Abstract
BACKGROUND
microRNAs (miRs) participate in many cardiac pathophysiological processes including ischemia/reperfusion (I/R)-induced cardiac injury. Recently, we and others observed that miR-494 was downregulated in murine I/R injured and human infarcted hearts. However, the functional consequence of miR-494 in response to I/R remains unknown.
METHODS AND RESULTS
We generated a mouse model with cardiac-specific overexpression of miR-494. Transgenic (TG) hearts and wild-type (WT) hearts from multiple lines were subjected to global no-flow I/R using the Langendorff system. TG hearts exhibited improved recovery of contractile performance over the reperfusion period. This improvement was accompanied by remarkable decreases in both lactate dehydrogenase release and the extent of apoptosis in TGs, compared to WTs. Also, myocardial infarction size was significantly reduced in TGs upon I/R in vivo, versus WTs. Similarly, acute overexpression of miR-494 in cultured adult cardiomyocytes demonstrated an inhibition of caspase-3 activity and reduced cell death upon simulated I/R. In vivo treatment with antagomiR-494 increased I/R-triggered cardiac injury relative to the administration of mutant antagomiR-494 and saline controls. We further identified that three pro-apoptotic proteins (PTEN, ROCK1 and CaMKIIδ) and two anti-apoptotic proteins (FGFR2 and LIF) were authentic targets for miR-494. Importantly, the Akt-mitochondrial signaling pathway was activated in miR-494-overexpressing myocytes.
CONCLUSIONS
Our findings suggest that although miR-494 targets both pro-apoptotic and anti-apoptotic proteins, the ultimate consequence is activation of the Akt pathway, leading to cardioprotective effects against I/R-induced injury. Thus, miR-494 may constitute a new therapeutic agent for the treatment of ischemic heart disease.
Keywords: microRNA, ischemia/reperfusion, myocardial infarction, cell death, Akt
Introduction
Ischemic heart disease (IHD), a leading cause of death worldwide, is the most common consequence of coronary artery disease (CAD).1, 2 While reperfusion of an occluded human coronary is effective to reduce overall mortality, it is now recognized that restoration of the blood flow through the previously ischemic myocardium can yield additional reperfusion injury including cardiomyocyte dysfunction and cell death.3 The cellular mechanisms underlying ischemia/reperfusion (I/R)-induced injury are complex and involve a multitude of signaling pathways and molecular players.4 Therefore, it would be rational to develop an effective pharmacological or genetic agent aimed at multiple molecular targets. MicroRNAs (miRs), a new class of ~22nt non-protein-coding single-strand RNAs, have emerged as regulators that control the expression of hundreds of proteins. 5 As a consequence, they may widely influence the signaling networks leading to pathological/physiological responses, such as myocardial I/R injury. Indeed, accumulating evidence has shown that miRs are involved in the process of cardiac remodeling and heart failure via regulation of numerous effectors within the same functional pathway.6, 7 Thus, miRs might be attractive nodal therapeutic targets for the treatment of heart disease.
One specific microRNA, miR-494, has recently garnered attention. It was initially identified as one of the upregulated miRs in human retinoblastoma tissues and Waldenström macroglobulinemia cells.8, 9 In addition, increased levels of miR-494 were observed in myocardial microvascular endothelial cells from type 2 diabetic Goto-Kakizaki rats compared with Wistar rats.10 However, miR-494 was downregulated in head and neck squamous cell carcinoma (HNSCC) primary tissue.11 Interestingly, overexpression of miR-494 suppressed the proliferation of HNSCC cells, whereas inhibition of miR-494 stimulated their growth.11 These data suggest that miR-494 may be involved in the initiation and progression of tumor pathogenesis. Recently, several miR profiling studies revealed that miR-494 was downregulated in human failing hearts and animal ischemic/hypertrophic hearts.12–14 However, in ex vivo I/R mouse hearts, we observed that miR-494 levels were significantly increased.15 Therefore, it would be very interesting to elucidate the possible roles of miR-494 in I/R-induced cardiac injury.
In the present study, we generated a mouse model with cardiac-specific overexpression of miR-494 to determine its functional significance in I/R-induced myocardial damage. In parallel, we acutely expressed miR-494 in adult rat cardiomyocytes by infection with a recombinant adenovirus that contains primary miR-494 DNA, and subjected these cells to simulated I/R challenge. For the first time, we demonstrate that increased miR-494 levels protect hearts against I/R-triggered injury; conversely, knockdown of endogenous miR-494 by antagomiR sensitized hearts to I/R-induced injury. Using multiple target-prediction software, Western-blot analysis and a luciferase reporter assay, we identified that miR-494 targeted not only pro-apoptotic proteins (PTEN, ROCK1, and CaMKIIδ), but also anti-apoptotic proteins (FGFR2 and LIF). Intriguingly, these pro-/anti-apoptotic proteins regulate a common downstream Akt-mitochondrial signaling pathway. Our study further revealed that the Akt signaling was activated in miR-494 transgenic hearts, which may contribute to its salutary effects on I/R. Thus, miR-494 may constitute a new therapeutic agent for the treatment of ischemic heart disease.
Materials and Methods
An expanded Materials and Methods section including regional ischemia in vivo, miR extraction and quantitative RT-PCR, global ischemia ex vivo, cardiac injury (necrosis and apoptosis) analysis, construction of adenovirus vectors, preparation of adult rat cardiomyocytes, simulated ischemia/reperfusion treatment, and western-blotting analysis is available in the online data supplement at http://circ.ahajournals.org. All animal protocols conformed to the Guidelines for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health, and were approved by the University of Cincinnati Animal Care and Use Committee.
Generation of a miR-494 Transgenic Mouse Model
Transgenic (TG) mice (FVB/N) were constructed by using a 372-bp DNA fragment containing murine primary miR-494 DNA under the control of the α-myosin heavy chain promoter (α-MHCp). Expression levels of miR-494 in TG hearts were detected by Northern blot, as previously described.15 U6 was used as a loading control to normalize expression levels. MiR-494 probe sequence: 5′-GAG GTT TCC CGT GTA TGT TTC A-3′; U6 probe sequence: 5′-GCA GGG GCC ATG CTA ATC TTC TCT GTA TCG-3′.
Luciferase Reporter Assay for miR-494 Targets
For luciferase reporter experiments, a ROCK1, PTEN, CAMKIIδ, FGFR2, LIF, or Api5 3′-UTR segment of ~120 bp and its respective mutant (all primer sequences are listed in the online supplemental data) were amplified by foot-print two-step PCR (supplemental Figure S1), and inserted into the pMIR-REPORT™ luciferase miRNA expression reporter vector (Ambion) (supplemental Figure S2). HEK293 cells were cotransfected in 12-well plates using DharmaFECT Duo Transfection Reagent (Thermo Fisher Scientific Inc) according to the manufacturer’s instructions, with 0.4 μg of the 3′-UTR luciferase reporter vector and 0.08 μg of the control vector pMIR-β-gal (Ambion, Inc.). Cell lysates were prepared 48-h later, luciferase activity was measured, using a Monolight 3010 luminometer (Pharmingen), and expressed as relative light units using a luciferase assay kit (Promega). β-galactosidase activity was measured with a commercially available kit (Promega). 3′UTR activity of each construct was expressed as the ratio of luciferase/β-galactosidase activity. All transfections were performed in triplicate from three independent experiments.
In Vivo Administration of AntagomiR-494
Chemically modified antisense oligonucleotides (antagomiR) have been used to inhibit microRNA expression in vivo. 15, 16 AntagomiRs were synthesized by Dharmacon (www.dharmacon.com). Sequences are 5′—gSaSgguuucccguguauguuSuScSaS—Chol-3′ (antagomiR-494); 5′—gSaSgguuucccguguuacugSgSaScS—Chol-3′ (antagomiR-494 mutant as a control). Lower case letters represent 2′-O-methyl-modified oligonucleotides, subscript ‘s’ represents a phosphorothioate linkage, ‘Chol’ represents linked cholesterol, and underlined letters are a mutated seed sequence. AntagomiR oligonucleotides were deprotected, desalted and purified by high-performance liquid chromatography. FVB/N male mice (6-week old) received either antagomiR-494, mutant antagomiR-494, or a comparable volume of saline (200 μl) through three consecutive daily injections via tail vein (3×40 mg/kg body weight). Global no-flow ischemia/reperfusion was performed on the third day after the final injection.
Statistical Analysis
Data are expressed as mean ± SD. ANOVA was conducted first across all investigated groups in measurement of myocardial function during ex vivo I/R, in which we performed Shapiro-Wilk test for the normality, and there was no evidence of deviation from normality for all variables. Then, Post hoc pairwise tests were performed with assessment of statistical significance after Bonferroni correction of p values. α level =0.05, only a p value from pairwise test for each time point less than 0.05/6=0.008 would be considered significant. P< 0.05 was considered statistically significant, when Student t-test was used for two-group comparisons in other figures.
Results
Expression Levels of miR-494 in Murine Hearts upon I/R in vivo and ex vivo
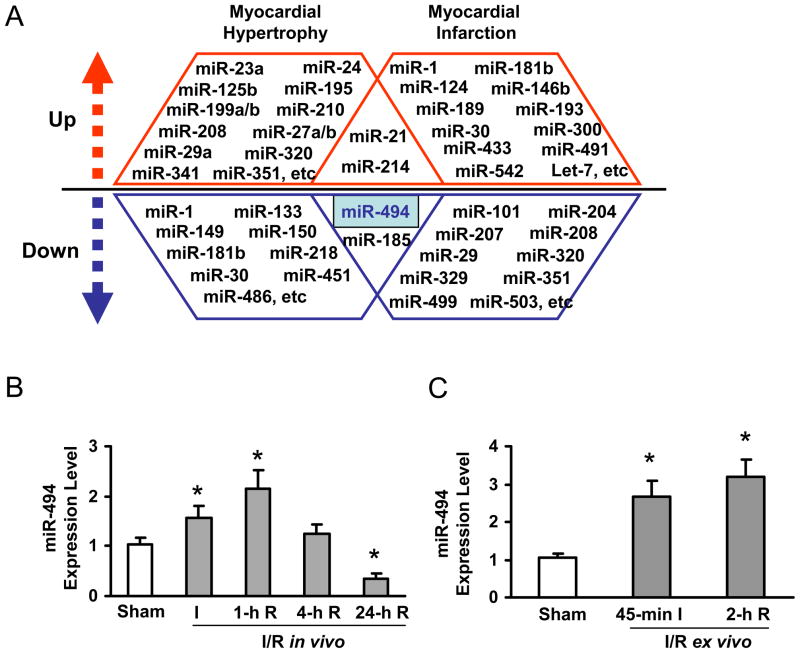
While several studies have reported the signature of miR expression profiles in diseased hearts, some miRs demonstrated conflicting alterations in their expression levels between ischemic and hypertrophic hearts.12–20 For example, levels of miR-1, miR-181b, and miR-30 are increased in ischemic hearts, but decreased in hypertrophic hearts (Figure 1A). Levels of miR-208, miR-320, and miR-351 are downregulated in ischemic hearts, but upregulated in hypertrophic hearts (Figure 1A). Nonetheless, several miRs demonstrated a similar alteration pattern, such as miR-21 and miR-214 (upregulated); miR-494 and miR-185 (downregulated), in both ischemic and hypertrophic hearts (Figure 1A). In the present study, we focused our measurements on miR-494 by stem-loop real-time PCR in both in vivo and ex vivo I/R hearts (Figure 1B, C and Supplemental Figure S3). Compared to a sham group, levels of miR-494 were increased by 1.6 ± 0.2-fold in 30-min LAD occluded hearts and kept increasing by 2.1 ± 0.4-fold at 1-h reperfusion. Then, miR-494 returned to basal levels at 4-h reperfusion and further decreased by 66% in the myocardium at 24-h reperfusion (Figure 1B). In ex vivo 45-min-ischemic hearts, miR-494 levels were increased by 2.6 ± 0.4-fold and further increased by 3.2 ± 0.5-fold after 2-h reperfusion (Figure 1C). These findings indicate that alteration of miR levels is dynamic and dependent on the physiological/pathological conditions. Our results suggest that down-regulation of miR-494 may be involved in I/R-induced myocardial damage, whereas transient upregulation of miR-494 may represent an adaptive response to I/R challenge.
Figure 1.
Expression levels of miR-494 were dynamically regulated in ischemic/reperfused hearts. (A) Summary of miR expression profiles in ischemic and hypertrophic hearts.12–20 (B) Dynamic alteration of miR-494 expression in the heart subjected to in vivo 30-min ischemia followed by reperfusion. (C) miR-494 was upregulated in ex vivo hearts subjected to 45-min ischemia followed by 2-h reperfusion. *, p<0.01 vs. sham controls, n=5 for Sham and I group, and n=3 for 1-h R, 4-h R and 24-h R groups. n=4 for 45-min I and 2-h R groups.
Generation and Characterization of miR-494 Transgenic Mouse Model
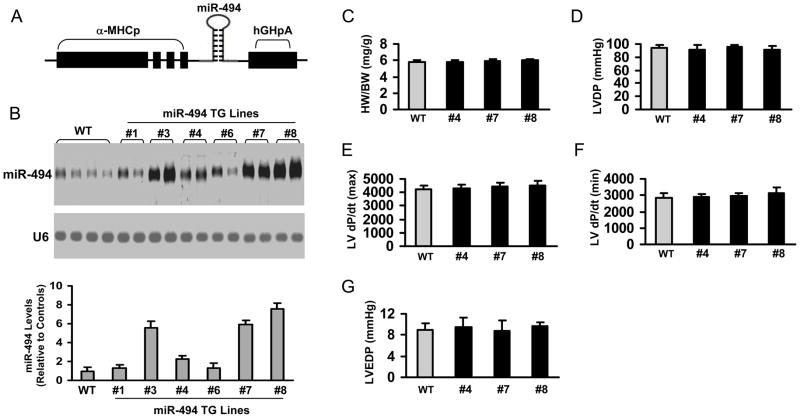
MiR-494 is predicted to have hundreds of potential targets (mouse miR-494 has 245 targets; human miR-494 has 360 targets, TargetScan 5.1, 2009, http://www.targetscan.org). More importantly, these assumed targets include both well-characterized pro-apoptotic (SOCS6, PTEN, ROCK1, CAMKIIδ, etc) and anti-apoptotic proteins (FGFR2, LIF, Api5, IGF1R, FGF7, etc). Therefore, it is necessary to evaluate the long-term and global consequences of miR-494 overexpression in adult hearts. We used a cardiac-specific promoter to drive the expression of a 372-bp primary miR-494 (Figure 2A). Northern blot analysis (Figure 2B) revealed that miR-494 was successfully overexpressed in the transgenic (TG) hearts from several lines (Lines #3, #4, #7 and #8) with a 2.3–7.5-fold expression. Thereafter, we selected Lines#4, #7 and #8 for further biochemical and physiological studies. MiR-494 TG mice from Lines#4, #7 and #8 showed normal heart weight/body weight ratio (Figure 2C) and normal cardiomyocyte cross-sectional area (Supplemental Fig. S4). The extent of fibrosis and capillary density was also no different between WTs and TGs (Supplemental Fig. S4). Cardiac function, assessed by Langendorff preparations, was normal in miR-494 TG mice (Figure 2D–G). Thus, increasing miR-494 in the myocardium did not alter normal cardiac growth and contractile function.
Figure 2.
Models of cardiac-specific overexpression of miR-494 showed no apparent cardiac morphological, physiological, or pathological abnormalities. (A) Diagram of miR-494 TG vector. (B) miR-494 expression was detected by Northern-blot. U6 was used as an internal control. (n=4). (C) The ratio of heart weight/body weight (HW/BW) was similar between WT and miR-494 TG mice. (p>0.1, n=14). (D–G) Myocardial contractile function (LVDP, ±dP/dt, and LVEDP) showed no difference between WT and miR-494 TG hearts. (n=12, WT; n=6 for each TG line; p>0.1 vs WT)
Overexpression of miR-494 Improves the Post-Ischemic Recovery of Cardiac Function
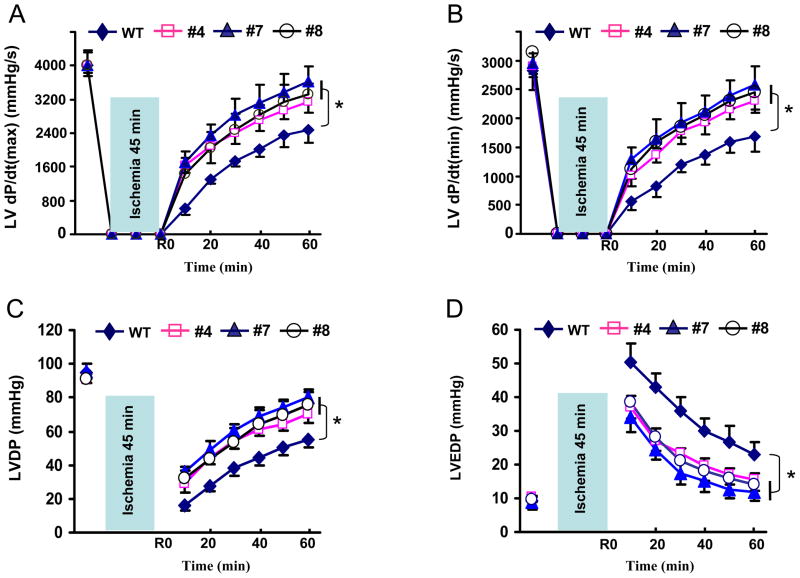
Next, we determined the effects of miR-494 overexpression on post-ischemic injury. To exclude the involvement of inflammatory components during reperfusion, we used an isolated perfused-heart preparation. Both TG and WT hearts were subjected to 45 minutes of global no-flow ischemia followed by 1 hour of reperfusion. Upon ischemia/reperfusion, TG hearts exhibited significantly better functional recovery than WT hearts (Figure 3), as determined by the rates of contraction (Figure 3A), relaxation (Figure 3B), and left ventricular developed pressure (LVDP, Figure 3C). In addition, the left ventricular end diastolic pressure (LVEDP) was significantly depressed in miR-494 hearts during reperfusion, compared with WT controls (Figure 3D). Together, these data reveal that increased levels of miR-494 (2.3–7.5-fold) are associated with improved cardiac functional recovery upon ex vivo ischemia/reperfusion. Notably, TG hearts with 2.3 ± 0.3-fold overexpression of miR-494 (Line#4) showed the effects similar to TGs with 5.9 ± 0.4-fold (Line #7) and 7.6 ± 0.6-fold (Line #8) overexpression of miR-494 in response to I/R, pointing to the complexity of miR dosage in vivo. However, these gene-dosage effects are consistent with others’ reports using transgenesis, e.g. TG hearts with 2.5-fold overexpression of Sirt1 exhibited similar resistance against oxidative stress to those with 7.5-fold overexpression.21
Figure 3.
MiR-494 hearts showed improved functional recovery (A–D) after 45-min ischemia followed by 1-h reperfusion. (n=12 for WT; n=6 for each TG line; *, p<0.008 vs. WT)
Overexpression of miR-494 Reduces I/R-Induced Cardiac Injury
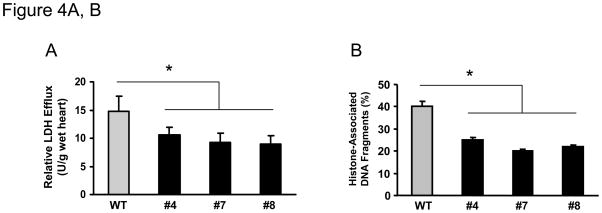
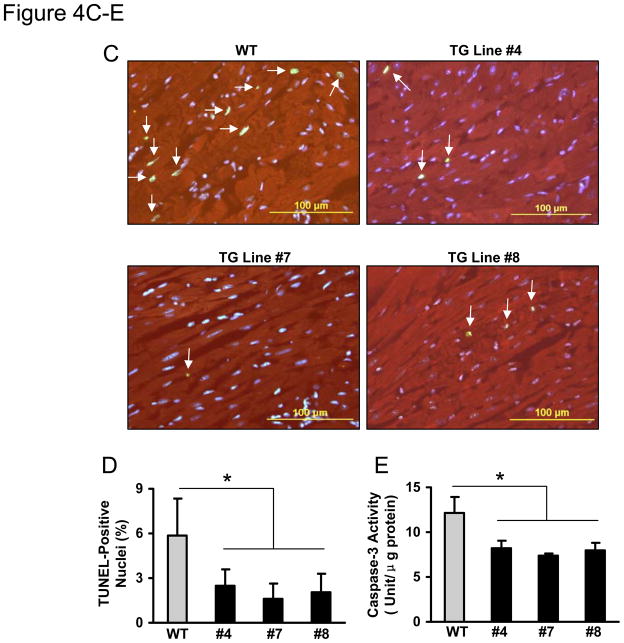
It is well appreciated that maintaining adequate numbers of myocytes is critical to the overall preservation of cardiac structural integrity and function following I/R.22–24 Given the markedly improved recovery of myocardial function in miR-494 hearts, we then determined the effects of miR-494 on post-ischemic cellular damage. The extent of necrotic and apoptotic cell death was examined after ex vivo ischemia for 45 minutes followed by 1 hour of reperfusion. Under basal conditions, release of lactate dehydrogenase (LDH), a biochemical marker for necrotic cell death, did not differ between miR-494-hearts and WT controls (data not shown). However, upon ischemia/reperfusion, LDH release was significantly suppressed by ~40% in miR-494 hearts, compared to WT hearts (Figure 4A). These results indicate that overexpression of miR-494 inhibits I/R–initiated cellular disruption in the myocardium. Furthermore, heart lysates from a subset of experimental animals were assayed for histone-associated DNA fragmentation, which exhibited a ~50% decrease in miR-494 hearts over WTs (Figure 4B). Additionally, the number of TUNEL-positive nuclei was significantly reduced in the ischemic/reperfused miR-494 myocardium (#4: 2.5±1.1%; #7: 1.6 ± 1.0%; #8: 2.1±1.3%), relative to WT controls (5.9 ± 2.4%, n=5, p<0.05, Figure 4C and D). Moreover, the activity of caspase-3 was decreased by ~40% in miR-494 hearts upon I/R, relative to WTs (Figure 4E). Therefore, all three assays (DNA fragmentation, TUNEL assay and caspase-3 activity) demonstrate that overexpression of miR-494 protects the myocardium against I/R-induced cellular damage.
Figure 4.
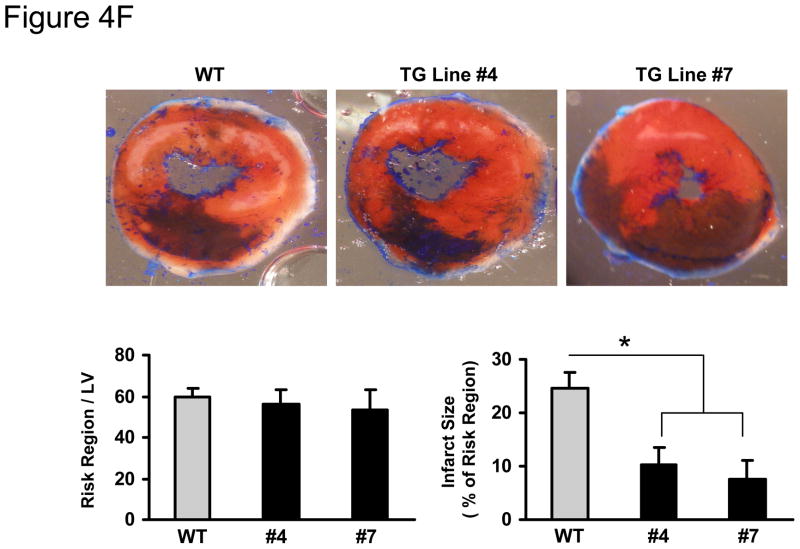
MiR-494 overexpression attenuated I/R-induced myocardial damage. (A) Total LDH in coronary effluent, collected during the first 10 min of reperfusion, was significantly decreased in miR-494 TG hearts, compared to WTs. (B) The degree of histone-associated DNA fragmentation, (C & D) the number of TUNEL-positive nuclei, and (E) caspase-3 activity were reduced in miR-494 hearts subjected to ex vivo 45-min ischemia followed by 1-h reperfusion. n=12, WT; n=6 for each TG line for LDH release assay; n=5 for TUNEL staining; n=6 for DNA fragmentation and caspase-3 activity assay; *, p<0.02 vs WT controls. (F) miR-494 overexpression greatly reduced in vivo myocardial infarct size after 30 min-LAD occlusion followed by 24 h-reperfusion. n=10 for WT, n=6 for TG Line #4, and n=11 for TG Line #7; *, p<0.001 vs. WT controls.
To further determine the consequence of increased miR-494 expression under in vivo conditions, we subjected WT and TG hearts to in vivo 30-min myocardial ischemia, via coronary artery occlusion, followed by 24-h reperfusion. We observed that the ratio of infarct-to-risk region was 10.2 ± 3.3% in Line #4 hearts (n=6) and 7.6 ± 2.4% in Line #7 hearts (n=11), compared to 24 ± 3.2% in WT hearts (n=10, p<0.001); whereas the region at risk was not significantly different between groups (Figure 4F). These results are consistent with our ex vivo findings that miR-494 has cardioprotective effects on I/R-induced injury.
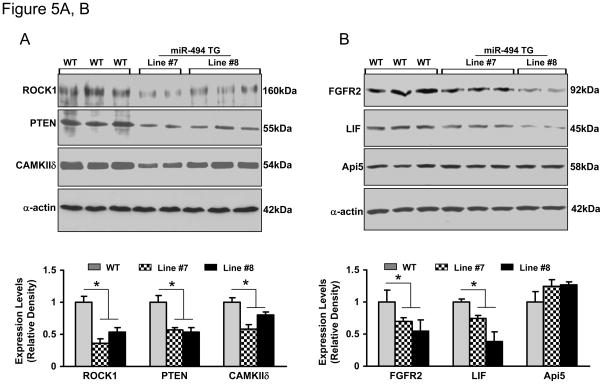
miR-494 Down-Regulates Expression of both Pro-Apoptotic and Anti-Apoptotic Proteins
To elucidate the potential mechanism(s) of miR-494-mediated cardioprotection, we assessed expression levels of miR-494 targets in TG hearts. Theoretically predicted by TargetScan, miRDB and PicTar, miR-494 has hundreds of potential targets which were surprisingly divided into two subgroups: pro-apoptotic and anti-apoptotic proteins. Considering the importance of these candidates in ischemic heart disease, we selected the following putative targets for Western blot analysis (pro-apoptotic: ROCK1, PTEN, and CAMKIIδ; anti-apoptotic: FGFR2, LIF, and Api5). As expected, the levels of proteins ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF were significantly decreased in miR-494 transgenic hearts, compared with those of WT hearts (Figure 5 and supplemental Figure S5). However, Api5 was not downregulated, although it is predicted to be a target of miR-494 by seed-sequence matching (Figure 5B and Supplemental Figure S6). Therefore, our findings suggest that some predicted targets are not authentic. Actually, accumulating evidence has suggested that perfect seed pairing may not necessarily be a reliable predictor for miR-mRNA interaction, as these bioinformatics tools do not consider mRNA secondary structures that may affect miR-target recognition.25
Figure 5.
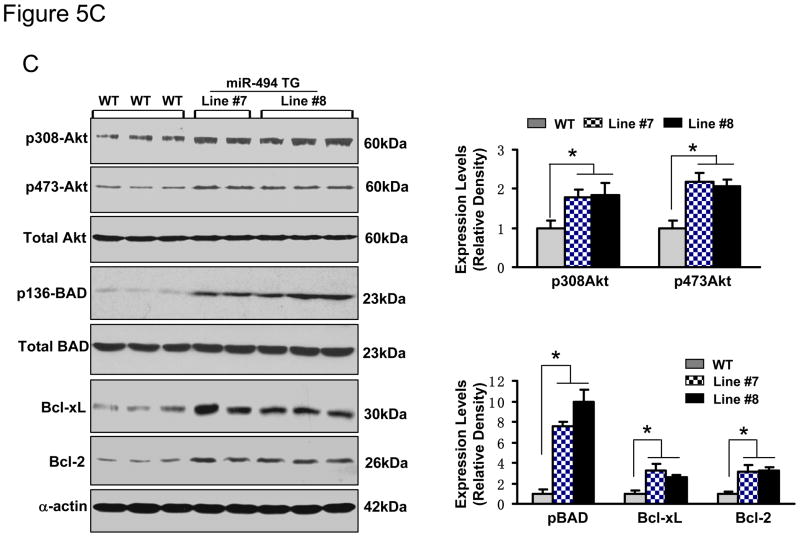
Overexpression of miR-494 downregulated protein levels of ROCK1, PTEN, CAMKIIδ, FGFR2 and LIF, but not Api5 (A and B). These six targets were predicted by TargetScan (Supplemental data). (C) The Akt-mitochondrial signaling was activated, as evidenced by increased phosphorylated Akt (S473), phosphorylated BAD (S136), Bcl-xL and Bcl-2 levels in miR-494 TG hearts. n=5; * p<0.001 vs. WT controls.
It is well documented that inhibition/downregulation of ROCK1, PTEN and CAMKIIδ by either pharmacological or genetic approaches, improves myocardial survival following I/R injury. Furthermore, these beneficial effects were mainly associated with activation of the pro-survival Akt pathway.26–29 On the other hand, both LIF and FGFR2 have been shown to be survival signal mediators in cardiomyocyte response against myocardial infarction.30–32 More interestingly, protective effects of LIF and FGFR2 were also related to upregulation of the Akt signaling. 30–32 Hence, we next examined whether the Akt signaling was activated or inhibited in miR-494 transgenic hearts. As shown in Figure 5C, while the total Akt was not altered, levels of phosphorylated Akt (T308 and S473) were increased by ~2-fold in miR-494 hearts, relative to WT controls. Accordingly, BAD, a downstream target of Akt, was greatly activated in miR-494 hearts, as evidenced by an 8–10-fold increase in levels of phosphorylated BAD. Furthermore, mitochondrial signaling molecules, Bcl-xL and Bcl-2, were upregulated in miR-494 transgenic hearts compared with WT samples (Figure 5C). Together, these findings reveal that, while miR-494 targets both pro-apoptotic kinases and anti-apoptotic growth factors/receptors, as indicated above, the ultimate consequence of miR-494 is activation of the Akt-mitochondrial signaling pathway in the myocardium.
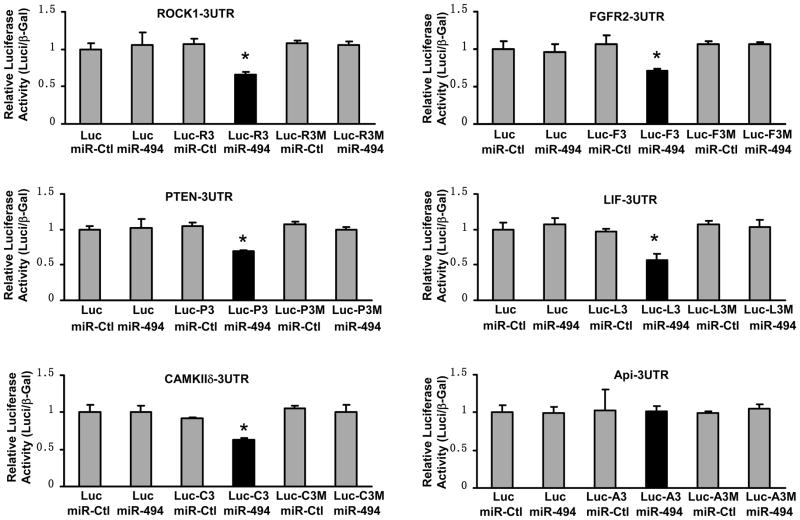
miR-494 Directly Targets at 3′ UTRs of both Pro-Apoptotic and Anti-Apoptotic Proteins
To further validate whether miR-494 directly recognizes 3′-UTRs of ROCK1, PTEN, CAMKIIδ, FGFR2, LIF and Api5, we generated luciferase reporter constructs harboring a segment of ROCK1, PTEN, CAMKIIδ, FGFR2, LIF, or Api5 3′UTR. Their respective mutant constructs contained a mutated seed sequence (Supplemental primer list and Figure S2). These luciferase reporter constructs were co-transfected into HEK293 cells with either miR-494 or miR control. β–gal vector was also co-transfected as an internal control. 48 hours after transfection, we conducted luciferase activity assays and observed that co-transfection of miR-494 strongly inhibited the luciferase activity from the reporter construct containing the 3′UTR segment of ROCK1, PTEN, CAMKIIδ, FGFR2, or LIF; whereas no effect was observed with their corresponding mutated constructs (Figure 6). This effect was specific, because there was no change in the luciferase reporter activity when a negative control miR was co-transfected with either reporter construct (Figure 6). Consistent with the above Western blot data, luciferase activity was not altered in cells co-transfected with miR-494 and the reporter vector of Api5-3′UTR (Figure 6). Collectively, these results indicate that transcripts of ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF may represent genuine targets of miR-494; whereas Api5, albeit predicted, appears to be a false target of miR-494.
Figure 6.
Luciferase-reporter assays showed that ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF were authentic targets of miR-494; however, Api5 was a false target of miR-494, albeit predicted by Target-Scan software. β-gal was used as transfection control. Similar results were observed in three additional, independent experiments (*, p< 0.003 versus miR mimic controls). Luc=plasmid luciferase; Luc-R3=plasmid Luc-ROCK1-3UTR; Luc-R3M= plasmid Luc-ROCK1-3UTR mutant; and so on.
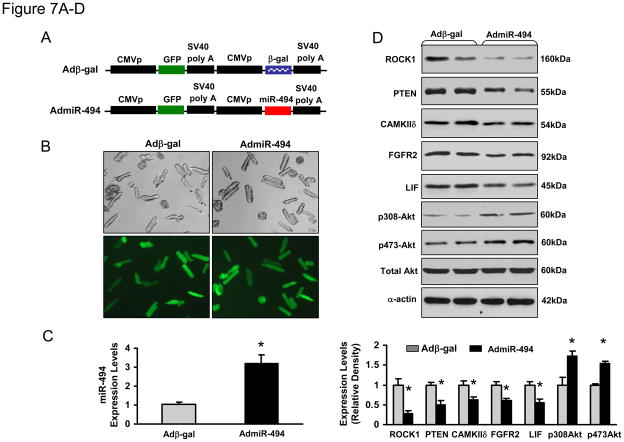
miR-494 Protects against Simulated I/R-induced Cardiomyocyte Death in vitro
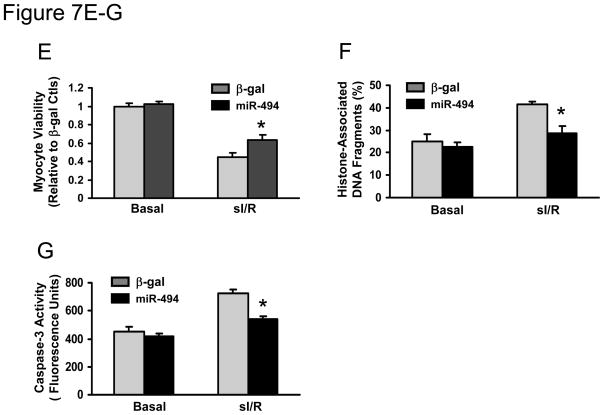
Considering that chronic overexpression of miR-494 in vivo may have compensatory effects, which influence the consequence of miR-494 in the ischemic myocardium, we performed further studies using ex vivo cultured adult cardiomyocytes, a well-controlled experimental setting. Cardiomyocytes were infected with AdmiR-494 and Adβ-gal (Figure 7A). Following infection for 48-h, we observed a nearly 95% infection efficiency(Figure 7B). Importantly, there were no apparent morphological alterations or differences in the number of adherent cells and rod-shaped cells between AdmiR-494- and Adβ-gal-infected groups (Figure 7B). Real-time PCR revealed that miR-494 was overexpressed by 3-fold in AdmiR-494-infected cardiomyocytes (Figure 7C), which resulted in the downregulation of ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF (Figure 7D). As a consequence, Akt was activated in miR-494-cells (Figure 7D). These data further validate the above in vivo results (Figure 5A and B; Figure 6). We next examined effects ofmiR-494 on cell survival upon 1-h simulated ischemia followed by 3-h reperfusion. Cell viability analysis showed that ectopic expression of miR-494 increased cell survival by 40 ± 3% relative to β-gal-cells (Figure 7E). Furthermore, upon I/R, AdmiR-494-infected myocytes exhibited a significant decrease in histone-associated DNA fragmentation, compared to Adβ-gal-infected myocytes (28 ±3% vs. 41±1%, Figure 7F). Consistently, caspase-3 activity in cell lysates upon I/R was significantly reduced in miR-494-cells, compared with controls (Figure 7G). Taken together, these data indicate that ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF are downregulated by acute expression of miR-494, contributing to attenuation of I/R-induced cell damage.
Figure 7.
Acute overexpression of miR-494 in adult cardiomyocytes protected against simulated I/R insults. (A) Constructs of AdmiR-494 and control. (B) More than 95% of cardiomyocytes were infected by AdmiR-494 and Adβ-gal. (C) Real-time PCR showed 3-fold overexpression of miR-494 in AdmiR-494-infected cells. (D) Acute overexpression of miR-494 downregulated protein levels of ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF. As a consequence, Akt was activated. α-actin was used as an internal control. (E) miR-494-cardiomyocytes were resistant to simulated I/R-triggered cell death. Overexpression of miR-494 reduced simulated I/R-triggered (F) histone-associated DNA fragmentation and (G) caspase-3 activity. n=5 hearts; *, p<0.008 vs. Adβ-gal control.
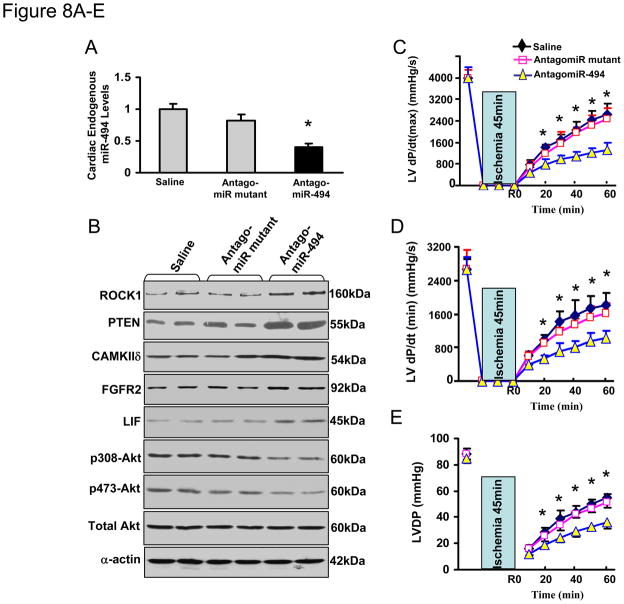
Knockdown of miR-494 Renders the Heart More Sensitive to I/R Injury
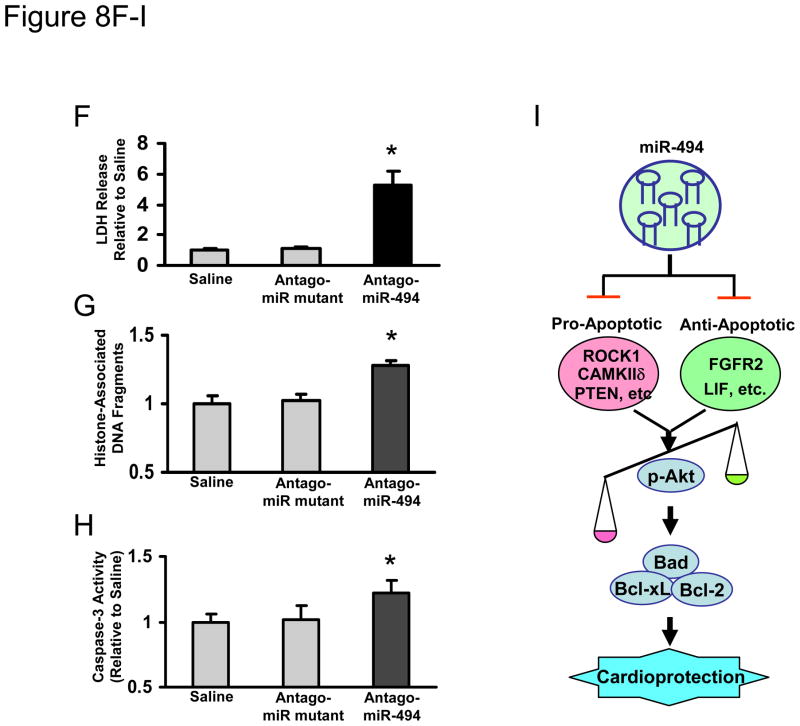
To recapitulate the in vivo setting that miR-494 is downregulated in the heart following I/R, we treated mice with an antagomiR-494 via three consecutive daily tail vein injection of cholesterol-modified antagomiR-494 (3×40 mg/kg). Mutated antagomiR-494 and saline were used as controls. Three days after the last administration of antagomiR-494, stem-loop RT-PCR showed a 60 ± 5% reduction of miR-494 expression in the heart tissue (Figure 8A). In contrast, the mutated antagomiR-494 had no effect on the expression level of miR-494 compared with the saline control (Fig. 8A). These results indicate that antagomiR treatment efficiently downregulates miR-494 expression in the heart, consistent with previous reports using antagomiRs. 15, 16 Consequently, administration of antagomiR-494, but not the antagomiR-494 mutant, yielded the upregulation of ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF in the myocardium, which ultimately reduced Akt activation (Fig. 8B). Furthermore, we observed that antagomiR-treated hearts exhibited significantly depressed functional recovery during ex vivo I/R (45 min/1h), evidenced by decreased rates of contraction (+dP/dt), relaxation (−dP/dt) and left ventricular developed pressure (LVDP) in antagomiR-494-hearts, relative to controls (Fig. 8C–E). In addition, treatment with antagomiR-494 led to 5.2 ± 0.9-fold increases in LDH release during reperfusion, relative to antagomiR-mutant- and saline-treated hearts (Fig. 8F). Similarly, knock-down of endogenous miR-494 by antagomiR increased cardiac DNA fragmentation and caspase-3 activity by 1.3 ± 0.02-fold and 1.2 ± 0.08-fold, respectively, compared with controls upon I/R (Fig. 8G & H). Collectively, these data indicate that reduced endogenous levels of miR-494 sensitize the heart to I/R-induced injury.
Figure 8.
Blockade of endogenous miR-494 by antagomiR-494 sensitized the heart to I/R-induced injury. (A) Levels of miR-494 were determined by real-time PCR (n=4, *, p<0.001 vs. saline- and mutant-treated controls). (B) Protein levels of ROCK1, PTEN, CAMKIIδ, FGFR2, and LIF were increased in antagomiR-494-treated hearts. Consequently, Akt was inactivated by reduced levels of phosphorylated Akt (T308 and S473). (C–E) AntagomiR-494 treatment suppressed cardiac functional recovery during ischemia/reperfusion, compared to saline- and mutant-treated controls (n=4, *, p<0.008). (F) Total LDH in coronary effluent, collected during the first 10 min of reperfusion, was significantly increased in antagomiR-494-treated hearts, compared to controls (n=4, *, p<0.001). (G) The degree of histone-associated DNA fragmentation and (H) caspase-3 activity were increased in antagomiR-494-hearts subjected to ex vivo 45-min ischemia followed by 1-h reperfusion. n=4; *, p<0.04 vs. saline- and mutant-treated controls. (I) Overexpression of miR-494 down-regulates both pro- and anti-apoptotic proteins, which ultimately tilts the balance of a downstream Akt-mitochondrial signaling pathway towards activation, leading to cardioprotection against I/R-induced injury.
Discussion
In recent years, tremendous effort has been made in utilizing a miR microarray screening approach to address the miR expression profiling in infarcted hearts from animal models and human patients.14–20 However, the role of a specific miR in ischemic heart disease is just emerging. In the present study, we center on elucidating the functional consequences of miR-494 in vivo, using gain-of-function and loss-of-function approaches. While miR-494 has been reported to be downregulated in the hypertrophic heart,18 chronic overexpression of miR-494 in vivo did not yield any cardiac morphological or pathological abnormalities (Fig. 2 and Supplemental Fig. S4). More excitingly, we observed that miR-494-overexpressing hearts were resistant to I/R-induced injury in vivo and ex vivo; whereas knockdown of endogenous miR-494 by antagomiR showed the opposite effect. Taken together, these data clearly demonstrate that increased levels of miR-494 render cardioprotection against I/R injury. Our study also suggests that down-regulation of miR-494 observed in human patients14 may be causally involved in the progression of heart failure, at least in part.
A prominent characteristic of miR-494 is that it potentially regulates both anti-apoptotic (e.g. CDK6, FGFR2, IGF2BP1, XIAP, HLF, LIF, API5, FGF7, IGF1R, HDGF, etc.) and pro-apoptotic (e.g. SOCS6, PTEN, ROCK1, PPARGC1α, CAMKIIδ, etc) proteins. Therefore, it is plausible that miR-494 may function as either pro-apoptotic or anti-apoptotic depending on the cellular environment and cell-specific expressed targets. For instance, in JHU-021 cancer cells, transfection of miR-494 induced cell death, indicating its pro-apoptotic role, 11 although the underlying mechanisms remain unknown. In cardiomyocytes, our results have implicated miR-494 as an anti-apoptotic microRNA via activation of the Akt signaling pathway. While we did verify that three pro-apoptotic proteins (PTEN, ROCK1, CAMKIIδ) and two anti-apoptotic proteins (FGFR2 and LIF) were bona fide targets of miR-494, the individual contribution of these targets to the downstream Akt pathway may be unequal. In miR-494-overexpressing cardiomyocytes, the impact of PTEN, ROCK1, and CAMKIIδ (either individually or in combination) on the Akt signaling may be more significant than those of FGFR2, LIF, or other anti-apoptotic targets, thereby tilting the balance of the Akt pathway towards activation (Fig. 8I). These interesting findings suggest that divergent targets of a miR may work unequally to balance a common signaling pathway and eventually affect its functional consequences.
In addition, we performed mRNA array analysis for miR-494 TG hearts and observed that a total of 685 mRNAs were significantly altered (upregulated: 292; downregulated: 393, supplemental Tables S1 and S2). Surprisingly, among these 685 mRNAs, only 6 are predicted to be targets of miR-494 by TargetScan. More intriguingly, mRNA levels of PTEN, ROCK1, CAMKIIδ, FGFR2 and LIF showed no alteration (supplemental Tables S3), while their protein levels were downregulated in miR-494-cardiomyocytes. The inconsistency between mRNA and protein levels suggests that the chronic miR-overexpressing model may have limitations when using conventional mRNA expression arrays to detect miR targets. Another possibility may be that miR-494 dampens target-protein levels mostly by impeding their translation, rather than by breaking down their messenger RNAs. Notably, a recent study using human bronchial epithelial cells33 showed that miR-494 regulated PTEN protein expression, but not mRNA levels, which is consistent with our present findings. To further identify the whole target network for miR-494, a new technique named SILAC (stable isotope labeling with amino acids in cell culture) 34, 35 will be an ideal tool; however, such analysis may fall outside the scope and intent of this study.
Currently, it is well accepted that inhibition of PTEN, ROCKs, or CaMKII has a major impact on improving myocardial survival following an I/R episode.26–29 While several pharmacological agents can effectively inhibit PTEN, ROCKs or CaMKII, these inhibitors may be toxic in physiological settings and their clinical applications may be restricted.36, 37 Thus, it would be very important to identify novel substitutes that orchestrate suppression of PTEN, ROCKs and CaMKII in the myocardium. In the present study, we clearly showed that overexpression of miR-494 reduced the levels of PTEN, ROCK1 and CaMKIIδ, which might work in concert to activate the Akt signaling, a critical survival pathway in the myocardium. However, it may be experimentally complicated to link PTEN, ROCK1 and CaMKIIδ together for elucidating mechanisms underlying the protective effects of miR-494 against I/R. Further studies are needed to ascertain the causal relationship between miR-494-modulated targets and their functional consequences.
In conclusion, to our best knowledge, this is the first report to demonstrate the protective role of miR-494 in I/R is mediated, at least in part, via activation of the Akt-mitochondrial signaling. The present observations expand our understanding of miRs associated with ischemic heart disease and may provide a basis for novel therapeutic strategies aimed at enhancing cardiomyocyte survival in the ischemic myocardium.
CLINICAL PERSPECTIVE.
MicroRNAs (miRs), a new class of non-protein-coding small RNAs, have emerged as regulators that control the expression of hundreds of proteins. As a consequence, they may widely influence the signaling networks leading to pathological or physiological responses such as myocardial ischemia/reperfusion-induced injury and ischemia preconditioning-elicited cardioprotection. While miR expression has been profiled in infarcted hearts from animal models and human patients, the role of a specific miR in ischemic heart disease is just emerging. Uncovering miRs as important regulators not only for single genes but also for whole gene networks has enormous therapeutic implications. In the present study, we discovered that increased levels of mature miR-494 rendered cardioprotection against ischemia/reperfusion (I/R)-induced injury, whereas knockdown of endogenous miR-494 by administration of antagomiR-494 sensitized hearts to I/R injury. Importantly, we identified that miR-494 targeted PTEN, ROCK1 and CAMKIIδ in cardiomyocytes, consequently, activated the Akt signaling pathway. These data suggest that down-regulation of miR-494 observed in human failing hearts may be causally involved in the progression of heart failure, at least in part. Therefore, systemically or locally administration of miR-494 may introduce the newest prospect for the management of ischemic heart disease.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Mr. Daniel Pietras, Drs. Jiang Qian, Tracy Pritchard, and Stela Florea for reading through the whole manuscript and making corrections. The authors also thank Dr. Evangelia G. Kranias for her suggestions and support.
Funding Sources: This study was supported by NIH grants HL-087861, HL087861-03S1 and University of Cincinnati Center for Environmental Genetics (CEG) grant #1007636 (Dr. G.-C. Fan).
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Topol EJ. Current status and future prospects for acute myocardial infarction therapy. Circulation. 2003;108:III6–13. doi: 10.1161/01.CIR.0000086950.37612.7b. [DOI] [PubMed] [Google Scholar]
- 2.Liao JK. Secondary prevention of stroke and transient ischemic attack: is more platelet inhibition the answer? Circulation. 2007;115:1615–21. doi: 10.1161/CIRCULATIONAHA.106.653741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross GJ, Auchampach JA. Reperfusion injury: does it exist? J Mol Cell Cardiol. 2007;42:12–8. doi: 10.1016/j.yjmcc.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Downey JM, Cohen MV. Why do we still not have cardioprotective drugs? Circ J. 2009;73:1171–7. doi: 10.1253/circj.cj-09-0338. [DOI] [PubMed] [Google Scholar]
- 5.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 6.Barringhaus KG, Zamore PD. MicroRNAs: regulating a change of heart. Circulation. 2009;119:2217–2224. doi: 10.1161/CIRCULATIONAHA.107.715839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res. 2008;103:1072–83. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, Xu J, Cheng JQ, Lin JY, Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 9.Roccaro AM, Sacco A, Chen C, Runnels J, Leleu X, Azab F, Azab AK, Jia X, Ngo HT, Melhem MR, Burwick N, Varticovski L, Novina CD, Rollins BJ, Anderson KC, Ghobrial IM. microRNA expression in the biology, prognosis, and therapy of Waldenström macroglobulinemia. Blood. 2009;113:4391–402. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36:181–8. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 11.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–7. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–70. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 13.Yang B, Lu Y, Wang Z. Control of cardiac excitability by microRNAs. Cardiovasc Res. 2008;79:571–580. doi: 10.1093/cvr/cvn181. [DOI] [PubMed] [Google Scholar]
- 14.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 15.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 21.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–21. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 22.Bialik S, Geenen DL, Sasson IE, Cheng R, Horner JW, Evans SM, Lord EM, Koch CJ, Kitsis RN. Myocyte apoptosis during acute myocardial infarction in the mouse localizes to hypoxic regions but occurs independently of p53. J Clin Invest. 1997;100:1363–72. doi: 10.1172/JCI119656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colucci WS. Apoptosis in the heart. N Engl J Med. 1996;335:1224–6. doi: 10.1056/NEJM199610173351610. [DOI] [PubMed] [Google Scholar]
- 24.Eefting F, Rensing B, Wigman J, Pannekoek WJ, Liu WM, Cramer MJ, Lips DJ, Doevendans PA. Role of apoptosis in reperfusion injury. Cardiovasc Res. 2004;61:414–26. doi: 10.1016/j.cardiores.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfrum S, Dendorfer A, Rikitake Y, Stalker TJ, Gong Y, Scalia R, Dominiak P, Liao JK. Inhibition of Rho-kinase leads to rapid activation of phosphatidylinositol 3-kinase/protein kinase Akt and cardiovascular protection. Arterioscler Thromb Vasc Biol. 2004;24:1842–1847. doi: 10.1161/01.ATV.0000142813.33538.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao W, Hu E, Tao L, Boyce R, Mirabile R, Thudium DT, Ma XL, Willette RN, Yue TL. Inhibition of Rho-kinase protects the heart against ischemia/reperfusion injury. Cardiovasc Res. 2004;61:548–558. doi: 10.1016/j.cardiores.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Ruan H, Li J, Ren S, Gao J, Li G, Kim R, Wu H, Wang Y. Inducible and cardiac specific PTEN inactivation protects ischemia/reperfusion injury. J Mol Cell Cardiol. 2009;46:193–200. doi: 10.1016/j.yjmcc.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Vila-Petroff M, Salas MA, Said M, Valverde CA, Sapia L, Portiansky E, Hajjar RJ, Kranias EG, Mundiña-Weilenmann C, Mattiazzi A. CaMKII inhibition protects against necrosis and apoptosis in irreversible ischemia-reperfusion injury. Cardiovasc Res. 2007;73:689–98. doi: 10.1016/j.cardiores.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Negoro S, Oh H, Tone E, Kunisada K, Fujio Y, Walsh K, Kishimoto T, Yamauchi-Takihara K. Glycoprotein 130 regulates cardiac myocyte survival in doxorubicin-induced apoptosis through phosphatidylinositol 3-kinase/Akt phosphorylation and Bcl-xL/caspase-3 interaction. Circulation. 2001;103:555–61. doi: 10.1161/01.cir.103.4.555. [DOI] [PubMed] [Google Scholar]
- 31.Zou Y, Takano H, Mizukami M, Akazawa H, Qin Y, Toko H, Sakamoto M, Minamino T, Nagai T, Komuro I. Leukemia inhibitory factor enhances survival of cardiomyocytes and induces regeneration of myocardium after myocardial infarction. Circulation. 2003;108:748–53. doi: 10.1161/01.CIR.0000081773.76337.44. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga S, Okigaki M, Takeda M, Matsui A, Honsho S, Katsume A, Kishita E, Che J, Kurihara T, Adachi Y, Mansukhani A, Kobara M, Matoba S, Tatsumi T, Matsubara H. Endothelium-targeted overexpression of constitutively active FGF receptor induces cardioprotection in mice myocardial infarction. J Mol Cell Cardiol. 2009;46:663–73. doi: 10.1016/j.yjmcc.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Jiang Y, Zhang H, Greenlee AR, Han Z. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life Sci. 2010;86:192–198. doi: 10.1016/j.lfs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 36.Noma K, Oyama N, Liao JK. Physiological role of ROCKs in the cardiovascular system. Am J Physiol Cell Physiol. 2006;290:C661–8. doi: 10.1152/ajpcell.00459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oudit GY, Sun H, Kerfant BG, Crackower MA, Penninger JM, Backx PH. The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J Mol Cell Cardiol. 2004;37:449–71. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.