Abstract
Although multiple genes have been identified as genetic risk factors for isolated, non-syndromic cleft lip with/without cleft palate (CL/P), a complex and heterogeneous birth defect, interferon regulatory factor 6 gene (IRF6) is one of the best documented genetic risk factors. In this study, we tested for association between markers in IRF6 and CL/P in 326 Chinese case–parent trios, considering gene–environment interaction for two common maternal exposures, and parent-of-origin effects. CL/P case–parent trios from three sites in mainland China and Taiwan were genotyped for 22 single nucleotide polymorphisms (SNPs) in IRF6. The transmission disequilibrium test was used to test for marginal effects of individual SNPs. We used PBAT to screen the SNPs and haplotypes for gene–environment (G × E) interaction and conditional logistic regression models to quantify effect sizes for SNP–environment interaction. After Bonferroni correction, 14 SNPs showed statistically significant association with CL/P. Evidence of G × E interaction was found for both maternal exposures, multivitamin supplementation and environmental tobacco smoke (ETS). Two SNPs showed evidence of interaction with multivitamin supplementation in conditional logistic regression models (rs2076153 nominal P = 0.019, rs17015218 nominal P = 0.012). In addition, rs1044516 yielded evidence for interaction with maternal ETS (nominal P = 0.041). Haplotype analysis using PBAT also suggested interaction between SNPs in IRF6 and both multivitamin supplementation and ETS. However, no evidence for maternal genotypic effects or significant parent-of-origin effects was seen in these data. These results suggest IRF6 gene may influence risk of CL/P through interaction with multivitamin supplementation and ETS in the Chinese population.
Introduction
Non-syndromic cleft lip with/without cleft palate (CL/P) is one of the most common birth defects, and Asians have higher prevalence rates compared to other racial groups (Mossey 2007). CL/P creates significant medical and psychological burdens for cases and their families (Christensen and Mortensen 2002). Recent epidemiological evidence also suggests that CL/P is associated with increased risk of overall mortality (Christensen et al. 2004). CL/P is considered to be a complex disease because both genetic and environmental risk factors contribute to its etiology (Lidral et al. 2008). Mutations of interferon regulatory factor 6 gene (IRF6) can lead to van der Woude syndrome (VWS), the most common Mendelian malformation syndrome which includes oral clefts as a hallmark feature (Kondo et al. 2002). Several genetic markers in IRF6 have also yielded evidence of linkage and linkage disequilibrium (LD) in studies of non-syndromic oral clefts, and have been associated with CL/P in many populations (Zucchero et al. 2004; Blanton et al. 2005; Ghassibé et al. 2005; Scapoli et al. 2005; Srichmthong et al. 2005; Vieira et al. 2007; Park et al. 2007; Jugessur et al. 2008; Jia et al. 2009; Huang et al. 2009; Marazita et al. 2009). However, there is inconsistency across studies and one study conducted in a Chinese sample failed to replicate association between IRF6 and CL/P (Pegelow et al. 2008; Tang et al. 2009; Paranaíba et al. 2009; Carter et al. 2009).
Gene–environment (G × E) interaction is biologically plausible when considering maternal environmental exposures and genes in the etiology of CL/P (Lidral et al. 2008). G × E interaction has been suggested for several genes associated with non-syndromic CL/P (van den Boogaard et al. 2008; Sull et al. 2009), although few studies have investigated this question for IRF6. In this study, we tested for association between single nucleotide polymorphisms (SNPs) in IRF6 and CL/P using 326 Chinese case–parent trios, while considering maternal genotypic effects and testing for G × E interactions with common maternal exposures [environmental tobacco smoke (ETS) and multivitamin supplementation].
Methods
Sample description
The “International Genetic Epidemiology of Oral Clefts” study is a multi-center, international family based study initiated in 2003 to investigate the genetic etiology of oral clefts. As part of this study, case–parent trios were recruited from three sites in mainland China (Weifang, Shandong Province; Wuhan, Hubei Province; Chengdu, Sichuan Province) and Taiwan. The majority of cases were infants seen during a surgical or postsurgical visit. All probands were examined for other congenital anomalies or major developmental delays, and were classified as having an isolated, non-syndromic CL/P. Research protocols were reviewed and approved by institutional review boards at each institution. After informed consent was obtained from parents, ethnicity and other data were obtained through structured interviews. Maternal exposure information, including cigarette smoking, ETS, multivitamin supplementation, and alcohol consumption was collected through direct interview of mothers. The proportion of infants exposed to maternal cigarette smoking and alcohol consumption was very low (around 1%), so only ETS and maternal multivitamin supplementation were analyzed. Environmental exposures were defined as being exposed from 3 months prior to pregnancy through the first trimester, except for ETS where exposure was defined as being exposed at any time from 3 months before or during entire pregnancy. Table 1 presents information on gender, family history and exposure among CL/P probands. As seen here, 93.5% of all cases had a negative family history. None of the parents of these CL/P cases were themselves affected. The proportion of exposure to ETS and multivitamin supplementation were 40.5 and 8.6%, respectively.
Table 1.
Characteristics of 326 Chinese non-syndromic cleft lip with or without cleft palate (CL/P) cases
| Number of probands | Cleft lip only | Cleft lip with cleft palate | Total |
|---|---|---|---|
| Gender | |||
| Male | 60 | 165 | 225 |
| Female | 36 | 65 | 101 |
| Family history | |||
| Sporadic | 87 | 218 | 305 |
| Familial | 9 | 12 | 21 |
| Exposure | |||
| Multivitamin supplementation | 9 | 19 | 28 |
| Environmental tobacco smoke | 45 | 87 | 132 |
SNP selection and genotyping
Single nucleotide polymorphisms in and around IRF6 on chromosome 1q32.3-q41 were identified from the literature, dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), and based on previous results in this study (Park et al. 2007). A set of 22 SNPs was chosen based on the criteria of high “design scores” as provided by Illumina, Inc. (San Diego, CA), high heterozygosity, and HapMap validation (http://www.hapmap.org/index.html.en). From these 22 selected SNPs, 1 SNP (rs12126910) was monomorphic and was dropped. The minor allele frequencies (MAF) of the remaining 21 SNPs are shown in Table 2. The last SNP, rs642961, was previously suggested to cause disruption of the binding site of a transcription factor AP-2α and to directly influence the risk of non-syndromic CL/P (Rahimov et al. 2008). Genomic DNA samples were prepared from peripheral blood lymphocytes by the protein precipitation method (Bellus et al. 1995). Primers for each SNP were synthesized using the Oligator technology by Illumina, Inc. as part of an oligo pool for the BeadLab 1000 system. A 4 μg aliquot of each genomic DNA sample was dispensed into a bar-coded 96-well microtiter plate at a concentration of 100 ng/μl and genotyped using the Illumina Golden-GateTM chemistry at the SNP Center of the Genetic Resources Core Facility, a part of the McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins School of Medicine. The average distance between neighboring markers was 1.795 kb (based on Build 36.1 of dbSNP). No Mendelian inconsistencies were found for these 21 SNPs. Two duplicates and four CEPH control DNA samples were included to evaluate genotyping consistency within and between plates and to insure correct plate orientation.
Table 2.
TDT analysis of 21 SNPs in IRF6 gene among 326 Chinese non-syndromic CL/P case–parent trios
| No. | SNP name | Positiona | MAF | TDT |
|||||
|---|---|---|---|---|---|---|---|---|---|
| T | NT | ORb | χ2 | P value | Corrected P valuec | ||||
| 1 | rs12083685 | 208018225 | 0.262 | 95 | 150 | 0.633 | 12.35 | 0.000442 | 0.009282 |
| 2 | rs12081405 | 208018889 | 0.263 | 95 | 149 | 0.638 | 11.95 | 0.000546 | 0.011466 |
| 3 | rs2076153 | 208020194 | 0.466 | 166 | 146 | 1.137 | 1.28 | 0.257518 | – |
| 4 | rs1044516 | 208026237 | 0.493 | 193 | 133 | 1.451 | 11.04 | 0.000890 | 0.018690 |
| 5 | rs599021 | 208027545 | 0.278 | 140 | 105 | 1.333 | 5.00 | 0.025347 | 0.532287 |
| 6 | rs17317411 | 208027937 | 0.039 | 22 | 27 | 0.815 | 0.51 | 0.475051 | – |
| 7 | rs2073485 | 208029417 | 0.400 | 112 | 191 | 0.586 | 20.60 | 0.000006 | 0.000126 |
| 8 | rs2235373 | 208030426 | 0.400 | 112 | 191 | 0.586 | 20.60 | 0.000006 | 0.000126 |
| 9 | rs674433 | 208031498 | 0.226 | 137 | 85 | 1.612 | 12.18 | 0.000483 | 0.010143 |
| 10 | rs595918 | 208033466 | 0.226 | 137 | 85 | 1.612 | 12.18 | 0.000483 | 0.010143 |
| 11 | rs17015218 | 208034538 | 0.068 | 40 | 43 | 0.930 | 0.11 | 0.741934 | – |
| 12 | rs2013162 | 208035307 | 0.496 | 118 | 206 | 0.573 | 23.90 | 0.000001 | 0.000021 |
| 13 | rs7552506 | 208036525 | 0.159 | 86 | 99 | 0.869 | 0.91 | 0.339184 | – |
| 14 | rs2236907 | 208038251 | 0.496 | 118 | 206 | 0.573 | 23.90 | 0.000001 | 0.000021 |
| 15 | rs2294408 | 208040172 | 0.496 | 118 | 206 | 0.573 | 23.90 | 0.000001 | 0.000021 |
| 16 | rs861019 | 208042009 | 0.278 | 139 | 103 | 1.350 | 5.36 | 0.020659 | 0.433839 |
| 17 | rs2073487 | 208043269 | 0.499 | 121 | 207 | 0.585 | 22.55 | 0.000002 | 0.000042 |
| 18 | rs3753517 | 208044721 | 0.341 | 110 | 186 | 0.591 | 19.51 | 0.000010 | 0.000210 |
| 19 | rs1005287 | 208047380 | 0.499 | 121 | 207 | 0.585 | 22.55 | 0.000002 | 0.000042 |
| 20 | rs17389541 | 208053795 | 0.090 | 49 | 61 | 0.803 | 1.31 | 0.252559 | – |
| 21 | rs642961 | 208055893 | 0.226 | 138 | 85 | 1.624 | 12.60 | 0.000364 | 0.007644 |
Non-italicized values indicates inferred LD blocks
T transmitted, NT not transmitted, OR odds ratio, CL/P cleft lip with or without cleft palate, TDT transmission disequilibrium test, LD linkage disequilibrium, SNP single nucleotide polymorphism
Based on NCBI Human Genome build 36.1
Odds Ratio of transmission of minor allele
P values after Bonferroni correction
Statistical analysis
Minor allele frequencies were computed among parents. Pairwise linkage disequilibrium (LD) was measured as r2 for all SNPs using the Haploview program (Barret et al. 2005), and pairwise LD was used to identify LD blocks. One independent SNP and 2 blocks of LD were identified, consisting of 3 and 17 SNPs, respectively. Clayton’s extension of the transmission disequilibrium test (TDT) incorporated into STATA 9 (Spielman et al. 1993; Cordell et al. 2004) was used on individual SNPs to test for evidence of linkage or LD. The family based association test program (FBAT, http://biosun1.harvard.edu/~fbat/fbat.htm) was used for haplotype analysis (Rabinowitz and Laird 2000). FBAT uses a score test statistic to compare expected genotype among offspring under the assumption of no association between observed genotype counts and the phenotype (Laird et al. 2000; Rabinowitz and Laird 2000) and can be extended to consider haplotypes of several SNPs (Horvath et al. 2004). We used a sliding window approach for haplotype selection which analyzes systematically adjacent SNP combinations of different sizes and locations without any assumption of LD structure.
The transmission asymmetry test (TAT) suggested by Weinberg et al. (1998) was used to examine potential parent-of-origin effects. The TAT is similar to the TDT but excludes matings between two heterozygotes (where transmission can be ambiguous). The TAT was stratified into separate allelic tests for fathers and mothers. The TRIMM package (Shi et al. 2007) was used to test for maternal genotypic effects separately from effects of the child’s genotype. The max_Z2 test implemented in TRIMM compares the genotypes observed in the case to those in a hypothetical “complement” child who would have inherited the parental alleles not transmitted to the case for each SNP to identify sets of SNPs showing evidence of transmission distortion. The test statistic is obtained by leveraging the difference in the number of risk alleles inherited by cases and these “complement” controls across all trios divided by its standard error. This approach preserves LD patterns among SNPs, and generates empirical P values by randomly permuting “case” versus “complement” pairs for each SNP. Under the assumption of complete exchangeability between parental haplotypes in the general population, comparing maternally versus paternally transmitted combinations of SNPs provides a test for maternal genotype (or haplotype) effects on risk to the child.
We also screened for possible G × E interaction between all SNPs in IRF6 and two common maternal exposures: ETS and multivitamin supplementation. In this analysis, we used the strategy proposed by Vansteelandt et al. (2008) where family based association tests are evaluated for individual SNPs while allowing for a potential G × E interaction in a 2 degree of freedom (df) test (Kraft et al. 2007), followed by a separate 1 df test for G × E interaction alone. This approach is implemented in the PBAT package (v3.6; http://www.biostat.harvard.edu/~clange/default.htm). The 2 df test examines the genetic effect of the SNP after taking into account the effects of G × E interaction, while the 1 df test investigates the effect of G × E interaction alone. PBAT can also perform haplotype analysis involving G × E interaction. In this analysis, we first used PBAT to screen for possible single SNP G × E interactions. We then used conditional logistic regression to estimate the odds ratios (OR) for being a case with or without environmental exposures to confirm the findings of PBAT.
Results
No SNPs showed evidence of deviating from HWE (data not shown). Figure 1 shows results of the FBAT analyses. Here −log10 (P values) are plotted over the physical distances for both single SNP (single dots) and haplotype (dots connected by lines). For clarity, only haplotypes with P values < 0.001 are plotted here. The lower panel of Fig. 1 shows LD across this region (as measured by r2). Among 21 SNPs in IRF6, 16 SNPs showed a statistically significant association with CL/P at a nominal value of P = 0.05. After Bonferroni correction, 14 SNPs remained statistically significant (Table 2). Three SNPs (rs2013162, rs2236907 and rs2294408) had P values = 10−6 (Fig. 1). Sliding windows of haplotypes consisting of two, three, four, and five SNPs were tested. A five-SNP haplotype consisting of rs674433, rs595918, rs17015218, rs2013162 and rs7552506 showed a highly significant P value (P = 1.86 × 10−5). However, this may be driven by the single SNPs. To minimize the influence of misclassification of syndromic forms of CL/P (i.e., undiagnosed or mild forms of VWS) and non-syndromic CL/P (Birnbaum et al. 2008), we also repeated this analysis excluding cases with any positive family history (21 trios) of oral cleft, but saw no substantial difference in results (data not shown).
Fig. 1.
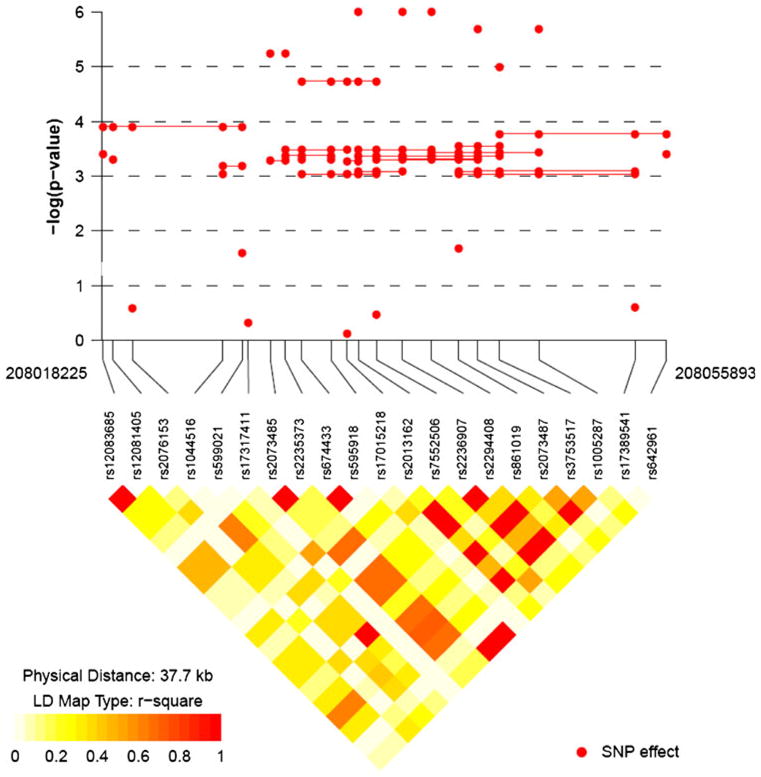
Significance of individual single nucleotide polymorphisms (SNPs) and sliding window haplotypes for the interferon regulatory factor 6 (IRF6) gene in 326 nonsyndromic cleft lip with or without cleft palate trios. The −log10 (empirical P value) for the overall test for an individual SNP (dots) and for sliding windows of haplotypes of two to five SNPs (dots connected by lines) is presented. Only haplotypes with P values < 0.001 are shown here. The plot was produced using snp.plotter (Luna and Nicodemus 2007)
Maternal genotypic effects for all 21 SNPs were investigated. Parent-of-origin effects were first investigated by stratifying informative transmissions (T) and non-transmissions (NT) by parental source in the TAT (Table 3). SNP rs1044516 showed excess maternal transmission of the minor allele (OR = 1.71, nominal P = 0.016), while two additional SNPs, rs2073485 and rs2235373, yielded excess maternal transmission of the major allele (OR = 0.51, nominal P = 0.004) based on the TAT. The TRIMM max_Z2 score was also used to test for maternal genotypic effects, however, these failed to yield significant results (nominal P > 0.05 for all markers).
Table 3.
Number of transmitted (T) or non-transmitted (NT) minor alleles in 326 CL/P trios
| No. | SNP name | Paternal |
Maternal |
Maternal genotypic effect |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TAT |
TAT |
||||||||||
| T | NT | P value | ORa | T | NT | P value | ORa | Z | P value | ||
| 1 | rs12083685 | 29 | 45 | 0.629 | 0.64 | 31 | 48 | 0.056 | 0.65 | 0.446 | 0.70 |
| 2 | rs12081405 | 29 | 44 | 0.079 | 0.66 | 31 | 48 | 0.056 | 0.65 | −0.507 | 0.65 |
| 3 | rs2076153 | 41 | 39 | 0.823 | 1.05 | 39 | 33 | 0.480 | 1.18 | 0.056 | 1.00 |
| 4 | rs1044516 | 44 | 32 | 0.169 | 1.38 | 53 | 31 | 0.016 | 1.71 | −0.162 | 0.91 |
| 5 | rs599021 | 46 | 36 | 0.269 | 1.28 | 47 | 36 | 0.227 | 1.31 | 0.475 | 0.69 |
| 6 | rs17317411 | 8 | 16 | 0.102 | 0.50 | 13 | 10 | 0.532 | 1.30 | −0.144 | 1.00 |
| 7 | rs2073485 | 35 | 50 | 0.104 | 0.70 | 27 | 53 | 0.004 | 0.51 | −0.054 | 1.00 |
| 8 | rs2235373 | 35 | 50 | 0.104 | 0.70 | 27 | 53 | 0.004 | 0.51 | −0.054 | 1.00 |
| 9 | rs674433 | 44 | 24 | 0.153 | 1.83 | 32 | 30 | 0.800 | 1.07 | −0.658 | 0.56 |
| 10 | rs595918 | 44 | 24 | 0.153 | 1.83 | 32 | 30 | 0.800 | 1.07 | 0.658 | 0.56 |
| 11 | rs17015218 | 15 | 21 | 0.317 | 0.71 | 21 | 16 | 0.411 | 1.31 | 0.553 | 0.66 |
| 12 | rs2013162 | 35 | 46 | 0.222 | 0.76 | 33 | 40 | 0.413 | 0.83 | −0.107 | 0.96 |
| 13 | rs7552506 | 30 | 37 | 0.392 | 0.81 | 33 | 27 | 0.439 | 1.22 | 0.400 | 0.75 |
| 14 | rs2236907 | 35 | 46 | 0.222 | 0.76 | 33 | 40 | 0.413 | 0.83 | −0.107 | 0.96 |
| 15 | rs2294408 | 35 | 46 | 0.222 | 0.76 | 33 | 40 | 0.413 | 0.83 | −0.107 | 0.96 |
| 16 | rs861019 | 45 | 36 | 0.317 | 1.25 | 47 | 36 | 0.227 | 1.31 | 0.536 | 0.64 |
| 17 | rs2073487 | 35 | 47 | 0.185 | 0.74 | 32 | 40 | 0.346 | 0.80 | 0.000 | 1.00 |
| 18 | rs3753517 | 26 | 48 | 0.011 | 0.54 | 30 | 54 | 0.009 | 0.56 | 0.230 | 0.87 |
| 19 | rs1005287 | 48 | 34 | 0.122 | 1.41 | 32 | 40 | 0.346 | 0.80 | 0.000 | 1.00 |
| 20 | rs17389541 | 20 | 27 | 0.307 | 0.74 | 23 | 24 | 0.884 | 0.96 | 0.193 | 0.93 |
| 21 | rs642961 | 44 | 23 | 0.010 | 1.91 | 32 | 30 | 0.799 | 1.07 | 0.593 | 0.60 |
OR (transmission) odds ratio of transmission of minor allele
Figure 2 shows results of screening for G × E interaction between these 21 SNPs in IRF6 and two exposures using PBAT. Here −log10(P values) are plotted over physical distance for both the 2 df test of G and G × E effects considered jointly (triangles) and the 1 df test for G × E alone (squares). SNPs showing evidence of linkage in the presence of LD without consideration of G × E are noted in bold along the X-axis. Most SNPs showed statistical significance in the 2 df test for G and G × E interaction, likely reflecting their marginal effects on risk.
Fig. 2.
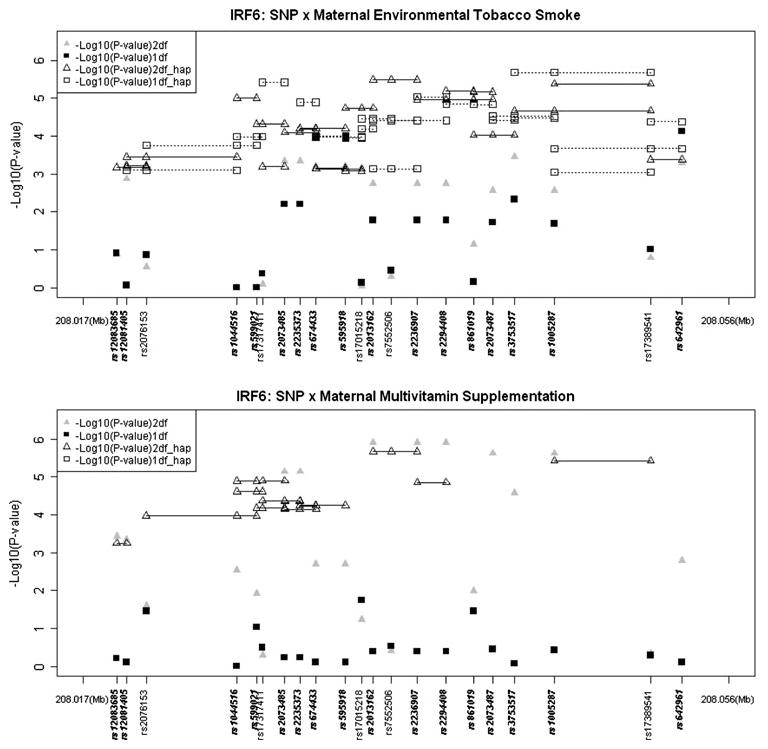
Testing for gene–environment interaction (G × E) for common maternal exposures in 326 CL/P case–parent trios from Chinese populations. Triangles represent the gene–environmental interaction 2 df test of G and G × E interaction, squares represent the 1 df test of G × E only. Haplotypes of 2- and 3-loci are connected by dashed lines (only nominally significant haplotypes P < 10−3). SNPs in bold showed significant P values (nominal P < 0.05) for genetic effects ignoring environmental exposures
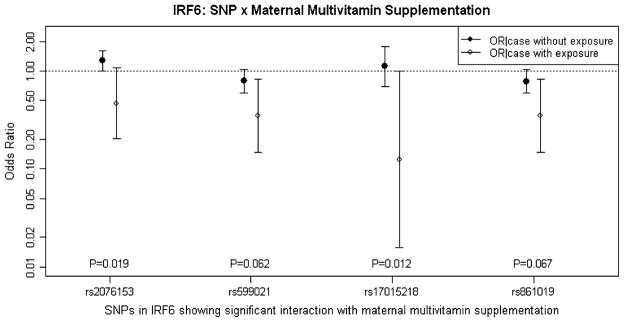
SNPs rs2076153, rs17015218 and rs861019 showed statistical significance in the explicit test of G × E interaction (with 1 df) for multivitamin supplementation when considered individually (rs2076153 nominal P = 0.035; rs17015218 nominal P = 0.018; rs861019: nominal P = 0.034) or within two- and three-SNP haplotypes. SNP rs599021 also yielded a marginally significant P value in the 1 df test for interaction with multivitamin supplementation (nominal P = 0.093). After screening for G × E interaction using PBAT, we used conditional logistic regression under an additive model to estimate odds ratios (OR) for these four SNPs. Figure 3 shows ORs and 95% confidence intervals (CI) for effects of gene–multivitamin supplementation interaction for these SNPs. Solid circles represent estimated ORs of being a case with a heterozygous genotype for subjects whose mother did not take multivitamin supplementation (for rs2076153 and rs17015218, the reference allele is the minor allele, but for rs599021 and rs861019, the reference allele is the major allele). Open circles represent the ORs of being a similar heterozygous case whose mother did take multivitamin supplementation. P values from the likelihood ratio test (LRT) are listed along the x-axis. Being heterozygous at rs2076153 among infants not exposed to maternal multivitamin supplementation was associated with a slightly increased risk (OR = 1.28, 95% CI = 1.01–1.63), but not among those whose mother took multivitamin supplementation (OR = 0.47, 95% CI = 0.20–1.09). The test for G × E interaction was also statistically significant (P = 0.019). Being exposed to maternal multivitamin supplementation in combination with the AG genotype at rs17015218 showed a modest, but significant, reduced risk of CL/P (OR = 0.13, 95% CI = 0.02–0.99). For haplotype G × E analysis using PBAT, 7 two-SNP haplotypes and 18 three-SNP haplotypes yielded significant P values in 1 df test of G × E interaction with multivitamin supplementation.
Fig. 3.
Odds ratios (OR) and 95% confidence intervals (CI) for estimated effects of gene–multivitamin supplementation interaction at four SNPs in IRF6. Odds ratios and CIs are drawn on a logarithmic scale. The ORs and 95% CI were obtained from conditional logistic regression using additive model. Filled circles represent the ORs of being a case with one copy of the risk allele and without exposure to maternal vitamin supplementation (for rs2076153 and rs7552506 the reference allele is the minor allele and for rs599021 and rs861019, the reference allele is the major allele). Open circles represent the odds ratios of being a case with one copy of the risk allele and being exposed to maternal vitamin supplementation. P values from the likelihood ratio test (LRT) with 1 degree of freedom testing the significance of gene–multivitamin supplementation interaction are showed for each SNP
SNP rs1044516 showed evidence of G × E interaction with ETS using the LRT. Being a heterozygote at rs1044516 (i.e., having the AC genotype) and being exposed to maternal ETS was associated with increased risk of CL/P (OR = 1.96, 95% CI = 1.38–2.78, LRT P = 0.041) compared to carrying this AA genotype and not being exposed to maternal ETS (OR = 1.22, 95% CI = 0.91–1.63). All two-SNP haplotypes involving this SNP showed evidence of both G effects and G × E interaction in the 2 df test using PBAT, and among these 17 two-SNP haplotypes yielded significant P values for G × E alone in the 1 df test (nominal P < 0.05). The two-SNP haplotype including rs17317411 and rs2073485 yielded the most evidence for G × E alone in the 1 df test (nominal P = 3.82 × 10−6). Twelve three-SNP haplotypes gave significant P values in the 1 df test for interaction with ETS (nominal P < 0.05). The most significant three-SNP haplotype included rs3753517, rs1005287 and rs17389541 (nominal P = 2.08 × 10−6).
Although marginal effects of rs2076153 (ignoring maternal exposures) were not statistically significant, this SNP showed evidence of a possible interaction with maternal multivitamin supplementation in both the 2 df test for G and G × E combined (nominal P = 0.026), and in the 1 df test for G × E alone (nominal P = 0.035).
Discussion
Our results showed evidence of linkage and LD for sixteen SNPs in the IRF6 gene and confirmed rs642961 was significantly associated with increased risk of non-syndromic CL/P (nominal P = 0.0004). Single SNP analysis using conditional logistic regression and haplotype analysis using PBAT suggested the IRF6 gene may also influence risk of CL/P through interaction with multivitamin supplementation and ETS.
The IRF6 gene is one of nine members of a family of transcription factors (IRFs) that share a highly conserved DNA-binding domain and a less conserved protein-binding domain (Taniguchi et al. 2001). One recent study found mice deficient in Irf6 have abnormal skin, limb and craniofacial development (Ingraham et al. 2006), and another study suggested IRF6 acts on the cell cycle to regulate mammary epithelial cell differentiation (Bailey et al. 2008). Recently, Rahimov et al. (2008) found one SNP (rs642961) in the enhancer region of IRF6 that disrupts a binding site for the transcription factor AP-2α, which may directly influence the risk of non-syndromic CL/P by altering IRF6 transcription. Our study in Chinese case–parent trios confirmed rs642961 was significantly associated with increased risk of non-syndromic CL/P (P = 0.0004).
Mutations in IRF6 lead to VWS, the most common Mendelian syndrome that includes CL/P. Most patients with VWS have an oral cleft and approximately 86% of VWS cases also have pits on the lower lip. Since the pits on the lower lip are often the only way to differentiate between non-syndromic oral cleft and VWS, there could be some misclassification between VWS and non-syndromic CL/P. An estimated one in 36 non-syndromic CL/P case will actually represent VWS if one parent is also affected (Jehee et al. 2009). Whether the association between IRF6 and non-syndromic CL/P is due to misclassification of syndromic and non-syndromic CL/P cases remains a subject of debate (Birnbaum et al. 2008). Excluding trios with any positive family could minimize the influence of this misclassification. Our analysis showed no substantial difference when we compared results of association analysis using all of 326 trios or only those trios without any positive family history of cleft. This result further validated the association between the markers in IRF6 and non-syndromic CL/P.
Gene–environmental (G × E) interactions have been suggested for several genes associated with non-syndromic CL/P (Vieira 2006; Sull et al. 2009). However, few studies to date have focused on whether G × E interaction influences the association between IRF6 and risk of CL/P. Our study suggests an interaction between maternal periconceptional multivitamin use and markers in IRF6. One SNP also showed possible G × E interactions with ETS. In this analysis, SNPs with evidence of gene effects ignoring exposures also showed evidence in the 2 df test of G and G × E when common maternal exposures were considered, especially in haplotype analysis. This 2 df test identifies SNPs influencing risk of disease when the effect of information on exposure is incorporated into a joint test. One SNP in IRF6 showing no significant association when exposure was ignored showed nominal significance in both the 2 df and 1 df tests when considering exposure to maternal multivitamin supplementation. This result illustrates the importance of considering possible G × E interaction in the etiology of CL/P.
Maternal vitamin use has been reported to be inversely associated with the risk of CL/P (Johnson and Little 2008) and potential interaction between candidate genes and vitamin supplementation has been suggested (Shaw et al. 1998). We found intriguing evidence of G × E interaction for maternal multivitamin supplementation at two SNPs in IRF6. These results suggest being exposed to maternal multivitamin supplementation and carrying certain genotypes at these two SNPs in IRF6 may lower the risk of the baby developing CL/P. Shi et al. (2008) summarized the findings of 19 studies investigating interaction between smoking and genes and noted these findings were not consistent across studies. In this analysis, we found evidence of G × E interaction for maternal ETS. A single SNP in IRF6 showed a significant interaction with ETS in the conditional logistic model and several two- and three-SNP haplotypes showed evidence of G × E interaction in PBAT analysis.
Maternal genotypic effects for non-syndromic CL/P have also been reported for several other candidate genes (CBS, RUNX2 and TGFA) (Rubini et al. 2005; Sull et al. 2008, 2009). In screening for parent-of-origin effects, we found suggestive evidence of excess maternal transmission for three of these SNPs (rs1044516, rs2073485 and rs2235373) using the TAT, however, none of these could be confirmed in formal tests for maternal genotypic effects.
The case–parent trio design is robust to population stratification, and provides a unique opportunity to investigate parent-of-origin effects (Cordell et al. 2004; Starr et al. 2005). The present study found some evidence of potential maternal over-transmission for markers in IRF6 and risk of non-syndromic CL/P. However, the evidence for association in the context of G × E interaction between markers in IRF6 and both maternal multivitamin use and ETS tended to be stronger. Maternal multivitamin supplementation, in particular, appears to reduce risk of non-syndromic CL/P in cases carrying certain genotypes. The exposure rate of maternal vitamin supplementation was different across different sites. The mothers in mainland China had much lower use of multivitamin supplementation compared to those in Taiwan (6.8 vs. 14.1%). If this observation can be confirmed, such a G × E interaction creates opportunities for an effective intervention to reduce the risk of non-syndromic CL/P, an important public health burden.
Acknowledgments
This research was supported by R21-DE-013707 and R01-DE-014581 from the National Institute of Dental and Craniofacial Research. TW is supported by the International Collaborative Genetics Research Training Program (ICGRTP), NIH D43 TW06176. We thank all participants who donated samples for this multi-center study of oral clefts, as well as the staff at each participating site and institution. We also thank the Smile Train Foundation for supporting cleft researches in China.
Contributor Information
Tao Wu, Email: twu@jhsph.edu, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA. Beijing University School of Public Health, Beijing, China.
Kung Yee Liang, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Jacqueline B. Hetmanski, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA
Ingo Ruczinski, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Margaret Daniele Fallin, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Roxann G. Ingersoll, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA. Johns Hopkins School of Medicine, Baltimore, MD, USA
Hong Wang, Beijing University School of Public Health, Beijing, China.
Shangzhi Huang, Peking Union Medical College, Beijing, China.
Xiaoqian Ye, Mount Sinai School of Medicine, New York, NY, USA. Key Laboratory for Oral Biomedical Engineering of Ministry of Education, Hospital and School of Stomatology, Wuhan University, Wuhan, China.
Yah-Huei Wu-Chou, Chang Gung Memorial Hospital, Taoyuan, Taiwan.
Philip K. Chen, Chang Gung Memorial Hospital, Taoyuan, Taiwan
Ethylin W. Jabs, Johns Hopkins School of Medicine, Baltimore, MD, USA. Mount Sinai School of Medicine, New York, NY, USA
Bing Shi, West China College of Stomatology, Sichuan University, Sichuan, China.
Richard Redett, Johns Hopkins School of Medicine, Baltimore, MD, USA.
Alan F. Scott, Johns Hopkins School of Medicine, Baltimore, MD, USA
Terri H. Beaty, Email: tbeaty@jhsph.edu, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA. Department of Epidemiology, School of Public Health, Johns Hopkins University, 615 N. Wolfe St., Baltimore, MD 21205, USA
References
- Bailey CM, Abbott DE, Margaryan NV, Khalkhali-Ellis Z, Hendrix MJ. Interferon regulatory factor 6 promotes cell cycle arrest and is regulated by the proteasome in a cell cycle-dependent manner. Mol Cell Biol. 2008;28:2235–2243. doi: 10.1128/MCB.01866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bellus GA, Hefferon TW, Ortiz de Luna RI, Hecht JT, Horton WA, Machado M, Kaitila I, McIntosh I, Francomano CA. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Reutter H, Lauster C, Scheer M, Schmidt G, Saffar M, Martini M, Hemprich A, Henschke H, Kramer FJ, Mangold E. Mutation screening in the IRF6-gene in patients with apparently nonsyndromic orofacial clefts and a positive family history suggestive of autosomal-dominant inheritance. Am J Med Genet A. 2008;146A:787–790. doi: 10.1002/ajmg.a.32219. [DOI] [PubMed] [Google Scholar]
- Blanton SH, Cortez A, Stal S, Mulliken JB, Finnell RH, Hecht JT. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137A:259–262. doi: 10.1002/ajmg.a.30887. [DOI] [PubMed] [Google Scholar]
- Carter TC, Molloy AM, Pangilinan F, et al. Testing reported associations of genetic risk factors for oral clefts in a large Irish study population. Birth Defects Res A Clin Mol Teratol. 2009 doi: 10.1002/bdra.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Mortensen PB. Facial clefting and psychiatric diseases: a follow-up of the Danish 1936–1987 Facial Cleft cohort. Cleft Palate Craniofac J. 2002;39:392–396. doi: 10.1597/1545-1569_2002_039_0392_fcapda_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Christensen K, Juel K, Herskind AM, Murray JC. Long term follow up study of survival associated with cleft lip and palate at birth. BMJ. 2004 doi: 10.1136/bmj.38106.559120.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Ghassibé M, Bayet B, Revencu N, Verellen-Dumoulin C, Gillerot Y, Vanwijck R, Vikkula M. Interferon regulatory factor-6: a gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur J Hum Genet. 2005;13:1239–1242. doi: 10.1038/sj.ejhg.5201486. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wu J, Ma J, Beaty TH, Sull JW, Zhu L, Lu D, Wang Y, Meng T, Shi B. Association between IRF6 SNPs and oral clefts in West China. J Dent Res. 2009;88:715–718. doi: 10.1177/0022034509341040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (IRF6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehee FS, Burin BA, Rocha KM, Zechi-Ceide R, Bueno DF, Brito L, Souza J, Leal GF, Richieri-Costa A, Alonso N, Otto PA, Passos-Bueno MR. Novel mutations in IRF6 in nonsyndromic cleft lip with or without cleft palate: when should IRF6 mutational screening be done? Am J Med Genet A. 2009;149A:1319–1322. doi: 10.1002/ajmg.a.32849. [DOI] [PubMed] [Google Scholar]
- Jia ZL, Li Y, Li L, Wu J, Zhu LY, Yang C, Chen CH, Shi B. Association among IRF6 polymorphism, environmental factors, and nonsyndromic orofacial clefts in western china. DNA Cell Biol. 2009;28:249–257. doi: 10.1089/dna.2008.0837. [DOI] [PubMed] [Google Scholar]
- Johnson CY, Little J. Folate intake, markers of folate status and oral clefts: is the evidence converging? Int J Epidemiol. 2008;37:1041–1058. doi: 10.1093/ije/dyn098. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Rahimov F, Lie RT, Wilcox AJ, Gjessing HK, Nilsen RM, Nguyen TT, Murray JC. Genetic variants in IRF6 and the risk of facial clefts: single-marker and haplotype-based analyses in a population-based case-control study of facial clefts in Norway. Genet Epidemiol. 2008;32:413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, et al. Mutations in IRF6 cause van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lidral AC, Moreno LM, Bullard SA. Genetic Factors and Orofacial Clefting. Semin Orthod. 2008;14:103–114. doi: 10.1053/j.sodo.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;15:774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, et al. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey P. Epidemiology underpinning research in the aetiology of orofacial clefts. Orthod Craniofacial Res. 2007;10:114–120. doi: 10.1111/j.1601-6343.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- Paranaíba LM, Bufalino A, Martelli-Júnior H, de Barros LM, Graner E, Coletta RD. Lack of association between IRF6 polymorphisms (rs2235371 and rs642961) and non-syndromic cleft lip and/or palate in a Brazilian population. Oral Dis. 2009 doi: 10.1111/j.1601-0825.2009.01627.x. [DOI] [PubMed] [Google Scholar]
- Park JW, McIntosh I, Hetmanski JB, et al. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegelow M, Peyrard-Janvid M, Zucchelli M, Fransson I, Larson O, Kere J, Larsson C, Karsten A. Familial non-syndromic cleft lip and palate–analysis of the IRF6 gene and clinical phenotypes. Eur J Othod. 2008;30:169–175. doi: 10.1093/ejo/cjm097. [DOI] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, et al. Disruption of an AP-2a binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubini M, Brusati R, Garattini G, et al. Cystathionine beta-synthase c.844ins68 gene variant and non-syndromic cleft lip and palate. Am J Med Genet A. 2005;136A:368–372. doi: 10.1002/ajmg.a.30812. [DOI] [PubMed] [Google Scholar]
- Scapoli L, Palmieri A, Martinelli M, Pezzetti F, Carinci P, Tognon M, Carinci F. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Wasserman CR, Murray JC, Lammer EJ. Infant TGF-alpha genotype, orofacial clefts, and maternal periconceptional multivitamin use. Cleft Palate Craniofac J. 1998;35:366–370. doi: 10.1597/1545-1569_1998_035_0366_itagoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Shi M, Umbach DM, Weinberg CR. Identification of risk-related haplotypes with the use of multiple SNPs from nuclear families. Am J Hum Genet. 2007;81:53–66. doi: 10.1086/518670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Wehby GL, Murray JC. Review on genetic variants and maternal smoking in the etiology of oral clefts and other birth defects. Birth Defects Res C Embryo Today. 2008;84:16–29. doi: 10.1002/bdrc.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Srichomthong C, Siriwan P, Shotelersuk V. Significant association between IRF6 820G->A and non-syndromic cleft lip with or without cleft palate in the Thai population. J Med Genet. 2005 doi: 10.1136/jmg.2005.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr JR, Hsu L, Schwartz SM. Assessing maternal genetic associations: a comparison of the log-linear approach to case-parent triad data and a case-control approach. Epidemiology. 2005;16:294–303. doi: 10.1097/01.ede.0000158223.98649.eb. [DOI] [PubMed] [Google Scholar]
- Sull JW, Liang KY, Hetmanski JB, et al. Excess maternal transmission of markers in TCOF1 among cleft palate case-parent trios from three populations. Am J Med Genet A. 2008;146A:2327–2331. doi: 10.1002/ajmg.a.32302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sull JW, Liang KY, Hetmanski JB, et al. Evidence that TGFA influences risk to cleft lip with/without cleft palate through unconventional genetic mechanisms. Hum Genet. 2009;126:385–394. doi: 10.1007/s00439-009-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Du X, Feng F, Long J, Lin Y, Li P, Liu L, Tian W. Association analysis between the IRF6 G820A polymorphism and nonsyndromic cleft lip and/or cleft palate in a Chinese population. Cleft Palate Craniofac J. 2009;46:89–92. doi: 10.1597/07-131.1. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, de Costa D, Krapels IP, Liu F, van Duijn C, Sinke RJ, Lindhout D, Steegers-Theunissen RP. The MSX1 allele 4 homozygous child exposed to smoking at periconception is most sensitive in developing nonsyndromic orofacial clefts. Hum Genet. 2008;124:525–534. doi: 10.1007/s00439-008-0569-6. [DOI] [PubMed] [Google Scholar]
- Vansteelandt S, Demeo DL, Lasky-Su J, Smoller JW, Murphy AJ, McQueen M, Schneiter K, Celedon JC, Weiss ST, Silverman EK, Lange C. Testing and estimating gene-environment interactions in family-based association studies. Biometrics. 2008;64:458–467. doi: 10.1111/j.1541-0420.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- Vieira AR. Association between the transforming growth factor alpha gene and nonsyndromic oral clefts: a HuGE review. Am J Epidemiol. 2006;163:790–810. doi: 10.1093/aje/kwj103. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Cooper ME, Marazita ML, Orioli IM, Castilla EE. Interferon regulatory factor 6 (IRF6) is associated with oral-facial cleft in individuals that originate in South America. Am J Med Genet A. 2007;143A:2075–2078. doi: 10.1002/ajmg.a.31884. [DOI] [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]