2) |
Well values for each plate (wvuntrt) were then corrected for background and normalized to 100% viability (wvuntrt_norm) by:
| Eq. (3) |
Similarly, for the plates treated with MMS, the median value of the plate (mMMS) was assumed to be equivalent to 65% viability of untreated cells and Thread (ThMMS) equivalent to 30% viability of untreated cells, based on examination of actual cell number after treatment in other experiments (data not shown). The calculated 100% viability in the absence of MMS treatment (100%MMS) was calculated by:
| Eq. (4) |
0% viability (0%MMS) was calculated by:
| Eq. (5) |
Well values for each MMS exposed plate (wvMMS) were then corrected for background and normalized to the calculated 100% viability (wvMMS_norm) by:
| Eq. (6) |
These data were either examined by themselves or then further normalized to account for inter-plate variation by quantile normalization.
cellHTS2
The R package cellHTS2 1 designed for RNAi genome-wide screen analysis was employed with the per plate normalization function normalizePlates set to median and the summary function set to mean. Z-scores were obtained, and the difference between z-scores of treatment and control were determined in order to rank genes. Only two replicates of each plate were used in this analysis.
Interquartile range normalization
The interquartile range (IQR) is the difference between the first and third quartiles of a data set. The IQR was used to measure the difference of each well value from the median for each plate 10. This method is insensitive to the presence of outliers in the data set. Genes were ranked by the difference between the IQR measurements of treatment and control.
Quantile normalization
Quantile normalization 6 is based on comparing quantiles between data sets, with no special allowance for outliers. Each of the 63 plate types was quantile normalized between replicates of untreated and between replicates of MMS treatment, with usually two replicates in each group. Following quantile normalization, “M,” the moderated t-statistic in log2 fold change, was calculated for each well, by which genes were ranked. This resulted in the moderation of well values across genes and shrunk toward a common value using a Bayesian model. This is therefore a log-odds Bayesian statistic that a gene differentially affects viability 9.
Background normalizations combined with quantile normalization
To improve normalization by quantile normalization, background subtraction and scaling were applied to the data before normalization. Two new sets of normalized data were generated: background subtraction followed by quantile normalization (BSQN) and scaling followed by quantile normalization (SQN). Our scaling method followed by quantile normalization is very similar to Robust Multichip Average (RMA) analysis 6, which is a standard for microarray analysis. Neither background subtraction nor scaling had an effect on IQR measurement. cellHTS2 includes its own background correction method.
Production of dsRNA
dsRNA were produced similarly to that previously described 8 with the following exceptions: primer sequences were obtained from the DRSC for amplicons to be validated, and oligonucleotides were purchased from Invitrogen (Carlsbad, CA). Primers contain the T7 promoter sequence at their 5′ end to allow in vitro transcription. RNA from adult Drosophila was reverse-transcribed using the Impromp-II Reverse Transcriptase system (Promega) to produce cDNA. This cDNA was amplified with amplicon specific primers and Taq polymerase (Roche, Indianapolis, IN) using conditions previously described 8. Products were resolved by agarose gel and gel purified using a 96-well gel isolation kit custom made by Invitrogen. A second round of PCR using the same amplicon specific primers was conducted to increase the amplification. In vitro transcription was then performed using the T7 Ribomax Express RNAi system (Promega), the dsRNA product purified using Millipore nucleic acid filter plates and recovered in 10mM Tris HCl (pH 7.0). The resuspended product was quantified using a Spectramax M5 spectrophotometer (Molecular Devices). Concentration of dsRNA was adjusted to 0.08 μg/μL, the quality of which was verified by agarose gel resolution.
Validation analysis
For validations, experiments were performed as described 8 with the following modifications: dsRNA were created in template 96-well plates and aliquoted into four distributed wells of a 384-well plate at a final quantity of 0.4 μg using a Precision XS robot (BioTek, Winooski, VT). A distributed pattern was chosen to reduce plate location effects. Control dsRNA were included on each plate, including four wells with dsRNA against Th to demonstrate function of dsRNA knockdown; 40 wells of non-targeting dsRNA against luciferase (Luc) to provide a control for the presence of dsRNA; four wells of dsRNA targeting GCLc, a gene required for viability following MMS exposure, as a positive control for treatment; and four wells containing a high level of MMS (0.12% w/v) resulting in 99% cell death to determine the background luminescence in the absence of any viable cells. MMS exposure and control experiments were performed in separate plates.
Luminescence values for each well were scaled similar to the screen data, except the value for high MMS was used as 0%. For data analysis, scaled well values were normalized by dividing by the median of scaled Luc values, where wvnorm is the normalized well value, and mlvs is the median Luc value:
| Eq. (7) |
T-tests (2 tailed, Type II) were performed between normalized quadruplicates to determine if there is a difference between control and treatment. In RNAi based screens, however, removal of certain genes can result in decreased viability without any treatment, i.e. Th. In order to account for reduced cell number under these circumstances and to determine if the reduced viability is significantly different from the expected 65% viability following MMS exposure, a second T-Test (2 tailed, Type II) was performed on Percent Control Viability (PCV) for each targeting dsRNA against the PCV of Luc. From this, significant hits were selected using a difference denoting death, i.e. 55% or less viability compared with Luc.
The normalized quadruplicate dataset for each dsRNA experiment was used to perform a T-test to compare untreated cells against MMS treatment to determine the significance. In order to determine PCV, the cell density from the experimental wells were calculated as follows, where ECV is the estimated control viability, EMV is the estimated MMS viability, cwvnorm is the control well value, and mwvnorm is the MMS well value:
| Eq. (8) |
| Eq. (9) |
| Eq. (10) |
Using the PCV for MMS treatment, a second T-test was performed to determine the significant difference between the PCV for each dsRNA against PCV with Luc dsRNA. A significant effect was deemed for any dsRNA that, following MMS treatment, resulted in 55% or less viability compared to 65% PCV with Luc dsRNA, with significance being P < 0.001. High confidence positives were those with significant P-values after both T-tests, while dsRNA with a significant P-value by only one of the T-tests was considered positive with moderate confidence.
Comparison of screen normalization methods using validated data
ROC curve analysis was performed and graphs generated using STATA (StataCorp LP, College Station, TX).
Results and Discussion
Use of genome-wide RNAi has become an important strategy to investigate functional genomics. Reliability of data generated from such genome-wide screens, however, is suspect due to confounding variables, such as the dynamic range and variation in the data, biological variations in the effectiveness of RNAi and other physical constraints associated with experimental set up 3, which often causes significant problems for the screeners to choose appropriate statistical method for analysis. Though several statistical parameters for the analysis of high-throughput RNAi screen have been published 1,3,12,13, the applicability of these statistical methods depends upon the experimental set up of screen, which could vary among screening centers and depend upon the experimental output. Here we describe a strategy using a combination of normalization and analysis methods to improve the selection of a large number of genes, as well a robust design for experimental validation of these genes for prospective studies.
We have used a Drosophila genomic library of dsRNA to determine the genes required for viability when exposed to MMS. This was done by comparing the data derived from RNAi viability screens performed with MMS treatment to that performed without treatment (original (Supplemental Table 1) or normalized and validated data (Supplemental Table 2) are also available at www.flyrnai.org and at gccri.uthscsa.edu/ABPublished_Data.asp). In order to determine which of these genes were required after treatment, we devised a validation scheme to confirm the authenticity of 806 dsRNA as conferring reduced viability after treatment, corresponding to 835 wells in the original library. We then compared several normalization strategies to determine the ability of each method to predict those dsRNA that actually validate as having an effect on viability. This paper describes novel usage of established normalization methodologies; none of these methods except cellHTS2 have previously been applied to RNAi screens. We have then adapted these methods to conduct an original cross-screen comparison of our continuous data to examine those genes required for viability after a damaging treatment.
RNAi genomic screens are performed in 384-well plates with each well containing a dsRNA targeting a single gene; with the first DRSC dsRNA library, 63 such plates are used to screen the entire genome. The labor-intensive preparative procedures and costs involved in both the library production and the screening procedures limit the number of biological replicates for any particular experiment. With only two replicate plates available from the DRSC for any one screen, variability in the results obtained is of significant concern. Intra-plate variations arise due to physical constraints of the plate; “edge effects” can result from both evaporation at the edge and differences in fluorescent or luminescent signal bleed through from neighbouring wells. Inter-plate variations tend to occur due to the sheer volume of plates required for the experiments and the time and effort required in their processing. As such, the performance of biological replicates often occurs at different times, especially when conducting cross-screen comparisons–this may cause variability due to the use of different batches of reagent or cells. These problems are further compounded by inconsistencies in the concentration of dsRNA present in each well and a lack of knowledge regarding the requirement for effective concentration of dsRNA to achieve complete knock-down of their respective target genes. Irrespective, the data generated from RNAi screens have led to significant advancements in science 5. In order to obtain the highest quality data from these genome-wide screens, we have attempted several different normalization methods either designed specifically for RNAi screens or borrowed from genome-wide expression analysis tools. Genes selected from these analyses were subjected to a robust validation methodology both to confirm the selection for future analyses and to examine the quality of the data normalization methods. We have applied several stringent criteria in the validation experiments to reduce the variations observed in the original screen and embedded controls that can be used to normalize experiments and to monitor their quality.
Experimental set-up of screen and validation
In order to evaluate normalization methods, we analyzed the validation experiments to correctly identify true positives and negatives. Here we validated the same dsRNA as were tested in the original screen in order to analyze screen normalization methods. Correct validation for genes identified as affecting the phenotype of interest, however, should be done with a dsRNA targeting a different site on the mRNA.
Validation experiments were performed using the same dsRNA amplicon present in original genome-wide screen. Unlike in the original DRSC library, which had an average of about 0.05 μg dsRNA per well with a range up to about 0.43 μg (DRSC, personal communication), we used a normalized concentration of dsRNA, 0.4 μg dsRNA per well, to eliminate false negatives due to incomplete knock-down. This concentration was chosen based on that needed to achieve an effect on viability after MMS treatment after knock-down of GCLc, a gene required after MMS (Figure 1a). The concentration of 0.45 μg provided the maximum effect on viability, whereas the lower concentrations did not, presumably due to incomplete knock-down. With a high level of dsRNA and a consistent concentration in every well, complete knock-down is assured for almost every gene targeted.
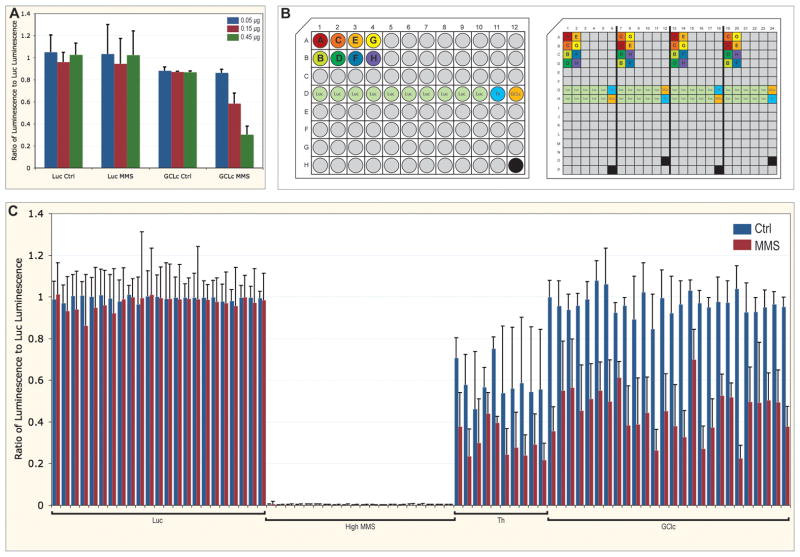
Figure 1.
Arrangement and analysis of validation experiments. A. Ratio of viability of cells with dsRNA targeting GCLc normalized to cells with non-targeting dsRNA against luciferase, with and without MMS treatment. Concentrations of dsRNA targeting GCLc were 0.05 (blue), 0.15 (red), and 0.45 μg (green). Error bars are standard deviation for quadruplicates. B. Layouts of the 96-well stock plate and 384-well assay plate. Unique dsRNA are filled grey, with eight of these labelled A through H and colored. Controls of dsRNA against luciferase (Luc, green), Thread (Th, light blue), and GCLc (orange) are also indicated. The black wells are empty. In the stock plate, each dsRNA is represented only once, except dsRNA targeting Luc. The assay plate is aliquoted using a robot with an eight-channel pipettor, and is comprised of four copies of the stock plate, with a distributed pattern to reduced edge effects (notice positioning of dsRNA A through H, this pattern is continued for all grey (unique) dsRNA). In the assay plate, a high concentration of MMS is added to the empty well. C. Ratio of viability of embedded controls in assay plates, with (red) and without (blue) MMS treatment, normalized to the median Luc value for each plate. Error bars are standard deviation for 40 replicates (Luc) or quadruplicates (high MMS, Th, and GCLc).
The original genome-wide RNAi screen available from the DRSC is only provided in duplicate, with each dsRNA unique on each plate. In our validation method, control and MMS treatment were performed simultaneously with quadruplicates of dsRNA present in the same plate, to lessen effects due to variation between biological replicates and avoid inter-plate variation (Figure 1b). Additionally, in an attempt to alleviate variation due to edge effects, validation experiments were set up with the quadruplicate dsRNA arranged in a distributed pattern within the plate (Figure 1b). The pattern is limited by the physical constraints of the robotics used, but was designed so that a single dsRNA would never be located at the edge in every instance.
Decreased cell growth in wells located on the edges of the plate is due to excessive evaporation of media and can result in edge effects. Despite maintaining 384-well plates in a humidified container, such edge effects are especially severe in plates located on top of a plate-stack. These edge effects can be diminished by placing either plates containing water or wet paper towels on top of the stack.
We included several controls in our validation experiments (Figure 1b). For viability assays, the DRSC screening plates provide the positive control of dsRNA targeting the anti-apoptotic gene Th, but it is in well B02, near the outside of the plate. As a negative control, dsRNA against GFP, not present in the Drosophila genome, is provided in well A02, on the plate edge. We found that this dsRNA against GFP had an effect after MMS treatment (data not shown) and discovered that the dsRNA had possible off-targets (data not shown). We therefore designed dsRNA against another gene not present in the genome, luciferase (Luc), to be used as a non-targeting control. In order to confidently normalize the plates to a dsRNA standard with truly no effect, dsRNA against Luc was included in 40 wells of each plate. To ensure the presence and functionality of dsRNA knock-down, dsRNA targeting Th also was included. It should be noted that we observed that dsRNA against Th did not result in the death of all cells (albeit it was most often the lowest luminescence value on the plate) and that the dsRNA to Th also resulted in increased sensitivity to MMS treatment. Because Th is inconsistent between control and treatment, it is not well suited for correction of background. Therefore, in order to normalize the plates to true background, a quadruplicate control of a high dose of MMS, sufficient to kill all cells, was embedded in the plate. We also included the positive control for treatment of dsRNA targeting GCLc, a gene that not required for viability itself but is required after MMS treatment (Figure 1a).
Quality metrics of validation experiments
Comparison of Luc dsRNA negative control values with high MMS positive control values from 17 different plates assayed on different days, the Z-factor 11 was calculated to be 0.705. Additionally, Luc dsRNA used in validation normalization had less than 10% standard deviation in untreated plates, while the MMS exposed plates for the same target dsRNA resulted in less than 20% standard deviation. Employing the described plate layout to decrease inter-plate variation was successful, as can be seen (Figure 1c). The background luminescence values obtained from the wells with high MMS were minimal and consistent across multiple plates. Wells containing dsRNA against the positive control Th and against the experimental control GCLc were also consistent across plates. Standard deviations are greater following normalization because data were divided by the median of Luc dsRNA values, while the observed standard deviation using raw values within any plate were far less (data not shown). By reducing inter- and intra-plate variation and normalizing to a large number of negative controls, results are more consistent and reliable.
Comparison of normalization methods
Genes that were confirmed from the validation experiments were used for a retrospective analysis comparing screen analysis methods. Seven methods of normalization or combination of normalizations were employed to normalize for inter-plate variation: background subtraction, scaling, cellHTS2, IQR measurement, quantile normalization, BSQN, and SQN. The data were subsequently compared between treatments to provide scores to rank genes required for viability, in each case, the normalized value for untreated control was subtracted from the normalized value following MMS exposure. After quantile normalization, an M score was calculated, which also serves this purpose.
This analysis is meant for analyzing data generated from RNAi screens yielding continuous data, such as a viability output, and not necessarily for other outputs, such as microscopy or reporter assays with internal controls. In particular, our analysis attempts to find the best methods to cross compare screens, such as a treatment and its untreated control, which inherently cannot be combined within the same well. Towards this end, researchers applying those methods we discuss that require values for percent viability after treatment or percent viability of a particular positive control, either treated or untreated, must empirically derive those values for themselves.
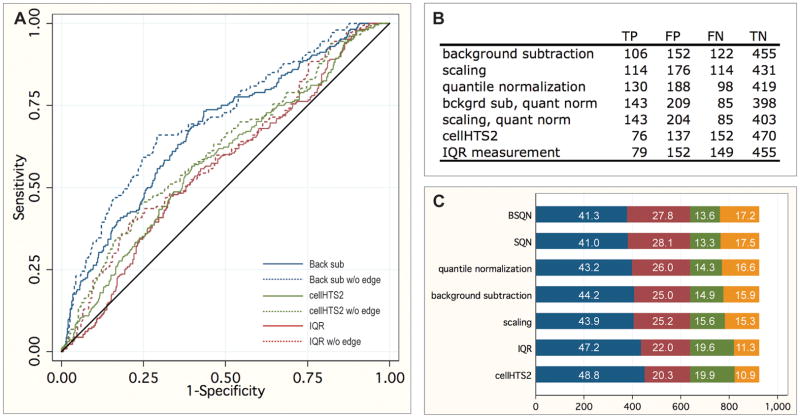
Using the data obtained from validation experiments, a retrospective analysis was performed to identify the best normalization method to identify the most true positive results. Ranked genes were plotted on an ROC curve using data generated by validation analysis as positives and negatives (Figure 2a) to determine the method yielding the best sensitivity and selectivity. Unfortunately, no single method excelled. BSQN and SQN were the best two methods, with areas under the curve 0.6758 and 0.6746, respectively, while IQR measurement and cellHTS2 were the worst two methods, with 0.5600 and 0.5826, respectively (Figure 2b). Because the normalizations and subsequent rankings were so poor, we chose the top 5%, 2.5%, 0.5%, and 0.25% ranked genes in the screen as the positive thresholds for each method to examine their individual validation rate. Figure 2c shows true and false positives and negatives for each method tested at the 5% threshold.
Figure 2.
Analysis of normalization methods. A. ROC curves for the best (background subtraction, blue) and worst (cellHTS2, green, and IQR measurement, red) normalization methods, both with (solid lines) and without (dashed lines) dsRNA located in the outer edge of the screening plate. The reference line is indicated in black. B. Area under curves for each method tested, both with and without dsRNA located in the outer edge of the screening plate. C. Number of true negatives (TN, blue), false positives (FP, red), false negatives (green), and true positives (orange) for each normalization method at thresholds of the top 5% ranked wells. Percentages of element are given.
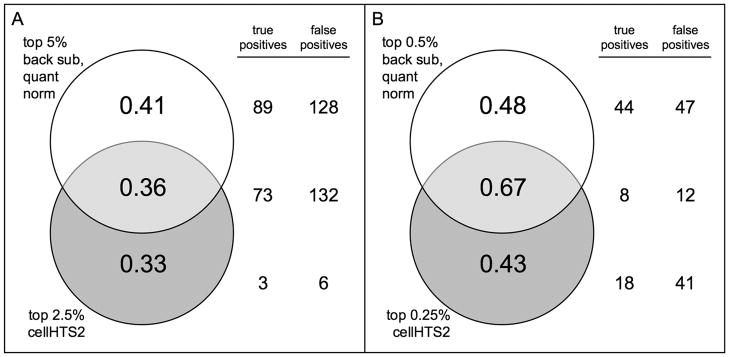
We wanted to determine the overlap within the top ranked wells after each normalization method. We expected that cellHTS2 and IQR measurement methods would have a good amount of overlap since the methods are based on a similar concept and found this true (data not shown). Additionally, we expected the quantile normalization methods would overlap in dsRNA amplicons ranked; we found this to be true (data not shown). We also found that wells ranked after background subtraction or scaling overlapped with amplicons ranked after any of the quantile normalization methods (data not shown). We therefore examined the overlap between dissimilar methods, namely BSQN and cellHTS2 (Figure 3). We found to obtain the highest ratio of true positives to total positives ranked, the combination of the top 5% following BSQN and the top 2.5% following cellHTS2 provided the most true positive amplicons with the least amount of false positives (Figure 3a). We currently recommend these cutoffs for examination of any continuous distributed dataset generated by RNAi screens to encompass the most reliable dataset for a systems level analysis. Alternatively, for a more selective study where only a few, top candidate genes are of interest, we advise using a higher stringency, combining the top 0.5% following BSQN and the top 0.25% following cellHTS2 (Figure 3b).
Figure 3.
Comparisons between normalization methods. Venn diagrams representing the ratio of true positives to total positives for A. the top 5% ranked wells after background subtraction followed by quantile normalization and the top 2.5% ranked wells after cellHTS2 and B. the top 0.5% after background subtraction followed by quantile normalization and the top 0.5% after cellHTS2. Total numbers of true and false positives overlapping for each method are given to the right.
We attempted to improve prediction by these normalization methods by examining the extent to which edge effects were having a negative impact on prediction. Indeed, ROC analysis indicates that by removing those genes represented in the wells at the outer edge of the 384-well plate from analysis, predictions improve greatly by every method (Figure 2a and b). Removing both the outermost wells of the plate and the next well in from the edges resulted in no further improvement of any method (data not shown). Amplicons located on edges are therefore more likely to result in false positives than amplicons located internally. It is therefore critical that when these amplicons are validated, they be distributed in the plate.
Overall, we have demonstrated that our validation method is extremely stringent, with an experimental design including use of consistent and effective quantity of dsRNA, performance of quadruplicates distributed within a single plate, and the embedding of multiple control features such as non-targeting dsRNA in a good portion of the wells in order to facilitate normalization between treatments (see Figure 1c for reproducibility between assays). Each of these points was addressed because of limitations of the screen experimental design, and thus we have been able to establish a stringent method by which to validate potential screen hits.
Unfortunately we were not able to determine a sensitive or selective normalization method, even though multiple methods were examined. Clearly, additional methods of normalization and ranking need to be developed as they have been for microarrays. An improved screen design would also reduce some of the issues. When the screening library is plated, those amplicons on the edge can be plated internally in the replicate plate to reduce edge effects (swapping quadrants). Additional controls for normalization could be included besides positive controls for the assay along with more replicates, although we acknowledge that this might be cost prohibitive. Finally, a standardized concentration of 0.4 μg of dsRNA per well would ensure more genes are fully knocked-down, thus reducing the number of false negatives at a given threshold. These points emphasize the importance of rigorous validation to follow up to any hits found by genome-wide RNAi screens of this nature.
Supplementary Material
Raw data from untreated control and MMS screening plates for each DRSC amplicon tested for replicates 1 through 4 (control) and 1 through 3 (MMS).
Normalized data values and rank for each method, along with whether the amplicon validated as being required after MMS treatment (validation_hit = 1) or not (validation_hit = 0). Normalizations reported: background subtraction (bs), background corrected (bc), quantile normalization (qn), background subtraction followed by quantile normalization (bsqn), background correction followed by quantile normalization (bcqn), interquartile range measurement (iqr), and cellHTS2
Acknowledgments
This work was supported by the NIEHS (K22-ES12264; A.J.R.B.). We thank the Drosophila RNAi Screening Center (flyrnai.org), which is supported by NIGMS (R01-GM067761). From UTHSCSA Center for Epidemiology and Biostatistics, we thank Gary Chisholm who performed initial statistical analyses, John Cornell and Daijin Ko for initial discussions, and Jonathan Gelfond for extensive discussion and manuscript review. From UTHSCSA GCCRI, we thank David Rodriguez for assistance with the figures.
Footnotes
Competing interests statement: The authors declared no competing interests.
Data availability: Raw data from the screen is available at WWW.FLYRNAI.ORG and GCCRI.UTHSCSA.EDU/ABPublished_Data.asp, and normalized data is available from GCCRI.UTHSCSA.EDU/ABPublished_Data.asp.
Contributor Information
Amy M. Wiles, Email: wiles@uthscsa.edu.
Dashnamoorthy Ravi, Email: dashnamoorth@uthscsa.edu.
Selvaraj Bhavani, Email: selvaraj@uthscsa.edu.
Literature Cited
- 1.Boutros M, Bras LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Consortium HF, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 3.Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- 4.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathey-Prevot B, Perrimon N. Drosophila genome-wide RNAi screens: are they delivering the promise? Cold Spring Harb Symp Quant Biol. 2006;71:141–148. doi: 10.1101/sqb.2006.71.027. [DOI] [PubMed] [Google Scholar]
- 8.Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc. 2007;2:2245–2264. doi: 10.1038/nprot.2007.250. [DOI] [PubMed] [Google Scholar]
- 9.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 10.Tukey JW. Exploratory data analysis. Reading, Mass: Addison-Wesley Pub. Co; 1977. [Google Scholar]
- 11.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89:552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, Holder DJ, Ferrer M, Espeseth AS. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics. 2006;7:299–309. doi: 10.2217/14622416.7.3.299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data from untreated control and MMS screening plates for each DRSC amplicon tested for replicates 1 through 4 (control) and 1 through 3 (MMS).
Normalized data values and rank for each method, along with whether the amplicon validated as being required after MMS treatment (validation_hit = 1) or not (validation_hit = 0). Normalizations reported: background subtraction (bs), background corrected (bc), quantile normalization (qn), background subtraction followed by quantile normalization (bsqn), background correction followed by quantile normalization (bcqn), interquartile range measurement (iqr), and cellHTS2