Abstract
Background/Aims
Limited access to large samples and independent replication cohorts precludes genome-wide association (GWA) studies of rare but complex traits. To localize candidate genes with family-based GWA, a novel exploratory analysis was first tested on 1,774 major histocompatibility complex single nucleotide polymorphisms (SNPs) in 240 DNA samples from 80 children with primary liver transplantation (LTx), and their biological parents.
Methods/Results
Initially, 57 SNPs with large differences (p<0.05) in minor allele frequencies were selected, when parents of children with early rejection (Rejectors) were compared with parents of Non-Rejectors. In hypothesis-testing of selected SNPs, the gamete competition statistic identified the minor allele G (ancestral allele T) of the SNP rs9296068, near HLA-DOA, as being significantly different (p=0.018) in parent-to-child transmission between outcome groups. Subsequent simple association testing confirmed over- and under-transmission of rs9296068 based on 1) the most significant differences between outcome groups, of 1,774 SNPs tested (p=0.002), and 2) allele (G) frequencies that were greater among Rejectors (51.4 vs. 36.8%, p=0.015), and lower among Non-Rejectors (26.8 vs. 36.8%, p=0.074), compared with 400 normal control Caucasian children. In early functional validation, a) Rejectors demonstrated significant repression of the first HLA-DOA exon closest to rs9296068, and b) Rejectors with the risk allele showed 3-fold greater intragraft content of B-lymphocytes, whose antigen-presenting function is inhibited selectively by HLA-DOA, compared with Rejectors without the allele.
Conclusions
The minor allele of the SNP rs9296068 is significantly associated with LTx rejection, and with enhanced B-lymphocyte participation in rejection, likely due to a dysfunctional HLA-DOA gene product.
Introduction
In children, liver transplantation (LTx) is usually performed for multiple congenital liver diseases, and results in highly variable outcomes (1). Acute cellular rejection (ACR) occurs in 50% and post-LTx lymphoma-like malignancy affects 2-10% (2). Genetic variants, exemplified most commonly by single nucleotide polymorphisms (SNPs) are a significant basis for individual variation (3). However, the majority of the >10 million catalogued SNPs do not alter gene function (4). Also, promising associations involving discrete SNPs are rarely reproduced (5-7). Population stratification, or over-representation of one ethnicity within an outcome group may allow a SNP representing this ethnicity to be viewed as the outcome-specific SNP (8). Family-based association studies can minimize stratification (9). In this design, parental genotype serves as the control, and transmission frequency of a SNP from heterozygous parents to affected offspring is compared with the expected 50% transmission frequency. Significant deviations reflect transmission disequilibrium, and are more likely to represent disease-specific variation than variations due to ethnicity. The known location of such a SNP is then used to identify a potential candidate gene, and a causal variant in proximity. Increasingly, genome-wide association (GWA) studies are being conducted, recognizing that most disease traits are complex, and likely originate from an interaction between multiple causal variants within multiple genes, and the environment.
Several limitations preclude application of GWA in transplantation, especially among pediatric LTx recipients. First, simultaneous testing of large numbers of SNPs necessitates stringent multiple-testing correction. Under these conditions, statistical power requirements mandate a large subject sample, or a two-step study in which candidate SNPs identified in a screening sample are replicated in a second step. Accruing a homogeneous validation cohort can take several years, because approximately 550 pediatric LTx are performed annually in the United States, and are distributed among >50 transplant centers (10). Secondly, the numbers of candidate SNPs in an association study may not exceed false-discovery thresholds. From such a limited list of candidates, any true-positive association may at best account for large effects.
We report for the first time, a novel, multi-step approach to candidate gene localization, which incorporates preliminary functional validation in the same test cohort. First, a two-tier, family-based association method identifies the candidate gene or locus. Next, transmission distortion in that locus is confirmed by allele frequency comparisons with a large control group. Because SNPs within non-coding regions can influence gene expression and splicing at distances of up to 100 kb, differential regulation and alternative splice variants of candidate genes is evaluated with probes specific to exons of whole gene transcripts in the first of two preliminary functional validation tests (11, 12). In the second, the candidate is characterized in diseased tissue with immunohistochemistry. Because our sample size is small, and consists of 80 case-parent-parent trios, our association method is based on the following biologic assumptions: A) If a SNP is strongly associated with an inherited trait, it should demonstrate differences in parental allele frequencies (PAF) between the two groups, B) In family-based association testing, a genetic variant that is associated with rejection/non-rejection should show over-transmission in one outcome group and under-transmission in the other in the gamete competition test (GC). The GC statistic evaluates transmission disequilibrium using full pedigree data, similar to the well-known Transmission Disequilibrium Test (TDT) (8), but is based on a likelihood ratio test (13). The GC statistic also handles missing data and allows haplotype-based analysis. C) In a confirmatory step, potential candidates should also demonstrate i) significant differences in allele frequency when the two outcome groups are compared with each other, and ii) evidence of overtransmission in one group and undertransmission in the other when compared individually, with a large cohort of 400 normal Caucasian children.
We asked whether rejection/non-rejection outcomes are associated with genetic variants in the major histocompatibility complex (MHC) region. The 1,774 MHC-SNPs evaluated here are a subset of 550,000 genome-wide SNPs recently characterized in our test population. MHC-SNPs have been analyzed separately at first, to explore the utility of our statistical method, which is based in part on screening/testing approaches for quantitative traits proposed by others, by applying it to a candidate region with known impact in organ transplantation (14-17). The results show that the minor allele G at the SNP locus rs9296068, which is significantly associated with rejection outcomes, localizes to the 5′ UTR (untranslated) region of the HLA-DOA gene. Differential regulation of HLA-DOA, which is selectively expressed in B-lymphocytes, is suggested by associated repression of the first HLA-DOA exon, and significantly higher B-lymphocyte content in allograft biopsies from Rejectors (18, 19).
Methods
Patient Population
All studies were performed with approval from the University of Pittsburgh’s Institutional Review Board. DNA was extracted from 3 ml whole blood samples from 80 primary pediatric LTx recipients ages 0-22 years, and their biological parents (240 DNA samples). All children received previously described Tacrolimus monotherapy after steroid-free induction with rabbit, anti-human thymocyte globulin (rATG, Genzyme, Cambridge, MA) (20). Children who experienced biopsy-proven ACR within the first 60 days after LTx were termed Rejectors. Normal controls consisted of 400 disease-free Caucasian children with no prior or ongoing use of imunosuppressants, or transplantation. In addition, all 400 samples were selected for the absence of large copy number variants that could suggest a disease association. This normal control population was recruited at the Center for Applied Genomics at the Children’s Hospital of Philadelphia (CAG-CHOP).
Genotyping
was performed with 550,000 genome-wide SNP loci with the HumHap550k SNP bead array (Illumina, San Diego, CA.). Analysis from this experiment was restricted to 1,813 SNPs covering 168 MHC genes on chromosome 6p, of which 1,774 passed our quality control criteria.
DNA extracted by the Gentra Purigene system (Minneapolis, MN) was activated, and SNPs were characterized using manufacturer’s protocol. The Illumina Beadstation GX software was used to extract genotype calls (Illumina, San Diego, CA.) (21).
Statistical Methods
A total of 1,813 SNPs from the MHC region of the HumHap550k SNP array (Illumina, San Diego, CA.) were identified for analysis. Genotype data from the array were merged with pedigree and phenotype data and quality controlled for the following: Mendelian errors (recoded to missing), Hardy-Weinberg equilibrium (SNPs with deviations at p<.001 removed), missing rate across patients (>10% removed), minimal minor allele frequency (SNPs with MAF <1% removed), and low genotyping rate for an individual (<10% removed in the simple association test) using the PLINK v0.99r software package (22). This filtering reduced the number of SNPs to 1,774 total. A two-sample proportions statistic (two-sided) was used to determine PAF differences (p-value cutoff <0.05) between parents of Rejectors and Non-Rejectors. This screening step yielded a subset of MHC-SNPs for subsequent hypothesis testing using the Gamete Competition (GC) statistic, as implemented in the software package Mendel v7.0.0 (23). In this approach, a segregation parameter τ is estimated by maximum likelihood from the family data, and a likelihood ratio test is then applied. For a two-allele locus with alleles 1 and 2, the probability that a heterozygous parent with genotype 1/2 transmits allele 1 is defined as Pr (1/2 -> 1)= τ1/( τ1+ τ2); the null hypothesis of Mendelian transmission corresponds to τ1 = τ2 = 1 (14). The transmission parameter τk is set to 1 for the most frequent allele k, while the remaining τk values (for all k not equal to the most frequent allele) are estimated by maximum likelihood. In our application, we assumed symmetric transmission: that is, an allele that is over-transmitted to Rejectors is assumed to be similarly under-transmitted to Non-Rejectors. Results are summarized as the p-value and chi-squared statistic (with 95% confidence bounds) for the PAF comparison, and the estimated segregation parameter (τ), the allele frequency, and the p-value of the GC statistic (see Appendix 1). Note that this screening/testing approach markedly reduces multiple testing problems because the PAF test (which only uses parental genotypes) is independent of the gamete competition test (which depends on how often specific alleles are transmitted to the children). Thus, since the PAF test alone is used for SNP selection, we only have to adjust for the actual number of GC tests conducted on the much smaller number of selected SNPs.
Confirmatory association testing with 400 normal Caucasian controls
Comparisons of MAF for significant MHC-SNPs were conducted, between Rejectors and normal controls, and between Non-Rejectors and normal controls (chi-square test).
Validating candidacy of the HLA-DOA locus with differential gene splicing patterns
The Affymetrix Human Exon 1.0 ST array was used to measure differential splicing patterns in archived RNA isolated from 29 of 80 children. Probe summaries for both the genes and exons were computed using the Affymetrix Power Tools (APT) software and ‘rma-sketch’ normalization method. The gene-level normalized intensities (NI) were computed with the MiDAS algorithm and both the splicing index (SI) (24) and Student’s t-test p-values (two-sided) were computed on the NI values in R (25). Principal components analysis (PCA) was calculated separately on exon-level and gene-level intensities and was used to remove 3 outlier samples, for a remaining total of 26 samples-11 Rejectors and 15 Non-Rejectors. To remove low expressed gene-level probes, those with values less than 3.5 (log2 scale) in >50% of the samples in either group were filtered out leaving 17,242 of the original 22,011 probes. Those gene-level probes that were highly differentially expressed between groups were also removed using a fold change threshold of log2 (10). This step accounts for the tendency for the gene-level probe set intensities in each group to be “disproportionately affected by background noise or saturation” (Affymetrix Technical notes). Exon-level probes were filtered based on APT’s detection above background (DABG) p-value greater than 0.05 in n-1 samples leaving 218,402 of the original 287,329 exon-level probes.
After gene and exon probe filtering, 7 gene-level probes (and 67 corresponding exon-level probes for the genes) within a window size of 200 kb that included the HLA-DOA gene on chromosome 6 were extracted and examined for differential splicing (positions 33,000,000-33,200,000). This was conducted to restrict the analysis to only the immediate region surrounding the HLA-DOA gene.
Relating intragraft content of B-lymphocytes, to rs9296068
Because the HLA-DOA gene inhibits class II antigen presentation selectively in B-cells, we asked whether B-cells were present in rejecting allograft biopsies, and whether the B-cell content of allograft biopsies varied with the presence or absence of the risk allele of rs9296068. Slides were re-cut from stored tissue blocks for 36 of 80 children, with 4μm thick tissue sections, stained in batches on a Ventana immunostainer using mouse monoclonal anti CD79a (DAKO M7050 at 1:100, Ventana CCI mild, 32 minutes at 32C,iView DAB detection), as well as their positive and negative biological staining controls for each batch. Hematoxylin counterstain was applied after DAB detection. The surface marker CD79a identifies cells of B-cell/plamacytoid lineage.
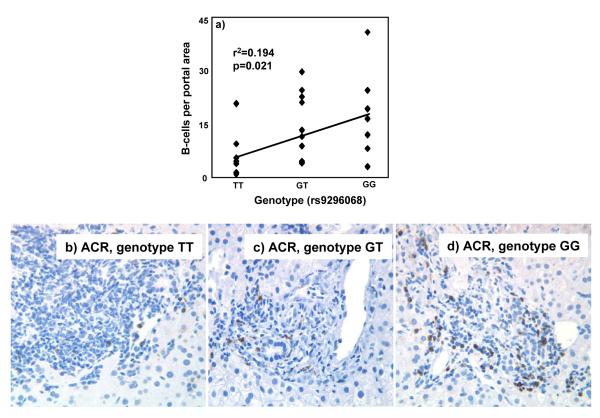
Cell counting was done blinded to outcome, and prior to histologic re-review of the tissues. Duplicate counts were done on each 10th case to assess variance. Portal and lobular regions of allografts were optically scored separately, at a magnification of ×400 and ×200, respectively. B-cell counts were expressed as total number per portal area, and total number per lobule (Figure 3).
Figure 3.
a). Significant correlation exists between B-lymphocyte content in liver grafts with ACR, and numbers of risk allele G at rs9296068 (r2=0.194, p=0.021). Accompanying micrographs show ACR, with portal areas containing brown, horseradish-peroxidase-stained CD79a+ B-lymphocyte/plasmacytoid cells. B-lymphocyte content is lowest, when the risk allele is absent (panel b), higher in heterozygotes (panel c), and highest when the risk allele is homozygous (panel d).
Results
Demographics
Early ACR occurred at mean±SD 32.5±4.2 days after LTx in 37 children who were termed Rejectors. Rejectors and Non-Rejectors (n=43) were similar with respect to age (median±SEM 6±1.1 vs 4.6±0.9 years, p=NS), male: female gender distribution (16:21 vs 21:22, p=NS), and etiology of liver failure requiring LTx (Table 1). The racial distribution in the Rejector and Non-Rejector groups was (Caucasian: African-American: Other=31:2:4 vs 42:0:1, respectively). Also, there were no differences between Rejectors and Non-Rejectors, in donor-recipient matching at the HLA-A, HLA-B, and HLA-DR loci (1.4±1.2 vs 1.8±1.2 antigens, p=NS), or disease severity as reflected in the Pediatric End-Stage Liver Disease score (PELD, 25.5±13.2 vs 24±14.2, P=NS). The sample size was too small to evaluate the effect of disease, age at transplant, and immunosuppression on rejection outcomes, or whether the highly associated SNP(s) were independent predictors of rejection outcome.
Table 1.
Etiology of liver disease leading to LTx in 80 children.
| Etilogy of liver disease | Rejectors | Non- Rejectors |
|---|---|---|
| Allagilles | 1 | 1 |
| Autoimmune Hepatitis | 3 | 0 |
| Biliary Atresia | 8 | 13 |
| BRIC: Benign Recurrent Intrahepatic Cholestasis |
0 | 1 |
| Budd-Chiari | 0 | 1 |
| Bylers Disease PFIC type I | 1 | 0 |
| Carolis Disease | 1 | 0 |
| Congenital Hepatic Fibrosis | 1 | 2 |
| Crigler-Najjar | 1 | 5 |
| Cystic Fibrosis | 1 | 1 |
| Fulminant Hepatic Failure | 2 | 2 |
| Giant Cell Hepatitis | 0 | 1 |
| Glycogen Storge Disease | 1 | 0 |
| Hepatic Fibrosis | 1 | 0 |
| Hepatoblastoma | 1 | 1 |
| Hepatocellular Carcinoma | 0 | 1 |
| Methylmalonic Acidemia | 1 | 0 |
| Maple Syrup Urine Disease | 7 | 8 |
| Ornithin Transcarbamylase Deficiency | 1 | 2 |
| Primary Sclerosing Cholangitis | 2 | 1 |
| Secondary Biliary Cirrhosis | 0 | 1 |
| Paucity of Bile Ducts | 1 | 0 |
| Tyrosenemia | 1 | 1 |
| Cirrhosis-unknown | 2 | 1 |
| Total | 37 | 43 |
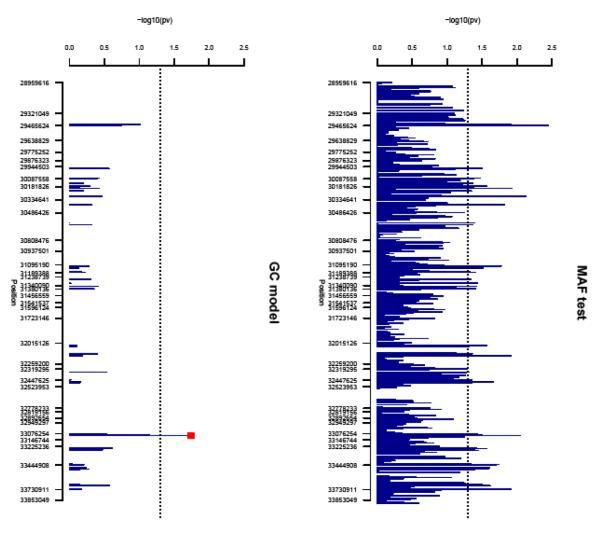
Fifty-seven SNPs passed our screening test due to large differences in MAF, when parents of Rejectors were compared with parents of Non-Rejectors (Appendix 1). When the GC statistic was applied to these 57 SNPs, only one SNP, rs9296068, in the 5′ flanking UTR of HLA-DOA demonstrated differences between groups with p<0.05 in parent-to-child transmission (p=0.0183, Table 2). Specifically, the minor allele G was transmitted more frequently to Rejectors, and less so to Non-Rejectors from biological parents. The physical position of the implicated SNP is 33096673 (Build 35). Figure 1 summarizes the results of the PAF comparison for the entire set of 1,774 MHC SNPs (upper panel), and the results of the GC test applied to the 57 selected SNPs for Rejectors and Non-Rejectors (lower panel).
Table 2.
Only one SNP, rs9296068 near the HLA-DOA gene, met criteria for selection and hypothesis testing in our two-tier approach. This SNP demonstrated a large difference (p=0.041) in MAF when parents of Rejectors were compared with parents of Non-Rejectors, as well as significant differences in parent-to-child transmission between Rejectors and Non-Rejectors, on the GC test (p=0.0183). Transmission data show more parents (n=19) transmitting the minor allele of rs9296068 to their Rejector offspring than parents who did not (n=12). The reverse was true among Non-Rejectors, whose parents were less likely to transmit this risk allele (7 transmissions vs 17 non-transmissions).
| HLA-DOA/ SNP=rs9296068 (physical position=33,096,673) | |
|---|---|
| Rejector vs. Non-Rejector MAF p value | 0.041 |
| Rejector vs. Non-Rejector GC p value | 0.018 |
| GC transmission parameter (τ2)† | 1.9 |
| Allele | 2 |
| Rejector parents | |
| MAF ‡ | 46.3% |
| Number transmitted | 19 |
| Number non-transmitted | 12 |
| Non-Rejector parents | |
| MAF‡ | 33.8% |
| Number transmitted | 7 |
| Number non-transmitted | 17 |
MAF=minor allele frequency
For the GC statistic, under the null hypothesis of Mendelian segregation, τk=1 for all k alleles (11)
Figure 1.
Upper panel shows −log10(p-values) for comparison of minor allele frequencies of 1,776 SNPs in parents of Rejectors, and parents of Non-Rejectors. Fifty seven SNPs show large differences (p<0.05, −log10(p-value)>1.3) in allele frequencies in this comparison and are shown in the lower panel. Only one of these 57 SNPs (lower panel) also shows significant differences in parent-to-child transmission when Rejectors were compared with Non-Rejectors in the gamete competition statistic. This SNP rs9296068, marked by the red square is located in the HLA-DOA gene.
In the confirmatory association testing step, 77 SNPs showed significant differences in allele frequencies when Rejectors were compared directly with Non-Rejectors (p≤0.05). Among these 77 SNPs, additional between-group comparisons showed 39 SNPs to be significantly different among Rejectors, and 19 SNPs to be significantly different among Non-Rejectors, when each group was compared separately with 400 normal controls (Appendix 2). In direct comparisons, the differences between Rejectors and Non-Rejectors were most significant (p=0.002) for the SNP rs9296068 (Table 3). When compared with normal controls, the minor allele (G) of rs9296068 was more commonly seen among Rejectors (36.7% vs 51.4%, p=0.015), but less commonly among Non-Rejectors (36.7 % vs 26.8%, p=0.074). Only one other SNP rs9276994 shows similar differences in distribution, when Rejectors are compared with Non-Rejectors (48.6% vs 28%, p=0.009), and when normal controls are compared either with Rejectors (37.7% vs 48.6 %, p=0.074) or with Non-Rejectors (37.7% vs 28%, p=0.083) (Table 4). In all, five of 14 top-ranked discriminatory SNPs localized to the 5′ flanking UTR of HLA-DOA (Table 4).
Table 3.
Confirmatory association testing shows that MAF for rs9296068 are greater among Rejectors, and less among Non-Rejectors, compared with a large cohort of 400 normal Caucasian children. Together, these results confirm parent-to-child over-transmission among Rejectors, and under-transmission among Non-Rejectors, observed in the GC test.
| Rejector vs. Controls | MAF‡ in Rejector children | 51.4% |
| MAF‡ in Controls | 36.8% | |
| x 2 | 5.90 | |
| P value | 0.015 | |
| Odds ratio | 1.82 | |
| Non-Rejector vs. Controls |
MAF‡ in Non-Rejector children | 26.8% |
| MAF‡ in Controls | 36.8% | |
| x 2 | 3.18 | |
| p value | 0.074 | |
| Odds ratio | 0.63 |
MAF=minor allele frequency
MAF differences are based on the samples used. For the unrelated case/control association shown in the table above, children are used to calculate allele frequencies. For the GC test in Table 2, trio information is used, resulting in minor differences in allele frequency.
Table 4.
Results of confirmatory association testing for SNPs, ordered by physicial position, which showed the most significant differences in MAF (p≤0.010) between Rejectors and Non-Rejectors (last column). Of 14 such SNPs, five are near HLA-DOA. Among them, rs9296068 and rs9276994 show MAF, which are greater among Rejectors (R), and less among Non-rejectors (NR), compared with 400 normal control Caucasian children (NC). Only rs9296068 shows differences in PAF between outcome groups, satisfying selection criteria for candidacy, while rs9276994 fails the PAF test.
| Allele | Frequency | Chi-square test p-values | ||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Physical location |
Closest Gene |
R | NR | NC | R vs NC | NR vs NC |
R vs NR |
| rs2517904 | 29,976,963 | HCG2P7 | 2.9% | 15.9% | 7.5% | 0.147 | 0.009 | 0.007 |
| rs3134879 | 30,123,884 | ZNRD1 | 26.6% | 48.7% | 41.1% | 0.023 | 0.192 | 0.007 |
| rs9366752 | 30,132,656 | ZNRD1 | 34.3% | 15.9% | 20.0% | 0.005 | 0.368 | 0.008 |
| rs9261441 | 30,199,737 | TRIM31 | 15.7% | 3.7% | 13.6% | 0.623 | 0.01 | 0.01 |
| rs1345229 | 30,290,374 | TRIM26 | 14.3% | 2.4% | 11.9% | 0.553 | 0.009 | 0.007 |
| rs1264583 | 30,401,462 | TRIM39 | 14.3% | 2.4% | 5.0% | 0.001 | 0.3 | 0.007 |
| rs1264581 | 30,405,484 | TRIM39 | 16.2% | 3.7% | 5.3% | 0.000 | 0.524 | 0.009 |
| rs3094097 | 30,741,854 | DHX16 | 14.3% | 2.4% | 9.8% | 0.228 | 0.028 | 0.007 |
| rs602875 | 32,681,607 | HLA-DRB1 | 42.9% | 22.0% | 23.8% | 0.000 | 0.707 | 0.006 |
| rs6457699 | 33,089,625 | HLA-DOA | 54.3% | 31.7% | 46.8% | 0.226 | 0.009 | 0.005 |
| rs9276994 | 33092233 | HLA-DOA | 48.6% | 28.1% | 37.8% | 0.075 | 0.083 | 0.009 |
| rs6933994 | 33095098 | HLA-DOA | 57.1% | 36.3% | 46.0% | 0.614 | 0.002 | 0.01 |
| rs9296068 | 33,096,673 | HLA-DOA | 51.4% | 26.8% | 36.8% | 0.015 | 0.074 | 0.002 |
| rs9277015 | 33,101,244 | HLA-DOA | 44.3% | 24.4% | 37.0% | 0.228 | 0.023 | 0.01 |
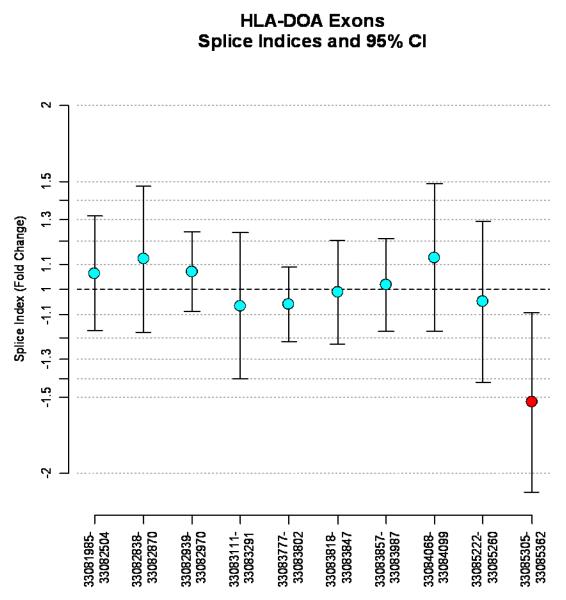
The first HLA-DOA exon is repressed in Rejectors
Following probe filtering (explained above) and retention of only those genes with at least one exon with p<0.05, two genes remained, HLA-DOA and HLA-DPA1 (Appendix 3). The first HLA-DOA exon between physical positions 33,085,305-33,085,362 was repressed among Rejectors, as suggested by −1.53-fold decreased expression, compared with Non-Rejectors (Figure 2). This repressed exon lies immediately adjacent to the 5′ flanking UTR of HLA-DOA, which contains the risk allele rs9296068 at position 33,096,673, and is ≈11.3 kb removed from the risk allele. In contrast, ≈47.8 kb, and three HLA-DPA exon transcripts separate rs9296068 from the discriminatory HLA-DPA exon at position 33,144,437-33,144,544, whose expression is −1.30-fold lower among Rejectors, compared with Non-Rejectors.
Figure 2.
Splicing index values, defined as the fold change between mean gene-level normalized exon intensities in the Rejector and Non-Rejector groups and 95% confidence intervals for the 10 exons that map to the HLA-DOA gene. The physical position ranges of each exon are represented on the x-axis and any exon point with a significant p-value (p<.05) is shaded red. Significant negative fold changes represent exon skipping or repression, while significant positive fold changes represent exon enrichment for the gene.
Intragraft B-cell content is higher in Rejectors with the risk allele of rs9296068 (Figure 3)
The risk allele (G) of rs9296068 was present in seven of 10 liver allograft biopsies from Non-Rejectors (allele frequency=0.4), and 19 of 26 Rejectors (allele frequency=0.5). B-cell content (median±SEM) was low at 1.1 cell per portal area in Non-Rejectors. Among Rejectors, the risk allele (G) was absent in seven, heterozygous in 12, and homozygous in seven. B-cell content in respective allograft biopsies from these Rejectors was 4±2.6 cells, 11±3.3 cells, and 16±4.7 cell per portal area, and showed significant correlation with the risk allele (r2=0.194, p=0.021, Figure 3). Also, differences between B-cell content were significant when Rejectors without the risk allele (n=7, 4±2.6 B-cells/portal area) were compared with Rejectors who were either homozygous or heterozygous for the risk allele of rs9296068 (n=17, 11.54±2.4 cells/portal area, p=0.022). The lobular areas contained an average of 0-3 B-cells per lobule in both Rejectors and Non-Rejectors, and showed no differences.
Subanalysis in Caucasian children to evaluate population stratification
The HapMap database shows rs9296068 (G) allele frequencies of 31% in Caucasians (CEU), and 67.8% in Yoruba (YRI) populations. To avoid false positive association in case-control comparisons, trios for two African-Americans and all six Non-Caucasians were excluded, five new trios from Caucasians children with LTx added, and both, the PAF comparison and simple case/control association were recalculated for 35 Rejectors, and 42 Non-Rejectors. Four SNPs adjacent to HLA-DOA remain discriminatory (p<0.05) in comparison of Rejectors with Non-Rejectors. Among them, rs6457699, rs9276994, and rs9296068 are ranked 3, 8 and 9 among the top ten SNPs, with all showing greater MAF among Rejectors and less among Non-Rejectors, compared with 400 normal Caucasian children (Appendix 4). In the PAF test, higher MAF among parents of Rejectors approached significance for rs6457699 (p=0.055) and rs9276994 (p=0.077) but not for rs9296068 (40 vs 34.7%, p=0.43). This observation suggests that the Rejector group is relatively underpowered to detect MAF differences for the rs9296068 SNP, even though the GC test confirms significant differences in parent-to child transmission between groups for its risk allele (p=0.004). Finally, both rs6457699, and rs9276994 are situated ≈4kb and ≈6kb upstream of the first HLA-DOA exon, and the GC test was significant for rs6457699 (p=0.04), implicating HLA-DOA once again (Appendix 4).
The GC test, unlike a case/control association, is based on distortions in allele transmission from parent to child, and so should be less sensitive to population substructure. It is of interest to assay whether or not transmission distortion occurs at the SNP of interest in the HapMap non-disease reference population (26). Therefore, the GC statistic was calculated for both alleles of rs9296068 for 30 YRI families (90 individuals), and 20 CEU families (90 individuals) using genotype and pedigree files from the Hapmap database. In both populations, the risk allele failed to show preferential transmission compared with its complementary allele, as suggested by GC p-values >0.05. Therefore, the null hypothesis of Mendelian transmission cannot be rejected. This sub-analysis supports our conclusions from GC testing of 72 Caucasian and 8 Non-Caucasian trios.
Discussion
Our multi-step approach identifies the HLA-DOA gene, and the B-lymphocyte in which it is exclusively expressed, as plausible candidates contributing to pediatric LTx rejection. Adding early functional validation to family-based association in the same dataset is especially useful, because it obviates the need for immediate replication in an independent cohort, whose accrual may take several years in the case of rare disease traits. This novel approach was motivated by failure when we first performed genome-wide association with the unmodified TDT applied to all 550,000 SNPs (on-going study and data not shown). All SNPs showing significant p-values failed to remain significant after Bonferroni correction for multiple hypothesis testing.
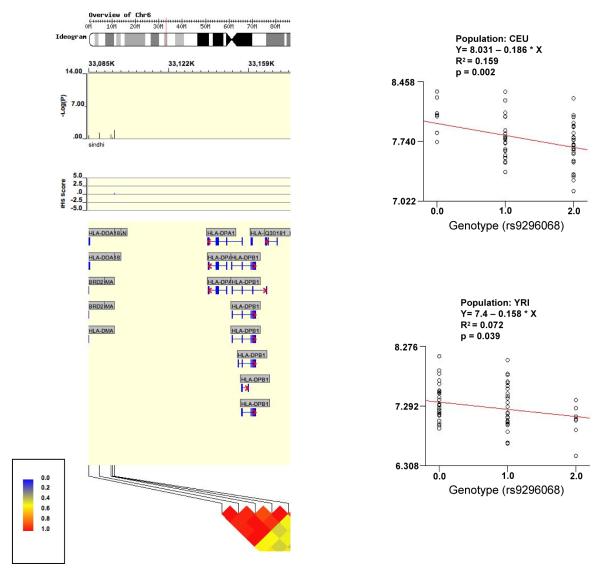
Because SNP reduction with PAF comparisons and the GC test are independent, the combined p-value for r9296068 is a multiple of values from both procedures (0.041*0.0183=0.00075). However, this would not be significant after multiple-testing correction for 1,774 SNPs. Therefore, transmission characteristics of rs9296068 were confirmed by showing that allele frequencies in Rejectors and Non-Rejectors were respectively, significantly greater (51.4 vs 36.8%, p=0.015), and less (26.8 vs 36.8%, p=0.074) than those in the normal control population of 400 Caucasian children (Table 3). Finally, in direct comparisons of allele frequencies between Rejectors and Non-Rejectors, the 5′ flanking UTR of HLA-DOA was represented by five SNPs among 14 top-ranked SNPs (p≤0.01) (Table 4), of which rs9296068 achieved the highest p-value (0.002), and another, rs9276994, also showed evidence of over-transmission in Rejectors (p=0.074), and under-transmission in Non-Rejectors (p=0.082), even though parental allele frequencies were similar. Significantly, all five highly-ranked SNPs, beginning with rs6457699 and ending with rs9277015, were present upstream, in an ≈11.6 kb segment, at a distance of ≈4-15 kb from the first HLA-DOA exon (Figures 2 and 3). Promoter function has been predicted for a highly conserved ≈10kb region immediately upstream of the first HLA-DOA exon, with Caucasians demonstrating a linkage disequilibrium (LD) block encompassing the rs9296068 locus (Figure 4) (27). Because 3 of 5 discriminatory SNPs localize to this putative promoter, altered transcription factor binding, and differential regulation of HLA-DOA gene expression can be postulated. Significant repression of the first HLA DOA exon among Rejectors in our study supports this view. Decreased HLA-DOA gene expression is also seen with increasing numbers of the risk allele of rs9296068 for both Caucasian and Yoruba populations in public databases (28).
Figure 4.
The HLA-DOA gene is transcribed from the minus strand. The upstream 5′ flanking UTR region defined by five discriminatory SNPs in simple association testing of Rejectors vs Non-Rejectors lies between rs9296068 to the right and rs6457699 to the left, and shows marked linkage disequilibrium. Rs6457699 lies ≈4kb from the first HLA-DOA exon, on the left. Rs9296068 is associated with allele-dependent decrease in HLA-DOA expresion among CEU and YRI in public databases (Source: WGAViewer).
We interpret our observed associations as suggestive of a causal role for the HLA-DOA gene, and for the B-lymphocyte, rather than a causal role for the SNP itself; however causality remains unproven until definitive studies identify a true causal variant, and the mechanism by which it might alter HLA-DOA gene expression and B-cell function. For the association itself, all statistical tests used sequentially in the current study are necessary because neither simple association testing nor family-based association testing generates, by itself, a list of SNPs larger than the expected false positive prediction (p=0.05 times 1,774 SNPs=89 false-positives). The HLA-DOA gene, and its adjacent region, is of interest for several reasons. Compared with one of its 3′ neighbors, HLA-DPB1, which is highly polymorphic, and can influence immunological outcomes in bone-marrow, corneal and renal transplantation, the HLA-DOA gene is relatively non-polymorphic, and inhibits class II antigen presentation in mature B-cells; but its role in organ transplantation is unknown (29-31, 19, 20). Our findings lead us to speculate that a missing exon transcript may produce a dysfunctional HLA-DOA gene product, which facilitates rejection by failing to inhibit antigen presentation by B-lymphocytes. The resulting increased B-lymphocyte participation in the rejection process is seen as nearly three-fold higher intragraft B-lymphocyte content in rejecting liver grafts in our study, when the minor allele G of rs9296068 was present, compared with rejecting allografts from children without this allele. Among pediatric LTx, prior supportive evidence also includes greater resistance of activated B-lymphocytes to immunosuppression, as well as a relative excess of B-lymphocytes during the risk period for early rejection (18, 32). B-lymphocyte-rich infiltrates, as well as B-lymphocyte-specific gene expression products have already been demonstrated in renal allografts with steroid-resistant acute cellular rejection (33). While our observations are preliminary, they suggest that the HLA-DOA gene and its vicinity be mapped further to identify novel causal variants.
We acknowledge several limitations. The heterogeneous diagnoses precipitating LTx are potential sources of stratification, although no single disease dominated either outcome group. For example, all 5 children with PSC carried the risk allele, with four experiencing rejection, despite statistically similar proportions of PSC in either group (4/37 Rejectors vs 1/43 Non-Rejectors, p=NS). Second, our SNP reduction step which relies on PAF comparisons, is biased by inclusion of eight Non-Caucasians trios, seven in the Rejector group. In accepting rs9296068 as a candidate, an illustrative subanalysis shows that our conclusions would be unchanged if an adequately powered Caucasian sample was available (Appendix 5). For these reasons, we have also relied on functional studies, coupled with allele-specific decrease in HLA-DOA expression in public databases for both CEU and YRI, to suggest biological relevance for the risk allele. We hope to relate differences in HLA-DOA exon repression to allelic variations at rs9296068 in a larger future sample. The small numbers of Rejectors (n=11) in whom HLA-DOA exon repression was shown, could not be divided further for adequately powered correlations with allelic variants in this study.
We conclude that our combination of methods can identify biologically relevant associations in small populations. We propose to validate our conclusions and address the primary power limitations of this pilot study by extending it to multicenter LTx populations.
Acknowledgments
Support: 5RO1AI073895-01, Children’s Hospital of Pittsburgh Research Foundation, and Hillman Foundation of Pittsburgh.
Special thanks: Timothy Billiar and Roger Oxendale.
Appendix 1
Appendix 1. p-values for parental allele frequency comparison, and for the GC statistic comparing prent-to-child transmission of SNPs between Rejectors and Non-Rejectors.
The minor allele for each snp is identified in the allele column, and the minor allele frequencies (MAF)s in both the parents and offspring for each group (NR and R) are provided for this allele.
For each SNP in the GC model, the transmission parameter tau k is set to 1 for the most frequent allele k, while the remaining tauk values (for all k not equal to the most frequent allele) are estimated by maximum likelihood.
| SNP | Closest Gene |
Distance to gene |
Position | SNP type | Parental allele frequency comparison, p- value |
Lower conf. Interval |
Higher conf. Interval |
Rejector Parents MAF |
Non-Rejector Parents MAF |
Rejector MAF | Non-Rejector MAF |
Rejector vs. Non-Rejector GC p value |
tauk | Allele frequency in group k |
Allele | distance to exon boundary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4713213 | OR5V1 | 0 | 29446941 | INTRONIC | 0.046 | 0.0027 | 0.2463 | 50.8% | 38.3% | 44.4% | 40.2% | 0.285 | 0.7749 | 44.0% | 1 | −9 |
| rs4598109 | OR5V1 | 0 | 29452683 | INTRONIC | 0.028 | 0.0146 | 0.2546 | 46.2% | 32.7% | 38.6% | 35.7% | 0.144 | 0.7032 | 38.8% | 1 | −9 |
| rs238883 | OR5V1 | 0 | 29454205 | INTRONIC | 0.033 | −0.2547 | −0.0115 | 38.6% | 52.0% | 43.1% | 46.4% | 0.097 | 1.5377 | 46.0% | 2 | −9 |
| rs238882 | OR5V1 | 0 | 29454342 | INTRONIC | 0.012 | 0.0324 | 0.2711 | 48.5% | 33.3% | 41.7% | 35.7% | 0.228 | 0.7473 | 40.7% | 1 | −9 |
| rs238872 | OR5V1 | 0 | 29459852 | INTRONIC | 0.040 | 0.0066 | 0.251 | 53.8% | 40.9% | 50.0% | 46.4% | 0.180 | 0.7316 | 47.2% | 1 | −9 |
| rs3094556 | OR5V1 | 0 | 29461387 | INTRONIC | 0.004 | 0.0571 | 0.2963 | 58.7% | 41.0% | 55.6% | 46.4% | 0.290 | 0.7794 | 49.7% | 2 | −9 |
| rs3094551 | OR5V1 | 0 | 29462778 | INTRONIC | 0.010 | −0.2759 | −0.0379 | 36.2% | 51.9% | 40.0% | 46.4% | 0.303 | 1.2708 | 44.4% | 1 | −9 |
| rs3094550 | OR5V1 | 0 | 29462788 | INTRONIC | 0.009 | −0.28 | −0.0399 | 36.6% | 52.6% | 40.0% | 47.6% | 0.382 | 1.2278 | 45.2% | 2 | −9 |
| rs2517817 | HLAH_HUMAN | 671 | 29967496 | DOWNSTREAM | 0.032 | −0.1501 | −0.009 | 4.6% | 12.5% | 2.9% | 13.8% | 0.273 | 0.6088 | 10.0% | 1 | −9 |
| rs7758512 | Q6ZU40_HUMAN | 0 | 30078568 | INTRONIC | 0.034 | 0.0049 | 0.1686 | 15.9% | 7.2% | 14.7% | 12.2% | 0.374 | 0.7223 | 12.1% | 2 | −9 |
| rs9261129 | Q6ZU40_HUMAN | 0 | 30087558 | INTRONIC | 0.042 | 0.0024 | 0.1605 | 14.4% | 6.3% | 14.3% | 11.9% | 0.405 | 0.735 | 11.8% | 2 | −9 |
| rs9261277 | ZNRD1 | 0 | 30139070 | INTRONIC | 0.044 | 0.0014 | 0.16 | 15.2% | 7.1% | 14.3% | 10.5% | 0.631 | 0.8399 | 11.5% | 2 | −9 |
| rs1264701 | TRIM31 | −4318 | 30174337 | DOWNSTREAM | 0.027 | −0.2051 | −0.0144 | 13.1% | 24.1% | 9.7% | 22.1% | 0.514 | 0.816 | 19.2% | 1 | −9 |
| rs3734838 | TRIM31 | 0 | 30188210 | NON_SYNONYMOUS_CODING | 0.042 | 0.0005 | 0.1417 | 11.5% | 4.4% | 10.0% | 2.3% | 0.719 | 1.1603 | 8.3% | 1 | −9 |
| rs9261441 | TRIM31 | 10891 | 30199737 | INTERGENIC | 0.012 | 0.019 | 0.1779 | 16.2% | 6.3% | 15.3% | 3.6% | 0.372 | 1.3678 | 10.8% | 1 | −9 |
| rs2022065 | TRIM10 | 0 | 30229439 | 3PRIME_UTR | 0.045 | 0.0021 | 0.224 | 34.1% | 22.8% | 35.7% | 23.2% | 0.638 | 1.1264 | 28.2% | 1 | 88 |
| rs1345229 | TRIM26 | 1191 | 30290374 | UPSTREAM | 0.008 | 0.0226 | 0.1713 | 14.2% | 4.5% | 13.9% | 2.4% | 0.341 | 1.4499 | 9.2% | 1 | −9 |
| rs9357097 | TRIM39 | −9500 | 30393100 | INTERGENIC | 0.015 | −0.2417 | −0.0275 | 19.2% | 32.7% | 16.7% | 32.1% | 0.480 | 0.8097 | 27.2% | 1 | −9 |
| rs2516649 | GNL1 | −16814 | 30604861 | INTERGENIC | 0.040 | −0.2333 | −0.0069 | 25.0% | 37.0% | 23.6% | 35.7% | 0.994 | 0.9958 | 31.2% | 2 | −9 |
| rs2844713 | GNL1 | 0 | 30627237 | INTRONIC | 0.043 | −0.2307 | −0.0052 | 26.8% | 38.6% | 27.8% | 36.0% | 0.476 | 1.1975 | 33.2% | 1 | −9 |
| rs4248154 | NM_001010909 | 44941 | 31110595 | INTERGENIC | 0.017 | −0.1853 | −0.02 | 8.1% | 18.4% | 11.1% | 20.2% | 0.522 | 0.7948 | 14.1% | 1 | −9 |
| rs2523849 | C6orf15 | −53952 | 31133030 | INTERGENIC | 0.031 | 0.0079 | 0.1963 | 22.4% | 12.2% | 24.3% | 13.1% | 0.734 | 1.1123 | 17.6% | 2 | −9 |
| rs2428514 | C6orf15 | −51487 | 31135495 | INTERGENIC | 0.044 | 0.0015 | 0.1804 | 19.9% | 10.8% | 20.0% | 12.2% | 0.910 | 0.9627 | 15.6% | 1 | −9 |
| rs2517403 | C6orf15 | −11994 | 31174988 | INTERGENIC | 0.048 | 0.0013 | 0.2352 | 41.0% | 29.2% | 42.9% | 34.5% | 0.669 | 0.9029 | 35.3% | 2 | −9 |
| rs2844635 | C6orf15 | −3522 | 31183460 | DOWNSTREAM | 0.040 | 0.0055 | 0.2384 | 41.0% | 28.9% | 42.9% | 35.0% | 0.603 | 0.8816 | 35.1% | 2 | −9 |
| rs9295957 | Q6H1K9_HUMAN | 11917 | 31265572 | INTERGENIC | 0.046 | −0.1814 | −0.0039 | 10.6% | 19.9% | 10.6% | 17.1% | 0.498 | 1.2478 | 15.6% | 1 | −9 |
| rs7745906 | 1C07_HUMAN | −32518 | 31311987 | INTERGENIC | 0.037 | −0.1749 | −0.0076 | 8.8% | 18.0% | 7.4% | 18.6% | 0.960 | 1.0194 | 14.2% | 1 | −9 |
| rs2074488 | 1C07_HUMAN | 524 | 31348410 | UPSTREAM | 0.038 | −0.156 | −0.0066 | 6.0% | 14.1% | 8.6% | 14.0% | 0.386 | 1.4316 | 10.6% | 1 | −9 |
| rs9366778 | 1C07_HUMAN | 29266 | 31377152 | INTERGENIC | 0.039 | −0.2425 | −0.0071 | 33.1% | 45.6% | 34.7% | 41.7% | 0.446 | 1.2061 | 40.3% | 1 | −9 |
| rs406936 | NP_008860.4 | 0 | 32041140 | INTRONIC | 0.048 | −0.1643 | −0.0031 | 8.1% | 16.5% | 5.6% | 15.0% | 0.780 | 0.9085 | 12.5% | 1 | −9 |
| rs454212 | NP_008860.4 | 0 | 32042351 | INTRONIC | 0.028 | −0.1715 | −0.0123 | 6.4% | 15.5% | 6.1% | 12.2% | 0.948 | 1.0266 | 11.3% | 1 | −114 |
| rs387608 | STK19 | 0 | 32049536 | INTRONIC | 0.048 | −0.1643 | −0.0031 | 8.1% | 16.5% | 5.6% | 15.0% | 0.780 | 0.9085 | 12.5% | 1 | −9 |
| rs2269429 | TNXB | 0 | 32137161 | NON_SYNONYMOUS_CODING | 0.044 | −0.1475 | −0.0044 | 5.2% | 12.8% | 5.6% | 9.8% | 0.400 | 1.3939 | 9.3% | 1 | −9 |
| rs204899 | TNXB | 0 | 32165605 | INTRONIC | 0.012 | −0.1571 | −0.0214 | 3.7% | 12.7% | 2.8% | 8.8% | 0.657 | 1.1994 | 8.6% | 1 | −9 |
| rs3115553 | C6orf10 | −14648 | 32353805 | INTERGENIC | 0.049 | 0.0004 | 0.2025 | 26.8% | 16.7% | 25.7% | 23.2% | 0.298 | 0.7368 | 21.9% | 1 | −9 |
| rs9268384 | C6orf10 | 0 | 32444564 | NON_SYNONYMOUS_CODING | 0.045 | −0.2416 | −0.004 | 33.8% | 46.1% | 35.7% | 43.9% | 0.962 | 0.9873 | 40.3% | 2 | 5 |
| rs4424066 | C6orf10 | 2096 | 32462406 | UPSTREAM | 0.039 | −0.2411 | −0.0078 | 30.6% | 43.0% | 26.4% | 37.8% | 0.798 | 1.0642 | 37.2% | 2 | −9 |
| rs3817973 | BTNL2 | −635 | 32469089 | DOWNSTREAM | 0.022 | −0.2518 | −0.0207 | 29.4% | 43.0% | 25.7% | 36.9% | 0.707 | 1.0954 | 37.0% | 1 | −9 |
| rs3793126 | BTNL2 | 0 | 32479597 | INTRONIC | 0.044 | −0.2128 | −0.0048 | 17.7% | 28.6% | 18.1% | 25.6% | 0.727 | 1.1156 | 23.8% | 2 | −9 |
| rs86567 | HLA-DOA | 0 | 33084737 | INTRONIC | 0.037 | −0.2434 | −0.0089 | 31.6% | 44.2% | 31.9% | 48.8% | 0.292 | 0.7825 | 37.8% | 2 | −9 |
| rs6457699 | HLA-DOA | 4258 | 33089625 | UPSTREAM | 0.031 | 0.0123 | 0.2547 | 53.7% | 40.4% | 52.9% | 30.5% | 0.072 | 1.5742 | 45.9% | 1 | −9 |
| rs6933994 | HLA-DOA | 9731 | 33095098 | INTERGENIC | 0.042 | 0.0053 | 0.2485 | 58.1% | 45.4% | 55.7% | 36.3% | 0.173 | 0.7236 | 49.2% | 1 | −9 |
| rs6457702 | HLA-DOA | 10660 | 33096027 | INTERGENIC | 0.009 | −0.2802 | −0.0399 | 33.3% | 49.3% | 37.5% | 52.5% | 0.757 | 0.9284 | 42.4% | 1 | −9 |
| rs9296068 | HLA-DOA | 11306 | 33096673 | INTERGENIC | 0.041 | 0.0053 | 0.2447 | 46.3% | 33.8% | 50.0% | 26.8% | 0.018 | 1.8513 | 38.9% | 2 | −9 |
| rs986521 | COL11A2 | 0 | 33244123 | INTRONIC | 0.038 | −0.2075 | −0.0074 | 16.2% | 26.9% | 15.7% | 30.0% | 0.243 | 0.7138 | 21.8% | 2 | −151 |
| rs9277932 | COL11A2 | 0 | 33249231 | INTRONIC | 0.028 | −0.25 | −0.0154 | 30.9% | 44.2% | 35.3% | 45.1% | 0.813 | 1.0611 | 38.0% | 1 | −26 |
| rs2855425 | COL11A2 | 0 | 33252351 | INTRONIC | 0.040 | −0.221 | −0.0067 | 20.9% | 32.3% | 18.6% | 32.9% | 0.336 | 0.7725 | 26.6% | 2 | −125 |
| rs6531 | RXRB | 0 | 33271429 | SYNONYMOUS_CODING | 0.036 | −0.2241 | −0.0088 | 20.6% | 32.2% | 18.6% | 32.9% | 0.342 | 0.776 | 26.4% | 2 | 28 |
| rs211474 | DAXX | 29822 | 33428591 | INTERGENIC | 0.045 | 0.0029 | 0.2401 | 46.3% | 34.2% | 43.1% | 32.1% | 0.926 | 1.0226 | 39.5% | 1 | −9 |
| rs211455 | KIFC1 | −31102 | 33436496 | INTERGENIC | 0.019 | 0.023 | 0.2572 | 45.7% | 31.7% | 38.9% | 29.1% | 0.824 | 0.9483 | 37.8% | 1 | −9 |
| rs211452 | KIFC1 | −28639 | 33438959 | INTERGENIC | 0.020 | 0.0223 | 0.263 | 52.2% | 38.0% | 43.1% | 34.9% | 0.624 | 0.8897 | 43.7% | 2 | −9 |
| rs211457 | KIFC1 | 0 | 33473618 | INTRONIC | 0.025 | −0.1556 | −0.0122 | 5.1% | 13.5% | 5.6% | 14.6% | 0.580 | 0.7843 | 9.5% | 1 | −166 |
| rs3116713 | PHF1 | 0 | 33490266 | NON_SYNONYMOUS_CODING | 0.025 | −0.1445 | −0.0116 | 3.7% | 11.5% | 4.2% | 12.2% | 0.537 | 0.7515 | 7.9% | 2 | 33 |
| rs211456 | ZBTB9 | 0 | 33497359 | INTRONIC | 0.041 | 0.006 | 0.2481 | 54.5% | 41.8% | 47.2% | 39.5% | 0.718 | 0.9181 | 46.7% | 1 | −9 |
| rs396746 | NP_997380.1 | 0 | 33665023 | INTRONIC | 0.032 | −0.1845 | −0.0101 | 10.1% | 19.9% | 13.9% | 20.7% | 0.709 | 1.1293 | 15.1% | 1 | −9 |
| rs169737 | ITPR3 | −12767 | 33684555 | INTERGENIC | 0.024 | −0.2185 | −0.0165 | 16.2% | 27.9% | 21.4% | 25.6% | 0.269 | 1.3651 | 22.0% | 1 | −9 |
| rs12529825 | ITPR3 | 0 | 33725381 | INTRONIC | 0.012 | −0.1571 | −0.0214 | 3.7% | 12.7% | 5.6% | 14.6% | 0.683 | 0.8395 | 8.5% | 2 | −9 |
Appendix 2
Appendix 2.
List of 77 SNPs with significant differences in allele frequencies between Rejectors (R, n=37) and Non-Rejectors (NR, n=43), between Rejectors and 400 normal control Caucasian children, and Non-Rejectors vs 400 normal control Caucasian children. When compared with normal controls, only rs9296068 shows allele frequencies which are greater in Rejectors, and less among Non-Rejectors.
rs926068 also shows the greatest differences in allele frequencies between Rejectors and Non-Rejectors, and is one of five SNPs representing the HLA-DOA gene for which allele frequency differences between groups (R vs NR) are≤0.01.
| Allele frequency | Rejector vs Non-Rejectors Chi-sq |
Rejectors vs normal controls Chi-sq |
Rejectors vs Normal Controls Chi-sq |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Closest Gene | Distance to gene |
SNP type | Position | Rejectors | Non-Rejectors | Normal Control | SNP rank | p-value | odds ratio | p-value | odds ratio | p-value | odds ratio |
| rs9296068 | HLA-DOA | 11306 | INTERGENIC | 33096673 | 51.40% | 26.80% | 36.70% | 1 | 0.002 | 2.888 | 0.0152 | 1.822 | 0.074 | 0.6311 |
| rs6457699 | HLA-DOA | 4258 | UPSTREAM | 33089625 | 54.30% | 31.70% | 46.70% | 2 | 0.005 | 2.558 | 0.226 | 1.353 | 0.009 | 0.5288 |
| rs602875 | HLA-DRB1 | 16048 | INTERGENIC | 32681607 | 42.90% | 21.90% | 23.80% | 3 | 0.006 | 2.667 | 0.0004 | 2.401 | 0.707 | 0.9003 |
| rs1264583 | HLAH_HUMAN | 10138 | INTERGENIC | 30401462 | 14.30% | 2.40% | 5% | 4 | 0.007 | 6.667 | 0.001 | 3.167 | 0.299 | 0.475 |
| rs3094097 | Q6ZU40_HUMAN | 0 | INTRONIC | 30741854 | 14.30% | 2.40% | 9.70% | 5 | 0.007 | 6.667 | 0.227 | 1.543 | 0.028 | 0.2314 |
| rs1345229 | TRIM26 | 1191 | UPSTREAM | 30290374 | 14.30% | 2.40% | 11.90% | 6 | 0.007 | 6.667 | 0.553 | 1.237 | 0.009 | 0.1855 |
| rs3134879 | TRIM39 | −1138 | UPSTREAM | 30123884 | 26.60% | 48.70% | 41.10% | 7 | 0.007 | 0.3807 | 0.023 | 0.519 | 0.192 | 1.363 |
| rs2517904 | DHX16 | 0 | INTRONIC | 29976963 | 2.90% | 15.80% | 7.50% | 8 | 0.007 | 0.1561 | 0.147 | 0.3618 | 0.009 | 2.317 |
| rs9366752 | C6orf12 | 0 | 3PRIME_UTR | 30132656 | 34.30% | 15.80% | 20% | 9 | 0.008 | 2.769 | 0.005 | 2.087 | 0.368 | 0.7536 |
| rs1264581 | TRIM39 | 0 | SYNONYMOUS_CODING | 30405484 | 16.20% | 3.70% | 5.30% | 10 | 0.009 | 5.082 | 0.0003 | 3.455 | 0.524 | 0.6799 |
| rs9276994 | HLA-DOA | 6866 | INTERGENIC | 33092233 | 48.60% | 28.10% | 37.70% | 11 | 0.009 | 2.423 | 0.075 | 1.557 | 0.083 | 0.6428 |
| rs9277015 | HLA-DOA | 15877 | INTERGENIC | 33101244 | 44.30% | 24.40% | 37% | 12 | 0.009 | 2.464 | 0.227 | 1.353 | 0.023 | 0.5493 |
| rs6933994 | TRIM31 | 10891 | INTERGENIC | 33095098 | 57.10% | 36.30% | 46% | 13 | 0.01 | 2.345 | 0.614 | 0.8808 | 0.002 | 2.065 |
| rs9261441 | HLA-DOA | 9731 | INTERGENIC | 30199737 | 15.70% | 3.70% | 13.60% | 14 | 0.01 | 4.91 | 0.623 | 1.184 | 0.01 | 0.2412 |
| rs2394255 | HLA-A | 9079 | INTERGENIC | 30057800 | 58.60% | 37.80% | 48.60% | 15 | 0.012 | 2.326 | 0.11 | 1.494 | 0.062 | 0.6423 |
| rs532098 | HCG9 | 3625 | DOWNSTREAM | 32686030 | 30.90% | 51.20% | 45.60% | 16 | 0.012 | 0.4255 | 0.019 | 0.5327 | 0.332 | 1.252 |
| rs9468829 | NP_003888.2 | 36906 | INTERGENIC | 30857212 | 21.40% | 7.30% | 12.10% | 17 | 0.012 | 3.455 | 0.024 | 1.994 | 0.204 | 0.5773 |
| rs16896742 | HLA-DRB1 | 20471 | INTERGENIC | 30030719 | 46.90% | 26.80% | 34.20% | 18 | 0.012 | 2.406 | 0.042 | 1.695 | 0.177 | 0.7043 |
| rs2516684 | RPP21 | 48325 | INTERGENIC | 30470974 | 17.10% | 4.90% | 13.10% | 19 | 0.014 | 4.034 | 0.345 | 1.369 | 0.031 | 0.3394 |
| rs12206499 | HCG9 | −5765 | INTERGENIC | 30045106 | 37.10% | 19.50% | 25.70% | 20 | 0.015 | 2.438 | 0.0377 | 1.709 | 0.219 | 0.7011 |
| rs6904029 | HCG9 | −809 | UPSTREAM | 30051046 | 37.10% | 19.50% | 25.70% | 21 | 0.015 | 2.438 | 0.039 | 1.704 | 0.215 | 0.699 |
| rs3823355 | HCG9 | 0 | NON_SYNONYMOUS_CODING | 30050062 | 37.10% | 19.50% | 25.90% | 22 | 0.015 | 2.438 | 0.041 | 1.693 | 0.207 | 0.6945 |
| rs6933546 | HLAH_HUMAN | 671 | DOWNSTREAM | 33103992 | 42.90% | 24.40% | 36.60% | 23 | 0.016 | 2.325 | 0.301 | 1.298 | 0.027 | 0.5582 |
| rs2517817 | HLA-DOA | 18625 | INTERGENIC | 29967496 | 2.90% | 14.50% | 7.90% | 24 | 0.016 | 0.1791 | 0.138 | 0.3548 | 0.048 | 1.982 |
| rs3869070 | Q6ZU40_HUMAN | 0 | INTRONIC | 30131847 | 55.90% | 36.60% | 44.70% | 25 | 0.018 | 2.196 | 0.077 | 1.564 | 0.156 | 0.7123 |
| rs2647044 | HLA-G | 23348 | INTERGENIC | 32775888 | 22.70% | 8.70% | 9.90% | 26 | 0.019 | 3.067 | 0.001 | 2.655 | 0.726 | 0.8655 |
| rs2975033 | BAT2 | 0 | INTRONIC | 29930240 | 35.70% | 18.70% | 25.60% | 27 | 0.019 | 2.407 | 0.066 | 1.612 | 0.176 | 0.6698 |
| rs9267522 | BAT2 | 0 | NON_SYNONYMOUS_CODING | 31711749 | 27.10% | 12.20% | 13% | 28 | 0.019 | 2.682 | 0.001 | 2.493 | 0.836 | 0.9295 |
| rs3117583 | BAT3 | 0 | INTRONIC | 31727555 | 27.10% | 12.20% | 13% | 29 | 0.019 | 2.682 | 0.001 | 2.493 | 0.836 | 0.9295 |
| rs3130618 | BAT4 | 0 | NON_SYNONYMOUS_CODING | 31740113 | 27.10% | 12.20% | 13% | 30 | 0.019 | 2.682 | 0.001 | 2.493 | 0.836 | 0.9295 |
| rs3115663 | HB25_human | 21592 | INTERGENIC | 31709822 | 27.10% | 12.20% | 13.10% | 31 | 0.019 | 2.682 | 0.001 | 2.466 | 0.812 | 0.9193 |
| rs4711207 | Q6ZU40_HUMAN | 0 | INTRONIC | 30113733 | 34.40% | 17.50% | 22.80% | 32 | 0.02 | 2.469 | 0.036 | 1.776 | 0.28 | 0.7193 |
| rs2040450 | RPP21 | 19669 | INTERGENIC | 30442318 | 16.20% | 4.90% | 12.70% | 33 | 0.022 | 3.763 | 0.42 | 1.321 | 0.037 | 0.3509 |
| rs4259245 | ITPR3 | 0 | INTRONIC | 33732199 | 27.10% | 45.10% | 35.70% | 34 | 0.022 | 0.4531 | 0.148 | 0.6695 | 0.093 | 1.478 |
| rs2107202 | TRIM40 | 0 | INTRONIC | 30213722 | 20 | 36.60% | 25.90% | 35 | 0.025 | 0.4333 | 0.279 | 0.7162 | 0.037 | 1.653 |
| rs915664 | DDR1 | −54188 | INTRONIC | 30902596 | 35.70% | 19.50% | 30.60% | 36 | 0.025 | 2.292 | 0.377 | 1.259 | 0.036 | 0.5492 |
| rs3129763 | HA25_HUMAN | −14209 | INTERGENIC | 32698903 | 38.60% | 21.90% | 22.90% | 37 | 0.025 | 2.233 | 0.003 | 2.11 | 0.84 | 0.9452 |
| rs3893464 | HCG9 | −7642 | INTERGENIC | 30043229 | 37.10% | 54.90% | 50.60% | 38 | 0.029 | 0.4859 | 0.03 | 0.5763 | 0.463 | 0.843 |
| rs9261301 | RNF39 | 0 | INTRONIC | 30149538 | 52.90% | 35.40% | 44.40% | 39 | 0.03 | 2.049 | 0.171 | 1.405 | 0.117 | 0.6859 |
| rs9267546 | LY6G6D | −1245 | UPSTREAM | 31781415 | 17.60% | 6.20% | 9.30% | 40 | 0.03 | 3.214 | 0.026 | 2.102 | 0.371 | 0.6541 |
| rs387608 | HCG9 | 0 | INTRONIC | 32049536 | 5.70% | 17.10% | 15% | 41 | 0.031 | 0.2944 | 0.033 | 0.3434 | 0.618 | 1.167 |
| rs406936 | TRIM39 | −9500 | INTERGENIC | 32041140 | 5.70% | 17.10% | 14.70% | 42 | 0.031 | 0.2944 | 0.037 | 0.3503 | 0.574 | 1.19 |
| rs9357097 | NP_008860.4 | 0 | INTRONIC | 30393100 | 17.10% | 32.50% | 30.90% | 43 | 0.031 | 0.4297 | 0.0155 | 0.4615 | 0.776 | 1.074 |
| rs2394250 | STK19 | 0 | INTRONIC | 30051635 | 51.40% | 34.10% | 41.60% | 44 | 0.031 | 2.042 | 0.111 | 1.485 | 0.189 | 0.7272 |
| rs2257914 | TRIM40 | 0 | 5PRIME_UTR | 30228542 | 10% | 23.20% | 17.50% | 45 | 0.032 | 0.3684 | 0.108 | 0.5238 | 0.203 | 1.422 |
| rs2021723 | TRIM10 | 0 | 3PRIME_UTR | 30211902 | 10% | 23.20% | 17.30% | 46 | 0.032 | 0.3684 | 0.119 | 0.533 | 0.182 | 1.447 |
| rs2844776 | TRIM26 | 0 | INTRONIC | 30279806 | 15.70% | 30.50% | 20.10% | 47 | 0.033 | 0.4251 | 0.374 | 0.74 | 0.028 | 1.741 |
| rs2395175 | HLA-DRA | −2621 | UPSTREAM | 32513004 | 4.30% | 14.60% | 14.20% | 48 | 0.033 | 0.2612 | 0.02 | 0.2714 | 0.907 | 1.039 |
| rs2187668 | HA25_HUMAN | 0 | INTRONIC | 32713862 | 20.60% | 8.50% | 8.60% | 49 | 0.034 | 2.778 | 0.0013 | 2.747 | 0.978 | 0.9888 |
| rs9267911 | TRIM31 | 0 | INTRONIC | 32313088 | 58.60% | 41.50% | 47.30% | 50 | 0.035 | 1.996 | 0.069 | 1.578 | 0.317 | 0.7908 |
| rs2523990 | NOTCH4 | 13266 | INTERGENIC | 30185208 | 58.60% | 41.50% | 47.40% | 51 | 0.035 | 1.996 | 0.0722 | 1.57 | 0.307 | 0.7868 |
| rs12212092 | HCG9 | −9631 | INTERGENIC | 30236421 | 12.90% | 3.70% | 6.30% | 52 | 0.036 | 3.885 | 0.037 | 2.19 | 0.338 | 0.5635 |
| rs6457109 | TRIM10 | 0 | NON_SYNONYMOUS_CODING | 30041240 | 18.60% | 7.30% | 9.90% | 53 | 0.036 | 2.889 | 0.0233 | 2.082 | 0.455 | 0.7205 |
| rs9277027 | HLAH_HUMAN | −5770 | INTERGENIC | 33106216 | 30% | 15.80% | 30.40% | 54 | 0.037 | 2.275 | 0.948 | 0.9824 | 0.006 | 0.4319 |
| rs2523809 | HLA-DOA | 20849 | INTERGENIC | 29957598 | 8.60% | 20.70% | 11.80% | 55 | 0.037 | 0.3585 | 0.424 | 0.7041 | 0.019 | 1.964 |
| rs375912 | NR_002139.1 | −14455 | INTERGENIC | 33124706 | 32.90% | 18.30% | 36.50% | 56 | 0.039 | 2.186 | 0.543 | 0.8514 | 0.001 | 0.3895 |
| rs2517930 | HLA-DPA1 | −15618 | INTERGENIC | 29853054 | 37.10% | 21.90% | 30.80% | 57 | 0.039 | 2.101 | 0.271 | 1.329 | 0.097 | 0.6325 |
| rs4713411 | NM_001010909 | 29501 | INTERGENIC | 31095155 | 28.10% | 44.90% | 43.40% | 58 | 0.04 | 0.4807 | 0.018 | 0.5114 | 0.796 | 1.064 |
| rs1264567 | TRIM40 | −11178 | INTERGENIC | 30474079 | 20% | 8.50% | 18.90% | 59 | 0.041 | 2.679 | 0.818 | 1.075 | 0.02 | 0.4011 |
| rs213213 | RPP21 | 51430 | INTERGENIC | 33291708 | 21.40% | 36.60% | 26.70% | 60 | 0.041 | 0.4727 | 0.332 | 0.7468 | 0.058 | 1.58 |
| rs1419675 | RING1 | 3232 | DOWNSTREAM | 30200686 | 21.40% | 36.60% | 25.90% | 61 | 0.041 | 0.4727 | 0.413 | 0.7813 | 0.037 | 1.653 |
| rs6910071 | C6orf10 | 0 | INTRONIC | 32390832 | 7.14% | 18.30% | 19.10% | 62 | 0.043 | 0.3436 | 0.0126 | 0.3253 | 0.855 | 0.9467 |
| rs86567 | HLA-DOA | 0 | INTRONIC | 33084737 | 31.40% | 47.60% | 38.10% | 63 | 0.043 | 0.5053 | 0.267 | 0.7439 | 0.095 | 1.472 |
| rs3734838 | RNF39 | 12015 | INTERGENIC | 30188210 | 10.30% | 2.40% | 8.50% | 64 | 0.044 | 4.59 | 0.622 | 1.229 | 0.052 | 0.2676 |
| rs6909253 | TRIM31 | −6114 | INTERGENIC | 30163622 | 52.90% | 36.60% | 37.30% | 65 | 0.044 | 1.943 | 0.01 | 1.889 | 0.906 | 0.9719 |
| rs9261394 | TRIM31 | 0 | NON_SYNONYMOUS_CODING | 30172541 | 52.90% | 36.60% | 38% | 66 | 0.044 | 1.943 | 0.015 | 1.829 | 0.801 | 0.9413 |
| rs1003581 | GABBR1 | 0 | INTRONIC | 29648183 | 9.10% | 21.20% | 19.90% | 67 | 0.045 | 0.3706 | 0.032 | 0.4019 | 0.778 | 1.085 |
| rs594223 | C6orf125 | 0 | INTRONIC | 33775943 | 21.40% | 9.80% | 17.90% | 68 | 0.045 | 2.523 | 0.46 | 1.253 | 0.063 | 0.4967 |
| rs9468692 | RNF39 | 1571 | UPSTREAM | 30227869 | 14.30% | 4.90% | 7.10% | 69 | 0.046 | 3.25 | 0.032 | 2.173 | 0.445 | 0.6685 |
| rs1150735 | TRIM10 | 0 | 3PRIME_UTR | 30153178 | 27.10% | 42.70% | 40% | 70 | 0.046 | 0.5003 | 0.034 | 0.5588 | 0.637 | 1.117 |
| rs1052486 | BAT3 | 0 | NON_SYNONYMOUS_CODING | 31718665 | 40% | 56.40% | 43.70% | 71 | 0.046 | 0.5152 | 0.008 | 1.934 | 0.987 | 0.9961 |
| rs1264701 | TRIM31 | −4318 | DOWNSTREAM | 30174337 | 10% | 21.90% | 22.40% | 72 | 0.048 | 0.3951 | 0.015 | 0.3842 | 0.921 | 0.9726 |
| rs1077393 | BAT3 | 0 | INTRONIC | 31718508 | 40% | 56.10% | 44.10% | 73 | 0.048 | 0.5217 | 0.01 | 1.899 | 0.969 | 0.991 |
| rs986521 | COL11A2 | 0 | INTRONIC | 33244123 | 15.70% | 29.30% | 20.10% | 74 | 0.048 | 0.4506 | 0.374 | 0.74 | 0.053 | 1.642 |
| rs9262138 | DHX16 | 0 | NON_SYNONYMOUS_CODING | 30735846 | 10% | 2.40% | 5.90% | 75 | 0.049 | 4.444 | 0.17 | 1.78 | 0.196 | 0.4005 |
| rs9267665 | ZBTB12 | 1087 | UPSTREAM | 31978835 | 10% | 2.40% | 4.30% | 76 | 0.049 | 4.444 | 0.029 | 2.503 | 0.43 | 0.5632 |
| rs7745906 | 1C07_HUMAN | −32518 | INTERGENIC | 31311987 | 7.35% | 18.30% | 13% | 77 | 0.05 | 0.3545 | 0.177 | 0.5311 | 0.182 | 1.498 |
Appendix 3
Appendix 3.
Summary statistics for differentially expressed exons between Rejectors and Non-rejectors
| Exon ID | Transcript ID | Exon fold change† | Exon p-value† | Transcript fold change‡ | Transcript p-value‡ | Transcript/SNP name | Exon start/SNP location | Exon stop |
|---|---|---|---|---|---|---|---|---|
| 2950312 | 2950307 | 1.063 | 0.555 | −1.083 | 0.326 | HLA-DOA | 33,081,985 | 33,082,504 |
| 2950313 | 2950307 | 1.122 | 0.397 | −1.083 | 0.326 | HLA-DOA | 33,082,838 | 33,082,870 |
| 2950314 | 2950307 | 1.069 | 0.372 | −1.083 | 0.326 | HLA-DOA | 33,082,939 | 33,082,970 |
| 2950317 | 2950307 | −1.065 | 0.638 | −1.083 | 0.326 | HLA-DOA | 33,083,111 | 33,083,291 |
| 2950322 | 2950307 | −1.060 | 0.395 | −1.083 | 0.326 | HLA-DOA | 33,083,777 | 33,083,802 |
| 2950323 | 2950307 | −1.011 | 0.905 | −1.083 | 0.326 | HLA-DOA | 33,083,818 | 33,083,847 |
| 2950324 | 2950307 | 1.017 | 0.84 | −1.083 | 0.326 | HLA-DOA | 33,083,857 | 33,083,987 |
| 2950325 | 2950307 | 1.127 | 0.383 | −1.083 | 0.326 | HLA-DOA | 33,084,068 | 33,084,099 |
| 2950326 | 2950307 | −1.048 | 0.748 | −1.083 | 0.326 | HLA-DOA | 33,085,222 | 33,085,260 |
| 2950327 | 2950307 | −1.532 | 0.0153 | −1.083 | 0.326 | HLA-DOA | 33,085,305 | 33,085,362 |
| untranslated | rs6457699 | 33,089,625 | ||||||
| untranslated | rs9276994 | 33,092,233 | ||||||
| untranslated | rs6933994 | 33,095,098 | ||||||
| untranslated | rs9296068 | 33,096,673 | ||||||
| untranslated | rs9277015 | 33,101,244 | ||||||
| 2950331 | 2950329 | 1.129 | 0.545 | −1.166 | 0.419 | HLA-DPA1 | 33,140,788 | 33,140,818 |
| 2950332 | 2950329 | −1.161 | 0.517 | −1.166 | 0.419 | HLA-DPA1 | 33,140,836 | 33,140,861 |
| 2950333 | 2950329 | −1.246 | 0.204 | −1.166 | 0.419 | HLA-DPA1 | 33,140,925 | 33,141,014 |
| 2950338 | 2950329 | −1.302 | 0.0128 | −1.166 | 0.419 | HLA-DPA1 | 33,144,437 | 33,144,544 |
| 2950340 | 2950329 | −1.114 | 0.31 | −1.166 | 0.419 | HLA-DPA1 | 33,144,774 | 33,144,804 |
| 2950341 | 2950329 | −1.014 | 0.875 | −1.166 | 0.419 | HLA-DPA1 | 33,144,844 | 33,144,876 |
| 2950342 | 2950329 | 1.030 | 0.792 | −1.166 | 0.419 | HLA-DPA1 | 33,144,929 | 33,144,963 |
| 2950343 | 2950329 | −1.114 | 0.479 | −1.166 | 0.419 | HLA-DPA1 | 33,144,969 | 33,144,999 |
| 2950345 | 2950329 | 1.057 | 0.565 | −1.166 | 0.419 | HLA-DPA1 | 33,145,413 | 33,145,477 |
| 2950346 | 2950329 | 1.346 | 0.113 | −1.166 | 0.419 | HLA-DPA1 | 33,145,573 | 33,145,641 |
| 2950348 | 2950329 | 1.035 | 0.723 | −1.166 | 0.419 | HLA-DPA1 | 33,149,238 | 33,149,262 |
Exon fold change, or splicing index, is calculated as the ratio of mean gene-level normalized intensities between rejectors and non-rejectors. Negative values indicate exon skipping or repression, whereas positive values indicate exon enrichment. The p-values are calculated using a Student’s two-sample t -test on gene-level normalized intensities. Orange cells indicate p<.05.
Transcript fold change is calculated as the ratio of the mean gene-level intensities between rejectors and non-rejectors. All exons within a transcript have identical gene-level intensities, but different exon-level intensities. The p-values are calculated using a Student’s two-sample t -test on gene-level intensities.
Appendix 4
Appendix 4.
Top-ranked SNPs with p<0.01 from simple association testing with Rejectors (n=35) vs Non-Rejectors (n=42), all of whom are Caucasians
| MAF | MAF | MAF | REJ vs. NONREJ | REJ vs. CONTROLS | NONREJ vs. CONTROLS | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rank | physical position | type | Closest gene | Distance to gene | Rejectors | Controls | Non-Rejectors | P-VALUE | OR | L95 | U95 | Chi-Sq P-value | OR | L95 | U95 | Chi-Sq P-value | OR | L95 | U95 |
| rs2975033 | 4 | 29930240 | INTERGENIC | HLA-G | 23348 | 36.36 | 25.62 | 14.86 | 0.0034 | 3.273 | 1.451 | 7.382 | 0.05727 | 1.659 | 0.9801 | 2.807 | 0.04007 | 0.5068 | 0.262 | 0.9804 |
| rs12206499 | 10 | 30045106 | INTERGENIC | HCG9 | −5765 | 36.36 | 25.69 | 16.22 | 0.0065 | 2.952 | 1.332 | 6.544 | 0.05914 | 1.653 | 0.9765 | 2.797 | 0.07133 | 0.5598 | 0.2957 | 1.06 |
| rs3823355 | 11 | 30050062 | UPSTREAM | HCG9 | −809 | 36.36 | 25.87 | 16.22 | 0.0065 | 2.952 | 1.332 | 6.544 | 0.06405 | 1.637 | 0.9675 | 2.77 | 0.06659 | 0.5545 | 0.2929 | 1.049 |
| rs6904029 | 12 | 30051046 | NON_SYNONYMOUS_CODING | HCG9 | 0 | 36.36 | 25.75 | 16.22 | 0.0065 | 2.952 | 1.332 | 6.544 | 0.06059 | 1.648 | 0.9738 | 2.788 | 0.06976 | 0.5581 | 0.2948 | 1.056 |
| rs2394255 | 2 | 30057800 | DOWNSTREAM | HCG9 | 3625 | 59.09 | 48.62 | 33.78 | 0.0027 | 2.831 | 1.423 | 5.631 | 0.1021 | 1.526 | 0.9166 | 2.542 | 0.01447 | 0.5391 | 0.3266 | 0.8901 |
| rs3869070 | 6 | 30131847 | INTRONIC | Q6ZU40_HUMAN | 0 | 56.25 | 44.75 | 32.43 | 0.0049 | 2.679 | 1.339 | 5.358 | 0.0755 | 1.587 | 0.9502 | 2.652 | 0.04098 | 0.5926 | 0.3572 | 0.9832 |
| rs9366752 | 1 | 30132656 | 3PRIME_UTR | C6orf12 | 0 | 33.33 | 20 | 12.16 | 0.0026 | 3.611 | 1.521 | 8.575 | 0.01061 | 2 | 1.165 | 3.433 | 0.1024 | 0.5538 | 0.27 | 1.136 |
| rs6909253 | 15 | 30163622 | INTERGENIC | RNF39 | 12015 | 54.55 | 37.25 | 32.43 | 0.0083 | 2.5 | 1.258 | 4.968 | 0.005529 | 2.021 | 1.22 | 3.35 | 0.4111 | 0.8086 | 0.4868 | 1.343 |
| rs9261394 | 16 | 30172541 | INTERGENIC | TRIM31 | −6114 | 54.55 | 38 | 32.43 | 0.0083 | 2.5 | 1.258 | 4.968 | 0.008155 | 1.958 | 1.181 | 3.245 | 0.344 | 0.7832 | 0.4716 | 1.301 |
| rs2523990 | 13 | 30185208 | INTRONIC | TRIM31 | 0 | 60.61 | 47.38 | 37.84 | 0.0071 | 2.527 | 1.278 | 4.997 | 0.0387 | 1.709 | 1.023 | 2.854 | 0.1156 | 0.6762 | 0.4143 | 1.104 |
| rs1345229 | 18 | 30290374 | UPSTREAM | TRIM26 | 1191 | 15.15 | 11.87 | 2.703 | 0.0086 | 6.429 | 1.354 | 30.53 | 0.4332 | 1.325 | 0.654 | 2.685 | 0.01625 | 0.2061 | 0.04976 | 0.8539 |
| rs9357097 | 7 | 30393100 | INTERGENIC | TRIM39 | −9500 | 15.15 | 30.95 | 36.11 | 0.0051 | 0.3159 | 0.1382 | 0.7224 | 0.006962 | 0.3984 | 0.1999 | 0.7937 | 0.3663 | 1.261 | 0.7619 | 2.087 |
| rs1264583 | 19 | 30401462 | UPSTREAM | TRIM39 | −1138 | 12.12 | 5 | 1.351 | 0.0095 | 10.07 | 1.224 | 82.82 | 0.01509 | 2.621 | 1.172 | 5.86 | 0.1556 | 0.2603 | 0.03527 | 1.921 |
| rs1264581 | 5 | 30405484 | SYNONYMOUS_CODING | TRIM39 | 0 | 14.06 | 5.29 | 1.351 | 0.0041 | 11.95 | 1.47 | 97.1 | 0.004299 | 2.93 | 1.356 | 6.329 | 0.1354 | 0.2453 | 0.03327 | 1.808 |
| rs3095150 | 17 | 31040511 | INTERGENIC | DPCR1 | 10534 | 45.45 | 42.8 | 24.32 | 0.0086 | 2.593 | 1.263 | 5.32 | 0.6759 | 1.114 | 0.6723 | 1.844 | 0.002013 | 0.4295 | 0.248 | 0.744 |
| rs6457699 | 3 | 33089625 | UPSTREAM | HLA-DOA | 4258 | 53.12 | 46.75 | 28.38 | 0.0031 | 2.86 | 1.414 | 5.786 | 0.3256 | 1.291 | 0.775 | 2.15 | 0.002381 | 0.4513 | 0.2672 | 0.7623 |
| rs9276994 | 8 | 33092233 | INTERGENIC | HLA-DOA | 6866 | 46.88 | 37.75 | 24.32 | 0.0055 | 2.745 | 1.332 | 5.658 | 0.1487 | 1.455 | 0.8726 | 2.426 | 0.02181 | 0.53 | 0.3058 | 0.9186 |
| rs6933994 | 14 | 33095098 | INTERGENIC | HLA-DOA | 9731 | 56.25 | 45.99 | 33.33 | 0.0072 | 2.571 | 1.282 | 5.156 | 0.7293 | 0.9134 | 0.5468 | 1.526 | 0.0007677 | 2.349 | 1.411 | 3.909 |
| rs9296068 | 9 | 33096673 | INTERGENIC | HLA-DOA | 11306 | 46.88 | 36.75 | 24.32 | 0.0055 | 2.745 | 1.332 | 5.658 | 0.1074 | 1.519 | 0.9105 | 2.533 | 0.0328 | 0.5532 | 0.3191 | 0.959 |
Parental Allele test and Gamete Competition (GC) test for the same snps as above
The 5′UTR flanking region of HLA-DOA is represented by 4 SNPs, of which rs6457699 lies roughly 4kb from the first HLA-DOA exon, and also shows differences in parental allele comparisons, which approach significance at p=0.055 (yellow cell).
rs9296068 is more frequent in parents of Rejectors, compared with parents of Non-Rejectors. However, differences are not statistically significant, due to reduced power in the Rejector group, and among Rejector parents, as a result of removing seven non-Caucasians, and adding 5 caucasian trios.
Appendix 5
| Parental Allele test | MAF-Parents | MAF-Parents | GC test | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rank | physical position | type | Closest gene | Distance to gene | Chi-Sq p-value | LCI | UCI | Rejectors | Non-Rejectors | P-VALUE |
| rs2975033 | 4 | 29930240 | INTERGENIC | HLA-G | 23348 | 0.08 | −0.01 | 0.21 | 30.95 | 21.05 | 0.075 |
| rs12206499 | 10 | 30045106 | INTERGENIC | HCG9 | −5765 | 0.22 | −0.04 | 0.19 | 31.75 | 24.34 | 0.0126 |
| rs3823355 | 11 | 30050062 | UPSTREAM | HCG9 | −809 | 0.19 | −0.04 | 0.19 | 32.03 | 24.34 | 0.026 |
| rs6904029 | 12 | 30051046 | NON_SYNONYMOUS_CODING | HCG9 | 0 | 0.22 | −0.04 | 0.19 | 31.75 | 24.34 | 0.026 |
| rs2394255 | 2 | 30057800 | DOWNSTREAM | HCG9 | 3625 | 0.42 | −0.07 | 0.18 | 47.62 | 42.11 | 0.004 |
| rs3869070 | 6 | 30131847 | INTRONIC | Q6ZU40_HUMAN | 0 | 0.46 | −0.07 | 0.18 | 45.16 | 40.00 | 0.014 |
| rs9366752 | 1 | 30132656 | 3PRIME_UTR | C6orf12 | 0 | 0.42 | −0.06 | 0.16 | 26.98 | 22.08 | 0.006 |
| rs6909253 | 15 | 30163622 | INTERGENIC | RNF39 | 12015 | 0.16 | −0.03 | 0.21 | 45.24 | 36.18 | 0.041 |
| rs9261394 | 16 | 30172541 | INTERGENIC | TRIM31 | −6114 | 0.19 | −0.04 | 0.21 | 45.97 | 37.33 | 0.039 |
| rs2523990 | 13 | 30185208 | INTRONIC | TRIM31 | 0 | 0.10 | −0.02 | 0.23 | 54.10 | 43.33 | 0.076 |
| rs1345229 | 18 | 30290374 | UPSTREAM | TRIM26 | 1191 | 0.01 | 0.02 | 0.18 | 14.52 | 4.61 | 0.35 |
| rs9357097 | 7 | 30393100 | INTERGENIC | TRIM39 | −9500 | 0.00 | −0.29 | −0.08 | 14.17 | 32.89 | 0.734 |
| rs1264583 | 19 | 30401462 | UPSTREAM | TRIM39 | −1138 | 0.26 | −0.03 | 0.11 | 8.73 | 4.67 | 0.138 |
| rs1264581 | 5 | 30405484 | SYNONYMOUS_CODING | TRIM39 | 0 | 0.25 | −0.03 | 0.11 | 9.68 | 5.33 | 0.0959 |
| rs3095150 | 17 | 31040511 | INTERGENIC | DPCR1 | 10534 | 0.35 | −0.06 | 0.18 | 39.52 | 33.33 | 0.008 |
| rs6457699 | 3 | 33089625 | UPSTREAM | HLA-DOA | 4258 | 0.06 | 0.00 | 0.25 | 52.42 | 40.13 | 0.045 |
| rs9276994 | 8 | 33092233 | INTERGENIC | HLA-DOA | 6866 | 0.08 | −0.01 | 0.23 | 44.44 | 33.33 | 0.063 |
| rs6933994 | 14 | 33095098 | INTERGENIC | HLA-DOA | 9731 | 0.12 | −0.02 | 0.23 | 55.56 | 45.27 | 0.074 |
| rs9296068 | 9 | 33096673 | INTERGENIC | HLA-DOA | 11306 | 0.40 | −0.07 | 0.18 | 40.32 | 34.67 | 0.004 |
Footnotes
Presented in part at the American Transplant Congress, May 7, 2007, San Francisco, CA.
NCBI Data: http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE11361 Accession No.: GSE11361
Conflict of interest: None. Study subjects informed.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin SR, Atkison P, Anand R, Lindblad AS, SPLIT Research Group Studies of Pediatric Liver Transplantation 2002: patient and graft survival and rejection in pediatric recipients of a first liver transplant in the United States and Canada. Pediatr Transplant. 2004;8(3):273–83. doi: 10.1111/j.1399-3046.2004.00152.x. [DOI] [PubMed] [Google Scholar]
- 2.Jain A, Mazariegos G, Kashyap R, Kosmach-Park B, Starzl TE, Fung J, Reyes J. Pediatric liver transplantation. A single center experience spanning 20 years. Transplantation. 2002;73(6):941–7. doi: 10.1097/00007890-200203270-00020. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. www.ncbi.nlm.nih/projects/SNP/snp_summary.cgi.
- 4.Ireland J, Carlton VEH, Falkowski M, Moorhead M, Tran K, Useche F, Hardenbol P, Erbilgin A, Fitzgerald R, Willis TD, Faham M. Large-scale characterization of public database SNPs causing non-synonymous changes in three ethnic groups. Human Genetics. 2006;119:75–83. doi: 10.1007/s00439-005-0105-x. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson IV, Pravica V, Sinnot PJ. Genetic regulation of cytokine synthesis: consequences on acute and chronic organ allograft rejection. Graft. 1998;1:15. [Google Scholar]
- 6.Daly AK, Day CP, Donaldson PT. Polymorphisms in Immunoregulatory genes: Toward Individualized Immunosuppressive therapy? Am J Pharmacogenomics. 2002;2(1):13–23. doi: 10.2165/00129785-200202010-00002. [DOI] [PubMed] [Google Scholar]
- 7.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. 2002. [DOI] [PubMed] [Google Scholar]
- 8.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. 2003. [DOI] [PubMed] [Google Scholar]
- 9.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 10. OPTN Annual Report 2006.
- 11.Hull J, Campino S, Rowlands K, Chan MS, Copley RRR, Taylor MSS, Rockett K, Elvidge G, Keating B, Knight J, Kwiatkowski D. Identification of Common Genetic Variation That Modulates Alternative Splicing. PLoS Genetics. 2007;3(6):e99. doi: 10.1371/journal.pgen.0030099. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. doi: 10.1038/nature06757. (Online) doi:10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinsheimer JS, Blangero J, Lange K. Gamete-competition models. Am J Hum Genet. 2000;66:1168–1172. doi: 10.1086/302826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ionita-Laza I, McQueen MB, Laird NM, Lange C. Genome-Wide Weighted Hypothesis Testing in Family-Based Association Studies, with an application to a 100K scan. The American Journal of Human Genetics. 2007;volume 81:000. doi: 10.1086/519748. (2007) preprint, May 10, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih MC, Whittemore AS. Tests for genetic association using family data. Genet Epidemiol. 2002;22(2):128–45. doi: 10.1002/gepi.0151. [DOI] [PubMed] [Google Scholar]
- 16.Van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, Demeo DL, Murphy A, Su J, Datta S, Rosenow C, Christman M, Silverman EK, Laird NM, Weiss ST, Lange C. Genomic screening and replication using the same data set in family-based association testing. Nat Genet. 2005;37(7):683–91. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 17.Sheldon S, Poulton K. HLA typing and its influence on organ transplantation. Methods Mol Biol. 2006;333:157–74. doi: 10.1385/1-59745-049-9:157. [DOI] [PubMed] [Google Scholar]
- 18.Sindhi R, Magill A, Abdullah A, Seward J, Tresgaskes M, Bentlejewski C, Zeevi A. Enhanced Donor-specific Alloreactivity occurs independent of immunosuppression in children with early liver allograft rejection. Am J Transplant. 2005;5:96–102. doi: 10.1111/j.1600-6143.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 19.Denzin LK, Sant’Angelo DB, Hammond C, Surman MJ, Cresswell P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science. 1997;278(5335):106–9. doi: 10.1126/science.278.5335.106. [DOI] [PubMed] [Google Scholar]
- 20.Roucard C, Thomas C, Pasquier MA, Trowsdale J, Sotto JJ, Neefjes J, van Ham M. In vivo and in vitro modulation of HLA-DM and HLA-DO is induced by B lymphocyte activation. Journal of Immunology. 2001;167(12):6849–58. doi: 10.4049/jimmunol.167.12.6849. [DOI] [PubMed] [Google Scholar]
- 21.Shen R, Fan JB, Campbell D, Chang W, Chen J, Doucet D, Yeakley J, Bibikova M, Garcia Wickham E, McBride C, Steemers F, Garcia F, Kermani BG, Gunderson K, Oliphant A. High-throughput SNP genotyping on universal bead arrays. Mutation Research. 2002;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. doi: 10.1086/519795. (in press) http://pngu.mgh.harvard.edu/purcell/plink/ [DOI] [PMC free article] [PubMed]
- 23.Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Amer J Hum Genetics. 2001;69(supplement):A1886. [Google Scholar]
- 24.Srinivasan K, Shiue L, Hayes JD, Centers R, Fitzwater S, Loewen R, Edmondson LR, Bryant J, Smith M, Rommelfanger C, Welch V, Clark TA, Sugnet CW, Howe KJ, Mandel-Gutfreund Y, Ares M., Jr Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37(4):345–59. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2007. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- 26.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao T, Chang LW, McLeod HL, Stormo GD. PromoLign: A Database for Upstream Region Analysis and SNPs. Human Mutation. 2004;23:534–539. doi: 10.1002/humu.20049. [DOI] [PubMed] [Google Scholar]
- 28.Ge D, Zhang D, Need AC, Martin O, Fellay J, Telenti A, Goldstein DB. WGAViewer: A Software for Genomic Annotation of Whole Genome Association Studies. Genome Res. 2008 doi: 10.1101/gr.071571.107. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loiseau P, Esperou H, Busson M, Sghiri R, Tamouza R, Hilarius M, Raffoux C, Devergie A, Ribaud P, Socie G, Gluckman E, Charron D. DPB1 disparities contribute to severe GVHD and reduced patient survival after unrelated donor bone marrow transplantation. Bone Marrow Transplantation. 2002;30(8):497–502. doi: 10.1038/sj.bmt.1703658. [DOI] [PubMed] [Google Scholar]
- 30.Munkhabat B, Hagihara M, Sato T, Tsuchida F, Sato K, Shimazaki J, Tsubota K, Tsuji K. Association between HLA-DPB1 matching and 1-year rejection-free survival in high-risk corneal transplantation. Transplantation. 1997;63(7):1011–6. doi: 10.1097/00007890-199704150-00018. [DOI] [PubMed] [Google Scholar]
- 31.Mytilineos J, Deufel A, Opelz G. Clinical relevance of HLA-DPB locus matching for cadaver kidney retransplants: a report of the Collaborative Transplant Study. Transplantation. 1997;63(9):1351–4. doi: 10.1097/00007890-199705150-00025. [DOI] [PubMed] [Google Scholar]
- 32.Talukdar A, AshokKumar C, Farrar J, Wilson P, Janakiramanan A, Tregaskes M, Sindhi R. Lymphocyte subset reconstitution in pediatric liver recipients induced with steroid-free rabbit anti-human thymocyte globulin. Pediatric Transplantation. doi: 10.1111/j.1399-3046.2007.00797.x. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O., Jr. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. New England Journal of Medicine. 2003;349(2):125–38. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]