Abstract
Genome-wide analyses in human lung adenocarcinoma have identified regions of consistent copy number gain or loss, but in many cases the oncogenes and tumor suppressors presumed to reside in these loci remain to be determined. Here we identify the “Downstream of tyrosine kinase” (Dok) family members Dok1, Dok2 and Dok3 as lung tumor suppressors. Single, double, or triple compound loss of these genes in the mouse results in lung cancer with penetrance and latency dependent on the number of lost Dok alleles, and which is associated with an aberrant expansion and signaling profile of alveolar type II cells and bronchioalveolar stem cells. In human lung adenocarcinoma, we identify DOK2 as a target of copy number loss and mRNA downregulation and find that DOK2 suppresses lung cancer cell proliferation in vitro and in vivo. Given the genomic localization of DOK2, we propose it as an 8p21.3 haploinsufficient human lung tumor suppressor.
DOK1, DOK2, and DOK3 are adaptor proteins that function in feedback loops to modulate tyrosine kinase signaling. p62dok(DOK1), the prototypical DOK family member, was cloned as the major phosphorylation substrate of the p210bcr/abl oncoprotein in Philadelphia chromosome-positive CML blasts1,2, and shown to be a substrate of many endogenous protein tyrosine kinases (PTKs). To date, six additional DOK family members, DOK2 to DOK7, have been identified in human and mouse3–7. DOK1, DOK2, and DOK3 comprise a closely related subfamily5, and over-expression of these adaptor proteins in cultured cells negatively regulates ERK4,8–10 and/or MYC11,12 downstream of RAS and PTKs. DOK1, DOK2, and DOK3 are substrates of dozens of critical PTKs, including EGFR10, PDGFR12, and c-kit1. Because tyrosine phosphorylation of DOK proteins is required for their function as signaling adaptors, DOK1, DOK2, and DOK3 are believed to function in negative feedback signaling loops that tightly modulate the duration and/or intensity of growth factor signaling.
We speculated that DOK genes might act as tumor suppressors in solid tissues as well as the hematopoietic compartment, where Dok1 and Dok2 can oppose BCR-ABL-driven leukemogenesis13,14. Using an in vivo genetic approach in the mouse, we discovered that Dok1, Dok2, and Dok3 act as tumor suppressors in the lung through their ability to regulate the biology of alveolar type II (AT2) cells and bronchioalveolar stem cells (BASCs). In addition, in a multifaceted analysis of human lung adenocarcinoma samples, we find frequent loss and downregulation of DOK2, consistent with a tumor suppressive role for DOK2 in human cancer.
RESULTS
Dok1, Dok2, and Dok3 single and compound knockout mice develop lung cancer
The biological functions of Dok1, Dok2, and Dok3 have been studied primarily in the hematopoietic system. To determine if these Dok genes might have functions in other tissues, we investigated the expression of Dok1, Dok2, and Dok3 mRNA in a panel of murine tissues. In addition to high expression in the spleen, the mRNAs of all three Dok family members were expressed at high levels in the lung (Supplementary Fig. 1a and ref. 3). Western blot analysis confirmed expression of Dok1, Dok2, and Dok3 protein in this tissue (Fig. 1a).
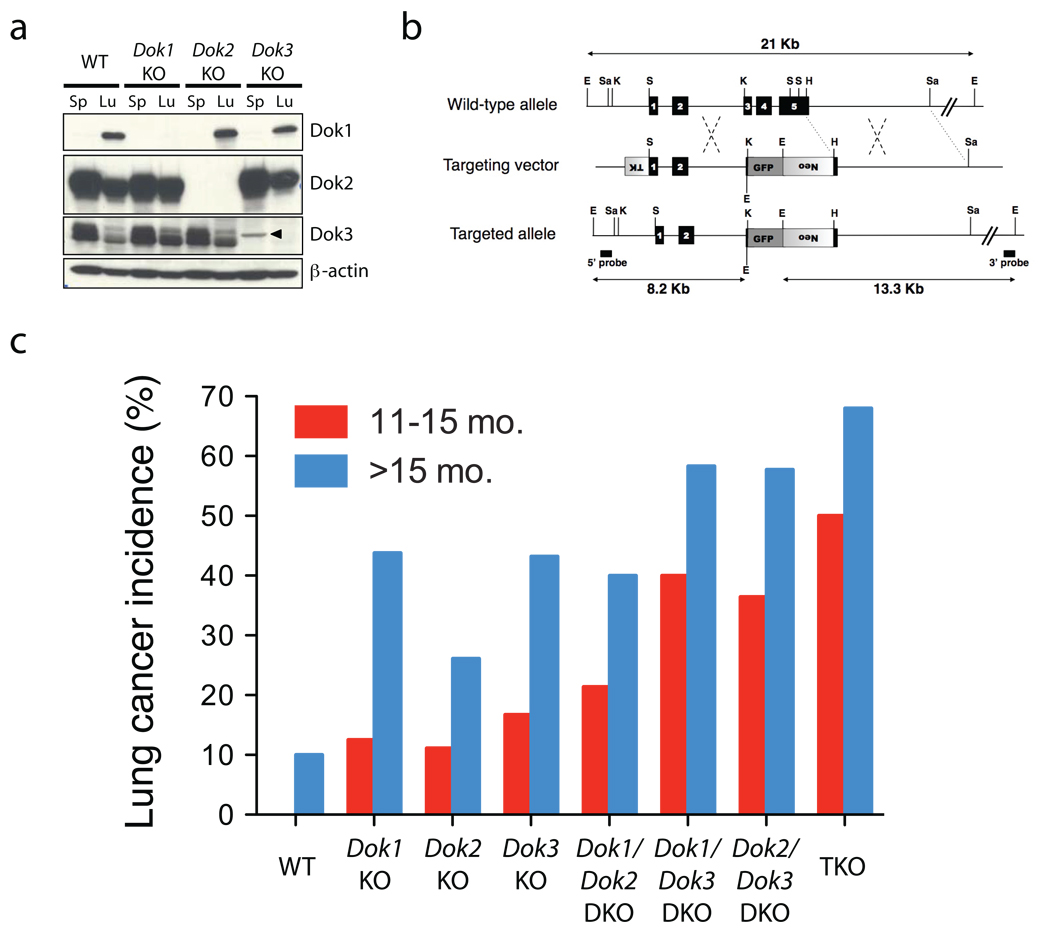
Figure 1. Dok1, Dok2, and Dok3 single and compound KO mice develop lung cancer.
(a) Immunoblot analysis of Dok1, Dok2, Dok3 and β-actin (loading control) proteins in cell lysates of splenocytes (Sp) or homogenized whole lung (Lu) from wild-type (WT), Dok1 KO, Dok2 KO, or Dok3 KO mice. An arrowhead indicates a non-specific band. Dok1 was detected in the spleen lysate at a longer exposure (not shown) and in our previously published work9. (b) Schematic map of the wild-type Dok3 locus (top), the targeting vector (middle) and the predicted targeted locus (bottom). The Dok3 genomic sequence is depicted as a line with solid boxes representing exons 1 to 5. Sequences from the pPNT plasmid are shown as boxes with lines, with shaded boxes representing the neomycin resistance cassette (neo), the HSV thymidine kinase (TK) cassette, or the GFP expression cassette, as indicated. The Dok3 genomic fragments used as probes for Southern-blot analysis are indicated (5’ probe, 3’ probe), as well as the expected fragments (arrows) following hybridization with the probes after digestion with EcoRI. EcoRI (E), SalI (Sa), HindIII (H), KpnI (K), and SmaI (S) sites are shown. (c) Lung adenocarcinoma incidence in Dok1, Dok2, and Dok3 single, double and triple KO mice. Animal numbers and statistics are summarized in Table 1.
We previously generated Dok1 and Dok2 KO mice for investigation of their hematopoietic phenotype9,14. Interestingly, when we analyzed the lungs of these animals, we discovered that many developed lung adenocarcinoma (see below). To further investigate the function of Dok family members as tumor suppressors, we generated Dok3 KO mutant mice using a homologous recombination targeting strategy in which a GFP cassette was inserted into the locus in-frame with the Dok3 gene (Fig. 1b and Supplementary Fig. 1b–e). Because no GFP expression was detected in the Dok3 KO lung or peripheral blood (Supplementary Fig. 1f), we concluded that no stable peptide is produced from the Dok3 locus in the targeted mice.
As in the case of Dok1 or Dok2 disruption8,9,13,14, Dok3 inactivation resulted in a lack of Dok3 protein expression without compensatory upregulation of other Dok proteins (see Fig. 1a). Dok3 KO mice were born in Mendelian frequencies and exhibited no gross hematopoietic defects (M.N., unpublished data). In standard methylcellulose colony forming assays, bone marrow (BM) progenitors from Dok3 KO mutants yielded numbers of erythroid and myeloid colonies comparable to wild-type sex-matched littermates (Supplementary Fig. 1g).
Examination of the lungs of the Dok3 KO, as well as the Dok1 and Dok2 single KO mice, revealed that all KO genotypes develop lung adenocarcinoma (Fig. 1c and Table 1, P < 0.05 by Fisher’s exact test of the overall incidence of lung adenocarcinoma at 11–25 mo. in each single KO genotype compared to wild-type). The similar amino acid sequences of the Dok proteins, as well as prior evidence in the hematopoietic compartment13,14, suggested that Dok1, Dok2, and Dok3 may exhibit partially redundant or overlapping functions. We therefore crossed the Dok3 KO animals with the Dok1 and Dok2 KO animals that we previously described9,14, to generate all compound double knock-out (DKO) and triple knock-out (TKO) mutants on a pure 129S1/SvImj genetic background9,14. Similar to the single mutants, the compound mutants were fertile and pathologically normal at birth. However, as early as 6 weeks after birth, 30% of TKO animals developed small lung tumors (n = 10). An analysis of 324 total mice at 11–25 months of age revealed that all single and compound mutant genotypes develop lung adenocarcinoma at moderate to high (24–64%) penetrance (Fig. 1c and Table 1). Lung adenocarcinoma incidence increased with each lost Dok allele, with the highest incidence in the TKO mice at all time points. In addition, both the Dok TKO and the Dok2/Dok3 DKO animals displayed a survival defect, with animals beginning to die at approximately 1 year (Supplementary Fig. 2).
Table 1.
Incidence of lung adenocarcinoma in cohorts of wild-type and Dok mutant mice
| 11–15 months |
> 15 months |
Overall (11–25 months) |
|||||
|---|---|---|---|---|---|---|---|
| Genotype | n with cancer/ total n |
% | n with cancer/ total n |
% | n with cancer/ total n |
% |
P value compared to WT& |
| WT | 0/13 | 0% | 3/30 | 10% | 3/43 | 7% | - |
| Dok1 KO | 1/8 | 13% | 14/32 | 44% | 15/40 | 38% | 0.001 |
| Dok2 KO | 1/9 | 11% | 12/46 | 26% | 13/55 | 24% | 0.0303 |
| Dok3 KO | 1/6 | 17% | 16/37 | 43% | 17/43 | 40% | 0.0006 |
| Dok1/Dok2 DKO | 3/14 | 21% | 8/20 | 40% | 11/34 | 32% | 0.0064 |
| Dok1/Dok3 DKO | 6/15 | 40% | 14/24 | 58% | 20/39 | 51% | <0.0001 |
| Dok2/Dok3 DKO | 4/11 | 36% | 15/26 | 58% | 19/37 | 51% | <0.0001 |
| TKO | 4/8 | 50% | 17/25 | 68% | 21/33 | 64% | <0.0001 |
For each Dok mutant genotype, the overall incidence of lung adenocarcinoma at 11–25 months of age was compared to that of the wild-type cohort using a Fisher's Exact test with a P value less than 0.05 considered significant.
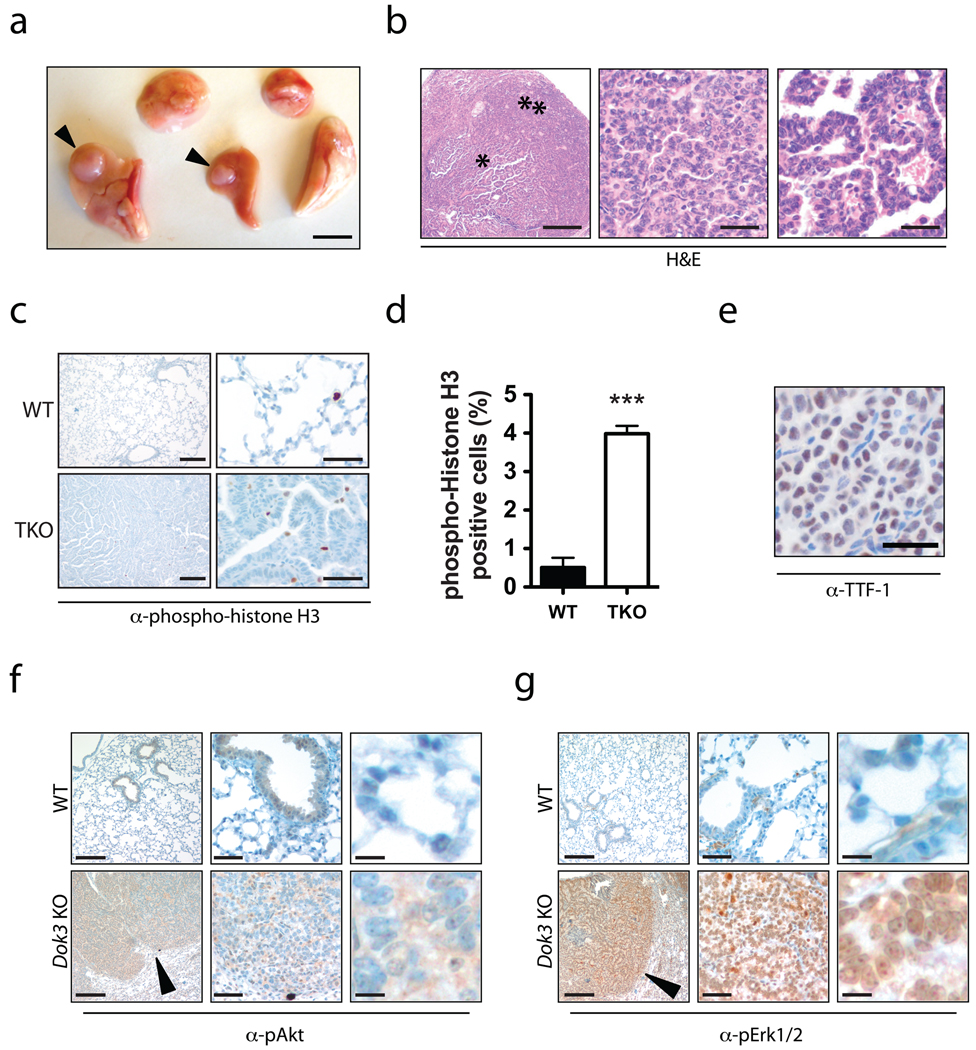
Dok KO lungs often contained multiple large tumor nodules (Fig. 2a). Hematoxylin and eosin (H&E) staining and pathological analysis showed that all cases were adenocarcinoma, often with papillary and solid growth patterns (Fig. 2b). Nuclei showed open chromatin, some with prominent nucleoli. Tumor cells exhibited nuclear inclusions and increased nuclear:cytoplasmic ratios with scattered cells showing nuclear pleomorphism. Mitotic figures and an elevated frequency of phospho-histone H3-positive mitotic cells were noted (Fig. 2c–d). Vascular invasion and multiple tumor nodules were observed. We did not find gross lymph node metastases, although lymph nodes were not systematically sampled. Inflammatory infiltrate composed of lymphocytes and macrophages was evident in many tumors ranging from mild to marked. Tumors were, in general, well differentiated rather than poorly differentiated. To assess if these tumors were primary and not secondary from another organ, we performed a standard15 immunohistochemical assessment using an antibody against TTF-1. All tumors analyzed were TTF-1 positive (Fig. 2e), indicating that the tumors were primary lung cancer.
Figure 2. Histopathology of lung tumors in Dok KO mice.
(a) Gross view of the five lung lobes from a Dok TKO mouse. Arrowheads indicate tumor nodules. Scale bar, 5 mm. (b) Left, H&E-stained adenocarcinoma with papillary features (star) and solid growth areas (double star) from a Dok2/Dok3 DKO lung. Scale bar, 500 µm. Middle, close-up of solid growth region. Scale bar, 50 µm. Right, close-up of papillary growth region. Scale bar, 50 µm. (c) IHC for phosphohistone H3 (brown) on lung tissue from age-matched wild-type (top panels) or Dok TKO (bottom panels) mice. Left panels, scale bar = 200 µm. Right panels, scale bar = 50 µm. (d) Quantification of phospho-histone H3 IHC shown in (c). Data shown is mean+SEM of three randomly selected tumor or normal fields. ***, P < 0.001 by two-tailed t-test. (e) TTF-1 IHC (brown) of a Dok1/Dok3 DKO lung tumor. Scale bar, 50 µm. (f) IHC for pAkt (Ser473; brown) on wild-type (top panels) or Dok3 KO lung tissue, showing cytoplasmic positivity in the Dok3 KO lung tissue. Left panels, scale bar = 200 µm. Middle panels, scale bar = 50 µm. Right panels, scale bar = 10 µm. An arrowhead indicates positive staining in the lung tumor region. (g) IHC for pErk1/2 (Thr202/Tyr204) in wild-type or Dok3 KO lung tissue showing predominantly nuclear staining. Left panels, scale bar = 200 µm. Middle panels, scale bar = 50 µm. Right panels, scale bar = 10 µm. An arrowhead indicates positive staining in the lung tumor region.
Given the role of Dok1, Dok2, and Dok3 in regulation of RTK and Ras signaling, we performed immunohistochemistry (IHC) with antibodies specific to phosphorylated Erk1/2 (pErk) and phosphorylated Akt (pAkt) to determine if loss of Dok expression in the lung could lead to abnormal Erk and/or Akt activation (Fig. 2f–g). Compared to wild-type lung tissue, tumors in Dok mutants exhibited moderate pAkt staining (Fig. 2f) and strong pErk staining (Fig. 2g).
Lung tumorigenesis in Dok mutants is preceded by an expansion of AT2 cells and BASCs
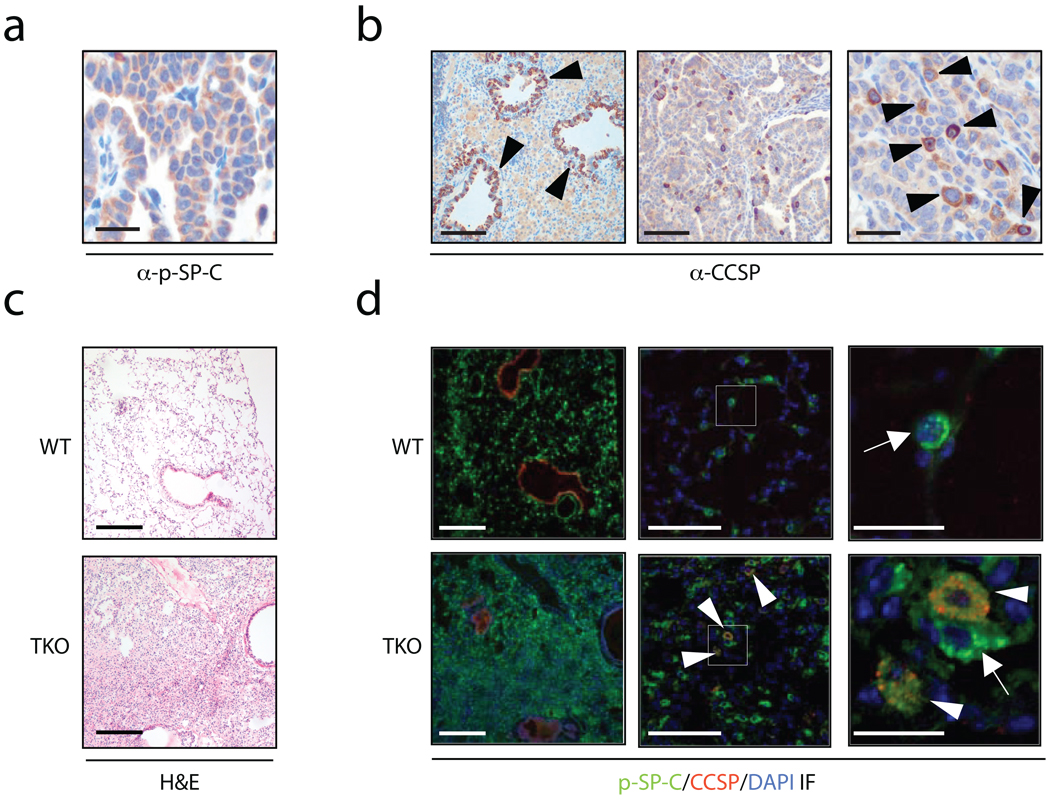
Recently it has been proposed that hallmark stem-cell qualities such as self-renewal, pluripotency, and resistance to injury, could promote and maintain tumorigenesis if gone awry16,17. In this regard, a putative stem cell population in the murine lung, BASCs, was identified and these same cells were connected to adenocarcinoma development18. To investigate the histogenesis of the Dok mutant lung tumors, immunohistochemistry (IH) was performed using antibodies against Clara cell secretory protein (CCSP) and pro-surfactant protein C (pSP-C), commonly used markers that distinguish between Clara cells and AT2 cells, respectively (Fig. 3a–b). Tumor regions were predominantly pSP-C positive (Fig. 3a). As expected, CCSP staining was seen in the cells lining the bronchioles (Fig. 3b, left panel). Tumor regions, however, were predominantly negative for CCSP (Fig. 3b, middle panel), with a few scattered cells showing positive staining of CCSP (Fig. 3b, right panel). This staining pattern suggests that the tumors likely arose either from AT2 cells or their progenitors, BASCs.
Figure 3. Hyperplasia and tumors in Dok KO mice consist of AT2 cells and BASCs.
(a) IHC for the AT2 cell marker pSP-C (brown) in a Dok1/Dok3 DKO tumor. Scale bar, 50 µm. (b) IHC for the Clara cell marker CCSP (brown) in a Dok1/Dok3 DKO tumor. Left panel, arrowheads indicate positive bronchiolar staining. Scale bar, 200 µm. Middle panel, scale bar = 200 µm. Right panel, arrowheads indicate scattered CCSP-positive cells in the tumor area. Scale bar, 50 µm. (c) H&E-staining of wild-type (upper panel) or Dok TKO (lower panel) lung tissue from 12-week old mice. Scale bar, 200 µm. (d) IF for pSP-C (green), CCSP (red), and DAPI (blue) on serial sections to those shown in panel (c). Left panels, scale bar = 200 µm. Middle panels, white boxes indicate the region magnified in the panels to the right. Scale bar, 50 µm. Right panels, close-up of the boxed region shown in the middle panels. Arrows indicate pSP-C+ AT2 cells. Arrowheads indicate pSP-C/CCSP double-positive BASCs. Scale bar, 10 µm.
BASCs are a unique subpopulation of the lung that express both the AT2 marker pSP-C and the Clara cell marker CCSP. We performed double immunofluorescence (IF) staining for these markers in the lungs of 12-week old wild-type and TKO mice. At this timepoint, the majority of TKO mice have not yet developed tumors, and a diffuse hyperplasia is often observed, as shown by H&E staining (Fig. 3c). Similar to the tumors in older TKO animals, hyperplastic regions in young TKO mice were composed primarily of pSP-C+ AT2 cells (Fig. 3d, left panels). Further examination of the TKO IF sections at high magnification, however, revealed a marked increase in the frequency of pSP-C/CCSP double-positive BASCs, which were not only located at their correct anatomic position, the bronchioalveolar duct junction (BADJ), but also aberrantly scattered throughout the TKO alveolar hyperplasia (Fig. 3d, middle and right panels). In contrast, in wild-type animals, BASCs were never found in the alveoli (Fig. 3d, middle and right panels).
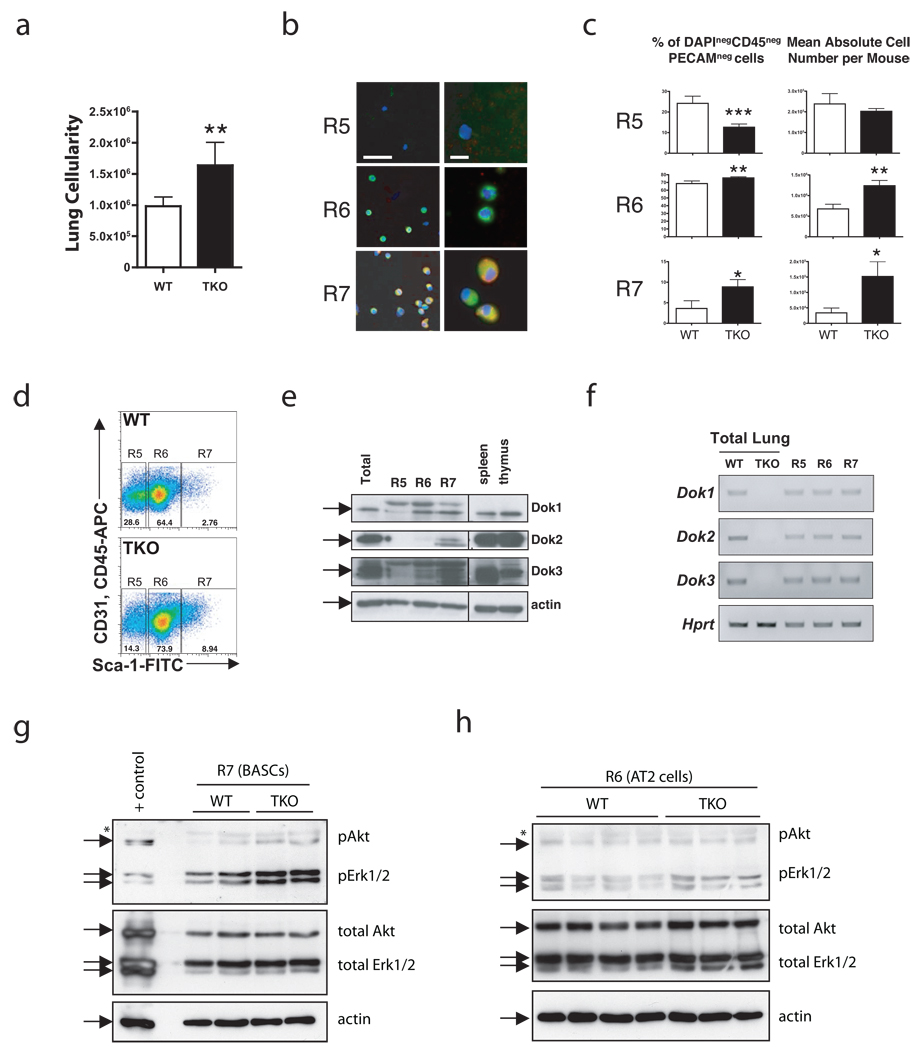
To quantify this phenotype and to determine if Dok genes were indeed expressed in these specific cellular compartments, we dissociated murine lung and used an established BASC flow cytometry sorting strategy18 (Methods and Supplementary Fig. 3a). Interestingly, cell dissociation from TKO lungs consistently yielded more cells than age-matched wild-type lungs, although the overall weight of the mice did not differ (data not shown). To ensure we were measuring pulmonary cells and not other cell types, the cellularity was estimated after using flow cytometry to exclude cells positive for the endothelial marker PECAM-1 and the hematopoietic marker CD45. This analysis confirmed that TKO animals had a significantly higher number of total lung cells than age-matched wild-type mice (Fig. 4a, P < 0.01).
Figure 4. Lung tumorigenesis in Dok TKO mice is preceded by an expansion of AT2 cells and BASCs.
(a) Cellularity of WT and TKO CD45negPECAMneg lung cells after dissociation, counting, and flow cytometry. Data shown is mean+SEM; **, P < 0.01 by two-tailed t-test, n = 5 WT and n = 4 TKO. (b) IF of the indicated populations after FACS. DAPI (blue), pSP-C (green), and CCSP (red). R5: Sca-1negCD45negPecamnegautofllo; R6, AT2 cells: Sca-1negCD45negPecamnegautoflhi; R7, BASCs: Sca-1posCD45negPecamneg. Left, scale bar = 50 µm. Right, scale bar = 10 µm. (c) Summary of percentages (left) and absolute numbers (right) of cell populations from 12-week old WT and TKO mice. Data shown are mean+SEM; *, P < 0.05, **, P < 0.01, ***, P < 0.001 by two-tailed t-test, n = 5 WT and n = 4 TKO. (d) Representative dot plot from flow cytometric analysis of WT and TKO CD45negPECAMneg cell populations using PECAM-APC, CD45-APC, and Sca-1-FITC antibodies. (e) Western blot of sorted cell populations. Cell lysates of splenocytes and thymocytes were used as controls. Arrows indicate Dok1, Dok2, Dok3 or β-actin. (f) RT-PCR of Dok1, Dok2, and Dok3 from WT and TKO unsorted lung, and WT R5, R6, and R7 fractions. Quantitative data is shown in Supplementary Figure 3b. (g) Western blot analysis of BASC lysates from 12-week old mice. 40,000 cells were pooled from 1–3 mice for each lane. Wild-type lung was used as a positive control (+ control). Arrows indicate the bands expected for each protein. A non-specific band is also indicated (*). (h) Western blot analysis of AT2 lysates. Each lane contains a sample from a different animal. Markings are as in (g).
Next, we used the BASC sorting strategy to quantify and purify three lung populations that are negative for PECAM-1 and CD45. The first population (referred to herein as R5) is comprised of mostly unidentified cells and Clara cells, whereas R6 and R7 contain AT2 cells and BASCs, respectively. Purity of each fraction was confirmed by immunofluorescence (IF) for pSP-C and CCSP (Fig. 4b). Consistent with the IF data, flow cytometric analysis showed an expansion of the Sca-1-positive BASC population (R7) and highly autofluorescent AT2 cell population (R6) in the TKO lungs compared to wild-type (Fig. 4c–d). The expansion involved both a significant increase in the percentage (Fig. 4c, left panels) and absolute number (Fig. 4c, right panels) of AT2 cells (P < 0.01) and BASCs (P < 0.05). The absolute number of R5 cells was not significantly altered in the TKO cohort (P = 0.24) indicating that the effect of Dok inactivation was specific to a subset of pulmonary cells.
To determine the expression of Dok genes in these cell populations, we used FACS to collect fractions from wild-type lungs for either protein or RNA isolation. Using Western blot and quantitative RT-PCR, Dok1, Dok2 and Dok3 protein and mRNA were detected in the BASC fraction (R7; Fig. 4e–f and Supplementary Fig. 3b).
We previously identified high levels of Akt and Erk phosphorylation in tumors from Dok mutant mice (see Fig. 2f–g). To further understand if Akt and Erk activation preceded tumor formation in Dok TKO mice, we isolated BASCs and AT2 cells from WT and Dok TKO mice and analyzed the level of Akt and Erk phosphorylation at the same timepoint used for the above analyses. BASCs (Fig. 4g) and AT2 cells (Fig. 4h) from Dok TKO mice exhibited higher levels of Erk phosphorylation than those isolated from wild-type mice. Furthermore, BASCs from TKO lungs showed markedly higher pAkt than BASCs from wild-type lungs. Thus Erk and Akt are activated in lung epithelial cells of Dok TKO mice prior to tumor formation, supporting a direct role for Dok family proteins in regulation of Erk and Akt signaling in the murine lung.
Together these data are consistent with a model of tumorigenesis whereby combinatorial Dok1, Dok2, and Dok3 inactivation leads to hyperactivation of Akt and Erk and an expansion of BASCs with subsequent differentiation into AT2 cells. While we cannot exclude that the tumors originate either from AT2 cells, or from both AT2 cells and BASCs, our analysis further associates an aberrant BASC expansion with preneoplastic and neoplastic lung transformations in the mouse (see Discussion).
Loss of DOK2 expression in human lung cancer
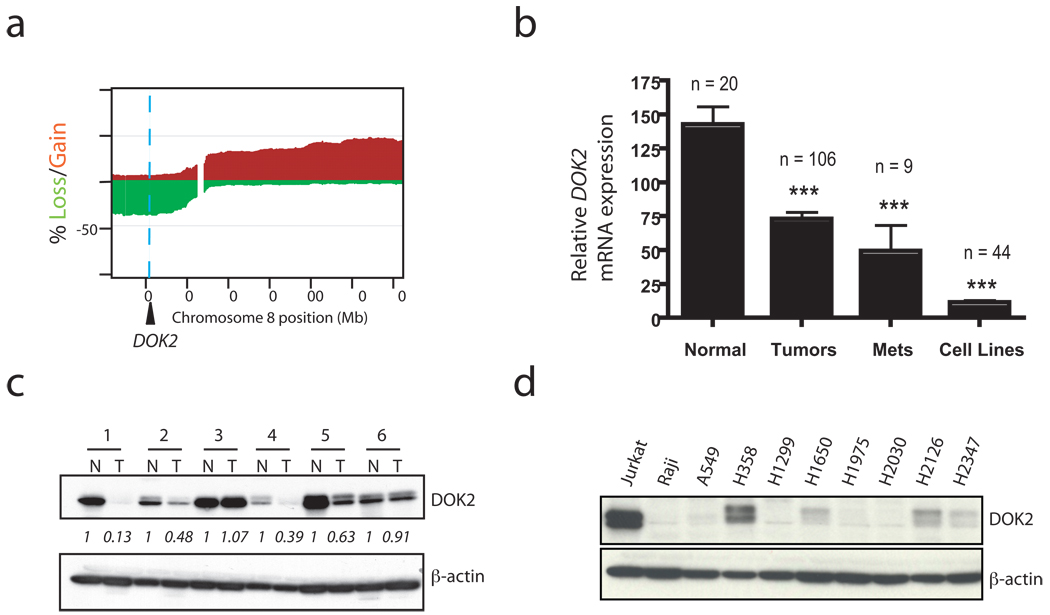
To assess the relevance of these findings to human lung adenocarcinoma, we evaluated the genomic status of DOK1, DOK2, and DOK3 in an array-based comparative genomic hybridization (aCGH) dataset19 of 199 primary human lung adenocarcinoma samples. Interestingly, the DOK2 gene is localized to chromosome 8p21.3, one of the most frequently deleted regions in human lung cancer19–22 and a region long hypothesized to contain one or more tumor suppressor genes21,22. In this dataset, 37% of tumor samples lost one copy of DOK2 (Fig. 5a) whereas DOK1 and DOK3, located at 2p13.1 and 5q35.3, were lost in 1.5% and 7% of cases, respectively. These data are broadly consistent with a recent genome-wide analysis of human lung adenocarcinoma that found frequent deletions of 8p (containing DOK2)20. To further validate the aCGH findings, we analyzed a SNP array of 21 primary human NSCLC tumors. Consistent with the aCGH analysis, one copy of DOK2 was lost in 33% (7 of 21) of the cases (Supplementary Fig. 4).
Figure 5. Loss of DOK2 expression in human NSCLC and functional data implicate DOK2 as a human lung tumor suppressor.
(a) Frequency of chromosome 8 copy number aberration determined by aCGH analysis of 199 primary human lung adenocarcinoma samples. Shown is the percentage of samples with loss (green) or gain (red) of a particular genomic locus on chromosome 8. Regional gain/loss was defined with a log2 ratio threshold of +/−0.15. A blue dashed line indicates the genomic position of DOK2. (b) DOK2 mRNA expression from a microarray of primary lung adenocarcinoma samples, lymph node metastases, cell lines, or normal lung (control). ***, P < 0.001 by two-tailed t-test. Data shown is mean + SEM. (c) Western blot of DOK2 protein or β-actin (loading control) in paired lysates from primary human lung tumors (T) and adjacent normal lung from the same patients (N). Relative abundance of DOK2 was quantified using ImageJ software. The numbers represent the ratio of DOK2 to actin expression after normalization by setting the value of each N sample to 1. (d) Western blot of DOK2 or β-actin (loading control) in human NSCLC cell lines. Jurkat and Raji cells were used as a positive and negative control, respectively. See Supplementary Figure 5d for the relative expression in these lines compared to normal, primary human lung tissue.
Next we evaluated the consequences of this genomic loss on DOK2 expression. To this end, we analyzed the mRNA expression of DOK1, DOK2, and DOK3 in primary and metastatic human lung cancer, lung cancer cell lines, and normal human lung using microarray-based expression profiling. In agreement with the aCGH data, expression analysis identified DOK2 as a target of significant mRNA downregulation in primary lung adenocarcinoma, lymph node metastases, and lung cancer cell lines compared to normal, non-cancerous human lung tissue (Fig. 5b). Unlike DOK2, no significant reduction of DOK1 or DOK3 mRNA expression was observed in the primary tumors, although expression of DOK3 was decreased in lymph node metastases compared to the normal lung (Supplementary Fig. 5a). DOK2 mRNA expression level correlated with copy number loss of DOK2, with significantly lower DOK2 expression in tumors with DOK2 loss than in those without DOK2 loss (Supplementary Fig. 5b). Further validating our finding of decreased DOK2 expression, a search of the microarray database Oncomine23 revealed two independent studies24,25 that confirmed the finding of DOK2 mRNA downregulation in human lung adenocarcinoma (Supplementary Fig. 5c). Based on the high rate of copy number alteration and mRNA downregulation of DOK2, but not DOK1 or DOK3, we focused on DOK2 in our further studies of the role of DOK family members in human lung cancer.
To assess the consequences of DOK2 mRNA downregulation on DOK2 protein levels in human lung cancer, we analyzed the protein level of DOK2 in primary lung cancer specimens and cell lines. We observed a reduction of DOK2 protein in tumor tissue from patients with lung cancer compared to the adjacent, non-involved normal lung of the same patients (Fig. 5c). Similarly, DOK2 protein expression in lung cancer cell lines was either low or undetectable (Fig. 5d and Supplementary Fig. 5d). Moreover, tumor tissue microarray (TMA) analysis of more than 80 lung tumors revealed low or undetectable expression of DOK2 protein in a substantial majority of samples, whereas the expression of DOK1 and DOK3 was usually maintained (Supplementary Fig. 5e).
DOK2 inhibits lung tumor proliferation in vitro and in vivo
The identification of DOK2 downregulation/loss in human lung cancer is consistent with a tumor suppressive role for DOK2 in the lung. To assess if DOK2 could indeed inhibit proliferation of lung cancer cells, we performed add-back experiments in which DOK2 was stably over-expressed in the lung cancer cell line H1299, which normally does not express detectable levels of DOK2. In a classical growth curve assay, cells expressing DOK2 had a significantly impaired growth rate compared to that of cells containing empty vector (Fig. 6a). Similar to the findings in the mouse setting, where Dok KO lung tumors (Fig. 2f–g) and BASCs (Fig. 4g–h) exhibited activation of Erk and Akt, H1299 cells without expression of DOK2 showed markedly higher serum-induced phosphorylation of Erk and Akt compared to those with forced expression of DOK2 (Fig. 6b). Next, a xenograft assay in immunocompromised mice confirmed that DOK2 expression suppresses the growth of H1299 cells in vivo (Fig. 6c–d). These functional data, together with the mouse genetics analysis and the observation that DOK2 is a target of frequent genomic loss and expression downregulation in human lung cancer, implicate DOK2 as a human lung tumor suppressor.
Figure 6. DOK2 suppresses lung cancer cell proliferation in vitro and in vivo.
(a) Growth curve analysis of H1299 cells with and without retroviral-mediated overexpression of DOK2. Relative cell number was determined after cell fixation and crystal violet staining using the optical density at 595 nm (OD 595). Triplicate wells were performed and the data shown is mean+SD of a representative experiment. The experiment was repeated five times. A Western blot, inset, confirms expression of DOK2. (b) Analysis of Erk and Akt activation in the same cells used for the experiment in panel (a). Cells were serum starved in medium containing 0.1% FCS for 12 hours, stimulated with 20% FCS for the times indicated (minutes), then lysed and used for Western blot analysis with antibodies against phosphorylated Erk1/2 (pErk) or phosphorylated Akt (pAkt). (c) Tumor volume measurements of tumors formed from subcutaneous injection of H1299 cells with or without DOK2 expression into the flanks of nude mice. The experiment was performed in triplicate and the data shown is mean+SD. A Western blot, inset, confirms expression of DOK2. (d) Picture of tumors formed by H1299 cells with or without expression of DOK2 taken at week 5 after cell injection. An arrowhead indicates the tumor mass in each panel.
Lung cancer susceptibility in Dok2 heterozygous mutants
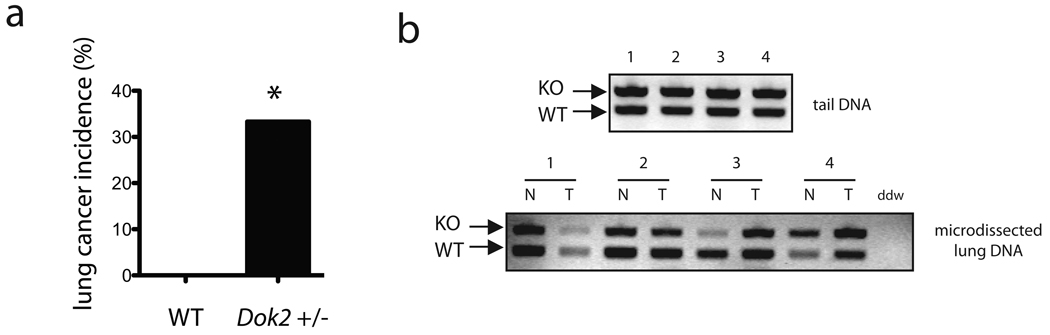
The high frequency of DOK2 heterozygous loss in human lung adenocarcinoma led us to analyze the impact of heterozygous Dok2 loss on lung tumorigenesis in the mouse. Interestingly, lung tumors were found in 4 of 12 Dok2 heterozygous mice at 15–19 months of age, compared to 0 of 13 wild-type mice at this age (P < 0.05 by Fisher’s exact test; Fig. 7a). To determine if Dok2 is a haploinsufficient gene or if tumor progression requires complete loss of Dok2, we used laser capture microdissection to isolate tumors from Dok2 heterozygous mice and then assessed the status of the Dok2 wild-type allele using allele-specific PCR primers. The wild-type Dok2 allele was retained in 4 of 4 tumors (Fig. 7b), indicating that loss of the wild-type allele is not required for tumor development, although formally we cannot at present exclude mutation or silencing of the remaining allele. These data are similar to the human cancer scenario, where we did not observe homozygous deletions of the DOK2 locus, suggesting that Dok2 is haploinsufficient in its tumor suppressive function. We conclude that Dok2 heterozygosity alone can promote lung cancer and human DOK2 should be considered a critical target of 8p loss in human lung adenocarcinoma.
Figure 7. Lung tumorigenesis in Dok2 +/− mice.
(a) Summary of lung tumor incidence in Dok2 +/− mice (n = 12) and wild-type controls (n = 13) at 15–19 months of age. *, P < 0.05 by two-tailed Fisher’s exact test. (b) PCR analysis of genomic DNA from tail DNA (top panel) or laser capture microdissected-lung cells (bottom panel) in four Dok2 +/− mice (#1–4). Tumor cells (T) or normal adjacent lung (N) on the same slide was microdissected and then used for DNA extraction and PCR analysis with Dok2 genotyping primers that distinguish between the KO allele (upper band) and wild-type allele (lower band).
DISCUSSION
In this work, we present a comprehensive analysis of the single and combinatorial functions of the DOK family members Dok1, Dok2, and Dok3 in lung tumor suppression. We find that either heterozygous loss (in the case of Dok2) or complete loss of a single Dok gene can promote tumorigenesis. In addition, Dok1, Dok2, and Dok3 have overlapping functions and can cooperate in lung tumor suppression, because combinatorial knockout of all three Dok genes strongly promotes lung tumor formation in vivo.
We elucidated the cell biological changes that result from Dok inactivation in vivo, specifically the perturbations of the AT2 and BASC cellular compartments that occur in the lungs of Dok TKO mice prior to frank tumorigenesis. BASC expansions are associated with lung tumorigenesis in a number of mouse models18,26–29, but the role of BASCs as cancer-initiating cells for lung cancer remains to be definitively established. Nonetheless, our data demonstrate that combinatorial loss of Dok1, Dok2, and Dok3 perturbs BASC and AT2 cell signaling pathways and homeostasis.
Although Dok single and compound mutant mice develop a range of immune30–34 and leukemia13,14 phenotypes, several pieces of evidence support a cell autonomous mechanism of lung tumorigenesis in Dok mutant mice. First, the kinetics of the CML-like myeloproliferative disorder and lung phenotype are different in Dok TKO mice, with the onset of lung cancer (as early as 6 weeks) preceding the onset of leukemia, in which the expansion of the myeloid compartment becomes apparent at approximately 1 year of age with incomplete penetrance (see Supplementary Fig. 6). Second, the immune phenotypes of the Dok133,34, Dok2, and Dok3 32KO animals are distinct, owing to the cell lineage-dependent expression of these genes in the hematopoietic compartment. In contrast, all three single KO genotypes develop lung cancer. Third, all three Dok genes are expressed in bronchioalveolar stem cells (BASCs) and these cells exhibit aberrant activation of Erk and Akt in vivo in Dok TKO mice. Fourth, enforced overexpression of DOK2 inhibits the growth of human lung cancer cells in vitro and after xenograft. Altogether, these facts indicate that Dok KO mice likely develop lung cancer due to a cell-autonomous deregulation of Erk and Akt signaling resulting from loss of Dok expression in lung epithelial and progenitor cells.
Importantly, we show that DOK2 is the target of frequent copy number loss in human lung cancer, and this genomic loss is accompanied by a downregulation of DOK2 mRNA expression, supportive of a tumor suppressive role for this gene. Moreover, heterozygous loss of DOK2 alone can promote lung tumorigenesis, as demonstrated by the fact that Dok2 heterozygous mice also develop lung cancer, and this occurs in the absence of LOH. These data suggest that DOK2 is a haploinsufficient human lung tumor suppressor.
Recent genome-wide analyses of copy number variation in human lung cancer have provided comprehensive descriptions of the genetic landscape of human cancer19,20,35. However, the success of this approach has been limited to loci subject to focal deletion and focal amplification, which represent only a portion of the genome20. Other regions of the genome, including 8p, are consistently targeted by broad-scale deletions20, and it is now believed that regions of broad losses may encompass multiple, rather than single, tumor suppressors19. Based on these observations, many groups have proposed that at least two to three tumor suppressors may reside on 8p, including at least one tumor suppressor in the 8p21.3 locus, and that the simultaneous compound loss of these tumor suppressors may be required for tumorigenesis19,36–39. In this context, our findings identify DOK2 as an 8p21.3 tumor suppressor, and the possible cooperative tumorigenic effect of compound haploinsufficiency of DOK2 and other putative 8p tumor suppressors, such as DUSP4,19 should be investigated (Supplementary Figure 7).
Our work emphasizes the key role of mouse molecular genetics in identifying and validating candidate tumor suppressor genes in broad-scale deleted regions. This in vivo approach in the mouse proves even more necessary and decisive when tumor suppressor haploinsufficiency and compound haploinsufficiency are suggested to represent the driving forces underlying human tumorigenesis.
Supplementary Material
ACKNOWLEDGEMENTS
For advice and discussion, we thank L.F. Cai, C. F. Kim, T. Motoi, R. Hobbs, J. Clohessy, T. Yung, A. Carracedo, K. Ito, Pandolfi lab members, B. Clarkson, members of the MSKCC Lung Cancer Oncogenome Group, and members of the DF/HCC Lung Cancer Research Program. We thank M. Asher, T. Matos, and A. Egia for histology services and immunohistochemistry. We thank the MSKCC, University of Iowa, and Dana Farber Cancer Institute flow cytometry core facilities for technical assistance. This work was funded by National Institutes of Health/NCI grants (CA-64593) to P.P.P., and Steps for Breath Fund by the Society of MSKCC and the Thomas G. Labrecque Foundation to M.N.
Footnotes
AUTHOR CONTRIBUTIONS
A.H.B., M.N., A.M. and P.P.P. designed and analyzed the experiments. B.S.T., C.B., W.L.G., and M.L. performed the human genetic studies. A.V. and N.D.S. analyzed the SNP array data. J.S., N.M., J.T.F., W.L.G., and M.L. coordinated human pathological sample acquisition and distribution. J.T.F. reviewed all mouse pathology. Some of the experiments were performed in the laboratory of P.B.R. A.H.B., M.N., and P.P.P. wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- 1.Carpino N, et al. p62(dok): a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 2.Yamanashi Y, Baltimore D. Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 3.Di Cristofano A, et al. Molecular cloning and characterization of p56dok-2 defines a new family of RasGAP-binding proteins. J Biol Chem. 1998;273:4827–4830. doi: 10.1074/jbc.273.9.4827. [DOI] [PubMed] [Google Scholar]
- 4.Cong F, Yuan B, Goff SP. Characterization of a novel member of the DOK family that binds and modulates Abl signaling. Mol Cell Biol. 1999;19:8314–8325. doi: 10.1128/mcb.19.12.8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm J, et al. Novel p62dok family members, dok-4 and dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J Cell Biol. 2001;154:345–354. doi: 10.1083/jcb.200102032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowder RJ, Enomoto H, Yang M, Johnson EM, Jr, Milbrandt J. Dok-6, a Novel p62 Dok family member, promotes Ret-mediated neurite outgrowth. J Biol Chem. 2004;279:42072–42081. doi: 10.1074/jbc.M403726200. [DOI] [PubMed] [Google Scholar]
- 7.Okada K, et al. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- 8.Yamanashi Y, et al. Role of the rasGAP-associated docking protein p62(dok) in negative regulation of B cell receptor-mediated signaling. Genes Dev. 2000;14:11–16. [PMC free article] [PubMed] [Google Scholar]
- 9.Di Cristofano A, et al. p62(dok), a negative regulator of Ras and mitogen-activated protein kinase (MAPK) activity, opposes leukemogenesis by p210(bcr-abl) J Exp Med. 2001;194:275–284. doi: 10.1084/jem.194.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones N, Dumont DJ. Recruitment of Dok-R to the EGF receptor through its PTB domain is required for attenuation of Erk MAP kinase activation. Curr Biol. 1999;9:1057–1060. doi: 10.1016/s0960-9822(99)80458-8. [DOI] [PubMed] [Google Scholar]
- 11.Suzu S, et al. p56(dok-2) as a cytokine-inducible inhibitor of cell proliferation and signal transduction. Embo J. 2000;19:5114–5122. doi: 10.1093/emboj/19.19.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao M, Janas JA, Niki M, Pandolfi PP, Van Aelst L. Dok-1 independently attenuates Ras/mitogen-activated protein kinase and Src/c-myc pathways to inhibit platelet-derived growth factor-induced mitogenesis. Mol Cell Biol. 2006;26:2479–2489. doi: 10.1128/MCB.26.7.2479-2489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda T, et al. Role of Dok-1 and Dok-2 in myeloid homeostasis and suppression of leukemia. J Exp Med. 2004;200:1681–1687. doi: 10.1084/jem.20041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niki M, et al. Role of Dok-1 and Dok-2 in leukemia suppression. J Exp Med. 2004;200:1689–1695. doi: 10.1084/jem.20041306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau SK, Luthringer DJ, Eisen RN. Thyroid transcription factor-1: a review. Appl Immunohistochem Mol Morphol. 2002;10:97–102. doi: 10.1097/00129039-200206000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 17.Dick JE, Lapidot T. Biology of normal and acute myeloid leukemia stem cells. Int J Hematol. 2005;82:389–396. doi: 10.1532/IJH97.05144. [DOI] [PubMed] [Google Scholar]
- 18.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 19.Chitale D, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009 doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wistuba II, et al. Allelic losses at chromosome 8p21-23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999;59:1973–1979. [PubMed] [Google Scholar]
- 22.Ohata H, et al. Deletion mapping of the short arm of chromosome 8 in non-small cell lung carcinoma. Genes Chromosomes Cancer. 1993;7:85–88. doi: 10.1002/gcc.2870070204. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su LJ, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stearman RS, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pei XH, Bai F, Smith MD, Xiong Y. p18Ink4c collaborates with Men1 to constrain lung stem cell expansion and suppress non-small-cell lung cancers. Cancer Res. 2007;67:3162–3170. doi: 10.1158/0008-5472.CAN-06-4517. [DOI] [PubMed] [Google Scholar]
- 28.Yanagi S, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Invest. 2007;117:2929–2940. doi: 10.1172/JCI31854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura JJ, et al. p38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet. 2007;39:750–758. doi: 10.1038/ng2037. [DOI] [PubMed] [Google Scholar]
- 30.Yasuda T, et al. Dok-1 and Dok-2 are negative regulators of T cell receptor signaling. Int Immunol. 2007;19:487–495. doi: 10.1093/intimm/dxm015. [DOI] [PubMed] [Google Scholar]
- 31.Shinohara H, et al. Dok-1 and Dok-2 are negative regulators of lipopolysaccharide-induced signaling. J Exp Med. 2005;201:333–339. doi: 10.1084/jem.20041817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng CH, Xu S, Lam KP. Dok-3 plays a nonredundant role in negative regulation of B-cell activation. Blood. 2007;110:259–266. doi: 10.1182/blood-2006-10-055194. [DOI] [PubMed] [Google Scholar]
- 33.Inoue A, Yasuda T, Yamamoto T, Yamanashi Y. Dok-1 is a positive regulator of IL-4 signalling and IgE response. J Biochem. 2007;142:257–263. doi: 10.1093/jb/mvm127. [DOI] [PubMed] [Google Scholar]
- 34.Kashiwada M, et al. Downstream of tyrosine kinases-1 and Src homology 2-containing inositol 5'-phosphatase are required for regulation of CD4+CD25+ T cell development. J Immunol. 2006;176:3958–3965. doi: 10.4049/jimmunol.176.7.3958. [DOI] [PubMed] [Google Scholar]
- 35.Tonon G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujiwara Y, et al. Evidence for the presence of two tumor suppressor genes on chromosome 8p for colorectal carcinoma. Cancer Res. 1993;53:1172–1174. [PubMed] [Google Scholar]
- 37.Emi M, et al. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res. 1992;52:5368–5372. [PubMed] [Google Scholar]
- 38.Macoska JA, et al. Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer Res. 1995;55:5390–5395. [PubMed] [Google Scholar]
- 39.Scholnick SB, et al. Chromosome 8 allelic loss and the outcome of patients with squamous cell carcinoma of the supraglottic larynx. J Natl Cancer Inst. 1996;88:1676–1682. doi: 10.1093/jnci/88.22.1676. [DOI] [PubMed] [Google Scholar]
- 40.Dobbin KK, et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005;11:565–572. [PubMed] [Google Scholar]
- 41.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.