Abstract
Purpose
Ureteral stents are susceptible to biofilm formation and crystal deposition, especially in stone formers. To identify proteins responsible for this accumulation, we compared conditioning film proteomes obtained from human ureteral stents with and without encrustation.
Materials and Methods
Twenty-seven Bard Inlay™ hydrophilic ureteral stents were removed after ureteroscopy. Stent encrustation was quantified by visual analog score 0 (none) to 4 (heavy) and further categorized as nonencrusted (scores 0 and 1; n = 22) or encrusted (scores 2, 3, and 4; n = 5). Stent conditioning film was sampled and digested with trypsin, and peptide tandem mass spectrometry data were acquired using liquid chromatography. After protein identification, unconditional exact tests were used to compare categorical variables versus encrustation outcome. Stone analysis and follow-up metabolic urine profiles were examined to identify additional risk factors for stent encrustation.
Results
More than 300 unique proteins with >95% confidence were identified. Proteins α-1 anti-trypsin, Ig kappa, IgH G1, and histone H2b and H3a were found to be highly associated with stent encrustation (p < 0.05), while Tamm-Horsfall protein and histone H2a were found to have a marginal association (p < 0.1). Patients with early stent encrustation were more likely to have mixed stone analysis (p = 0.03) and low urinary volumes (p < 0.01).
Conclusion
Immunoglobulins and Tamm-Horsfall protein are common urinary proteins that appear to nonselectively bind early onto ureteral stent surfaces. Histones, nuclear DNA-condensing proteins, likely contribute to stent encrustation because of their unique net positive charge and may represent a potential clinical target for encrustation prevention.
Introduction
The use of ureteral stents within the urinary tract can be problematic because of biofilm formation and build-up of mineral salts along the stent surface, a process termed “encrustation.” Prolonged encrustation may lead stent fracture, obstruction of the stent lumen and renal unit, urosepsis, and even loss of renal function.1,2 Kidney stone patients, the most likely population requiring ureteral stents, have been shown to be at the highest risk for encrustation, sometimes occurring within days of stent placement.1–4 In spite of advances in biomaterials and surface coatings, no stent completely resists biofilm formation and encrustation because of a complex protein adsorption cascade that occurs along the stent surface within minutes of implantation.5–7
The first monolayer of biological molecules that absorb onto any medical device within the human body is called the “conditioning film.”8 Depending on the surface characteristics of the device, a range of material, including proteins, lipids, and polysaccharides, may be deposited. Protein deposition is of particular interest as it involves a dynamic process of smaller, partially charged proteins rapidly adsorbing onto the biomaterial surface followed by other larger, more strongly charged proteins—a phenomenon known as the Vroman effect.9,10 If one could identify stent conditioning film proteins that allow for further protein, urinary salt, or perhaps even bacterial binding, then new targets could potentially be explored for biomaterial design.
Multiple proteins from complex solutions, such as those in conditioning biofilms, may be identified using a “comprehensive, bottom-up,” mass spectrometry (MS)–based approach.11 This approach entails proteolytic digestion of all proteins within a sample into smaller peptide fragments of predicted arrangement based on enzyme specificity. These peptides are then discretely dissociated into peptide fragments using MS/MS (tandem MS) technology. Experimental fragment ion masses are then matched to theoretical fragment ion masses using sophisticated database software, and candidates for protein identification are reported. To explore early protein layers, we qualitatively compare the proteomes of conditioning film from human ureteral stents with and without encrustation using an MS-based approach. We hypothesize that a pattern of protein expression will be identified that may be further explored for encrustation prevention.
Materials and Methods
Stent and conditioning film collection
After upper urinary tract manipulation of renal or ureteral stones and placement of a Bard Inlay ureteral stent, 27 consecutive patients were enrolled in an Institutional Review Board–approved study. On the day of stent removal, voided urine samples were collected for routine urinalysis and culture, and patient demographic data recorded. Ureteral stents were removed under sterile conditions with a cystoscope and grasper. Encrustation scoring, modified from Keane et al,4 was quantified by the following visual analog score: 0 (no visible stent biofilm), 1 (visible stent biofilm), 2 (bladder coil encrustation), 3 (<50% of entire stent encrusted), and 4 (>50% of stent encrusted). Stents were washed in sterile saline for 1 minute to remove surface debris, placed in a sterile container, and transported to a −80°C freezer on dry ice. Once all stents were collected, biofilm was removed by methodically scraping the entire length of the stent using a curved spatula and immediately placed in 400 μL of 0.1% 3-cholamidopropyl-dimethylammonio-1-propanesulfonate, 0.5% sodium dodecyl sulfate, 20 mM tetraethylammonium bromide buffer, and standard Sigma protease cocktail inhibitor. Protein was quantified by modified colorimetric Bradford protein assay and dried down in a vacuum dried in a ThermoSavant speed vacuum. Fourier transform infrared spectroscopy stone analysis was performed on stone samples at ARUP® Laboratories (Salt Lake City, UT). Patients were considered “mixed” stone formers if the dominant mineral content was <70%. Metabolic 24-hour urine profiles, obtained 1 month after stent removal, were analyzed through Litholink® (Chicago, IL).
In-solution protein digestion and clean-up
Protein pellets were reconstituted in 20 μL of 8 M urea and 10 mM of dithiothreitol and allowed to solubilize on ice for 4 hours. Samples were vortexed and centrifuged at 12,500 rpm for 10 minutes. Soluble aliquots were incubated at 37°C for 1 hour. After centrifugation, an equal volume of 50 mM ammonium bicarbonate/30 mM iodoacetamide was added to each sample and incubated in the dark at room temperature. Trypsin was reconstituted to a concentration of 0.1 μg/μL with 25 mM ammonium bicarbonate/5 mM calcium chloride and added in a 1:20 (trypsin:total protein) ratio to each sample. Samples were incubated overnight at 37°C. Digests were then placed at −80°C for 1 hour and vacuum-dried in a speed vacuum. Dried digests were reconstituted in 0.2% formic acid, poured into Oasis®MCX extraction cartridges (Waters, Milford, MA), and gravity-filtered through the cartridge to remove trypsin, salts, and buffers.
Protein identification by liquid chromatography–MS/MS
Tryptic peptides were analyzed by reversed-phase, high-performance liquid chromatography using C18 capillary trap column in-line with a homemade fritless C18, 75-μm ID, 12-cm long, 5-μm particles, 200 Å pores online with a with microESI-QSTAR (ABI) quadrapole time of flight mass spectrometer as described previously.12 Gradient elution profile was modified to the following: 0 to 35 minutes, 5% to 30% acetonitrile (ACN); 35 to 45 minutes, 30% to 80% ACN; 45 to 50 minutes, 80% to 95% ACN; and 50 to 60 minutes, 95% ACN. Peptide elution pattern was assessed upon inspection of total ion chromatoGram traces. Protein extraction efficiency was presumed efficient, and trypsin digest sample was presumed compatible with liquid chromatography–MS if the total ion chromatoGram elution pattern showed a normal increase in MS signal intensity during gradient elution followed by a drop in signal back to baseline. Tandem mass spectra were searched using Protein Pilot™ 2.0 software by previously described parameters.13 Protein identifications with confidence limits ≥95% were considered significant. Proteins with <95% confidence or with only one peptide are not reported in this study. All peptide MS/MS spectra from reported proteins were manually inspected.
Statistical analysis
Unconditional exact tests (Pearson's Chi-square) were conducted to compare the encrustation outcome to demographics, stone types, metabolic urinary profiles, and qualitative stent proteins. Variables with p-values ≤ 0.05 were considered significantly associated with encrustation, while those with p-values between 0.05 and 0.1 were considered marginally significant.
Results
Patient characteristics
Patients in nonencrusted (n = 22) or encrusted (n = 5) groups were similarly matched by age, sex, and indwelling stent time (Table 1). No urine culture was positive at the time of stent removal. Patients with stent encrustation were more likely to be mixed stone formers (p = 0.03), whereas the nonencrustation group had a trend toward calcium oxalate mineral content (p = 0.14, Table 1). Low urinary volumes (p < 0.01) were more common within stent encrustation group, while a marginally significant protective trend was seen for nonencrusted patients who were hyperuricosuric (p = 0.06). We had one patient (age 43) with mixed stone type who was found to have encrustation along the entirety of her stent on the day of stent removal (postoperative day 10). She subsequently had low urinary volume (<1 L), extreme hypocitraturia (<100 mg/day), and hypercalciuria of 345 mg/day on metabolic work-up.
Table 1.
Demographics, Stone Type, and Metabolic Studies of Patients with or without Stent Encrustation
| Characteristic | Nonencrusted (n = 22) | Encrusted (n = 5) | p-Valuea |
|---|---|---|---|
| Male, n (%) | 8 (36) | 2 (40) | NS |
| Age, mean (SD) | 47 (11) | 56 (5) | NS |
| Days indwelling (SD) | 8 (2) | 8 (2) | NS |
| Visual encrustation scale, n | |||
| 0 | 4 | – | – |
| 1 | 18 | – | – |
| 2 | – | 3 | – |
| 3 | – | 1 | – |
| 4 | – | 1 | – |
| Stone type, n | |||
| Calcium oxalate | 14 | 1 | 0.14 |
| Mixed | 2 | 3 | 0.03 |
| Calcium phosphate | 3 | – | NS |
| Urate | 1 | – | NS |
| None | 1 | 1 | NS |
| 24-hour urine,bn (%) | |||
| Hypocitraturia (<450 mg) | 11 (50) | 3 (60) | NS |
| Hyperuricosuria (>800 mg) | 11 (50) | 0 | 0.06 |
| Low volume (<1.5 L) | 6 (27) | 5 (100) | 0.01 |
| Hypercalciuria (>250 mg) | 5 (23) | 1 (20) | NS |
| Hyperoxaluria (>40 mg) | 3 (14) | 2 (40) | 0.22 |
| pH (5.0–8.0) | 6.11 | 5.74 | NS |
| None performed | 6 (27) | 0 | NS |
Actual value reported if p < 0.25, otherwise NS.
Several individuals had multiple abnormalities. Normal values are based on standard urinary metabolic guidelines and represented by mg or L/day.
NS = nonsignificant.
Proteins
Average total protein recovered after mixed cation exchange was 30.4 micrograms (range 2–167), and all samples were suitable for MS analysis by examination of total ion chromatogram pattern. Approximately 312 unique proteins with >95% confidence were identified from stent samples, with an average of 28 proteins identified per stent (range 4–63). Table 2 lists the 40 most commonly identified proteins by descending order based on frequency of identification from each stent sample. Thirty-three proteins were identified from three stent samples, 79 from two samples, and 157 proteins from single stent samples. Theoretical protein isoelectric points (pI) and molecular weights (kDa) were calculated using the on-line pI software tool ExPASy Proteomics server (www.expasy.ch/tools/pi_tool.html).
Table 2.
Top 40 Proteins Recovered from Ureteral Stent Surfaces
| Protein | na | Accessionb | Isoelectric point | kDa | Coveragec |
|---|---|---|---|---|---|
| Hemoglobin β chain | 17 | 56749856 | 6.7 | 16.0 | 92 |
| Hemoglobin α chain | 16 | 57013850 | 8.7 | 15.3 | 97 |
| Albumin | 15 | 23243418 | 6.1 | 66.9 | 52 |
| Calgranulin B | 14 | 56205191 | 5.7 | 13.1 | 87 |
| Fibrinogen β chain | 13 | 7924018 | 8.5 | 55.9 | 23 |
| Vitronectin/Protein S | 13 | 88853069 | 5.6 | 54.3 | 10 |
| Annexin A1 | 12 | 54696654 | 6.6 | 38.7 | 9 |
| Calgranulin A | 11 | 30583595 | 6.5 | 10.8 | 77 |
| Fibrinogen γ chain | 11 | 70906439 | 5.4 | 51.1 | 49 |
| Tamm-Horsfall protein | 10 | 137116 | 5.1 | 69.8 | 21 |
| α1 Anti-trypsin | 9 | 28966 | 5.4 | 51.7 | 13 |
| β-Actin | 9 | 93121215 | 5.3 | 41.7 | 11 |
| Calgranulin C | 9 | 8574384 | 5.8 | 10.6 | 87 |
| Histone H2b | 9 | 9863669 | 10.3 | 13.9 | 42 |
| Azurocidin | 8 | 62739619 | 9.8 | 26.9 | 7 |
| Defensin α-1 | 8 | 85567619 | 6.5 | 10.2 | 19 |
| Fibrinogen α chain | 8 | 458555 | 5.7 | 95 | 8 |
| Cathepsin G | 7 | 179915 | 11.2 | 28.8 | 18 |
| Eosinophilic cationic protein | 7 | 1139050 | 10.3 | 18.4 | 12 |
| Cofilin | 7 | 9739169 | 7.7 | 18.7 | 8 |
| Heat shock 27 kDa protein | 7 | 86278454 | 6.0 | 26.3 | 13 |
| IgH G1 | 7 | 15779222 | 8.6 | 52.5 | 9 |
| Lactoferrin | 7 | 640200 | 8.5 | 78.2 | 38 |
| Transferrin | 7 | 94717618 | 6.8 | 77.1 | 21 |
| Antithrombin Iii | 6 | 999514 | 6.0 | 48.9 | 4 |
| Histone 1 | 6 | 9863664 | 11.0 | 11.4 | 71 |
| Complement C3 | 5 | 40786791 | 6 | 187 | 3 |
| Histone H3a | 5 | 559902 | 11.1 | 15.4 | 86 |
| Ig kappa light chain | 5 | 21669427 | 6.2 | 25.5 | 41 |
| Keratin 1 | 5 | 7331218 | 8.1 | 66.0 | 19 |
| Keratin 10 | 5 | 119581085 | 5.1 | 63.3 | 13 |
| α-1 Antichymotrypsin | 4 | 1340142 | 5.5 | 49.8 | 7 |
| Annexin A2 | 4 | 30962862 | 7.6 | 38.6 | 26 |
| Apolipoprotein A-I | 4 | 90108666 | 6.8 | 26.7 | 17 |
| Carbonic anhydrase I | 4 | 999580 | 6.6 | 28.7 | 9 |
| Glyceraldehyde dehydrogenase | 4 | 53734502 | 8.6 | 36.1 | 24 |
| Haptoglobin | 4 | 83699647 | 6.1 | 45.2 | 35 |
| Histone H2a | 4 | 74752099 | 10.9 | 14.2 | 57 |
| Lectin, mannose-binding 2 | 4 | 62896777 | 6.5 | 40.2 | 13 |
| Prothrombin | 4 | 67624831 | 5.7 | 70.0 | 8 |
Number of stents from which protein was identified (listed in decreasing order).
National Center for Biotechnology Information protein database found at www.ncbi.nlm.nih.gov/entrez-search.
Maximum % of the protein coverage by peptide.
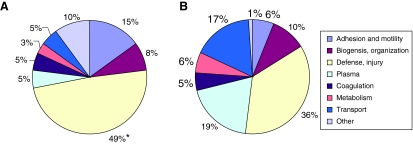
Proteins from encrusted stents (Fig. 1A) and nonencrusted stents (Fig. 1B) were then grouped based on the protein function. Function was determined by the classification system of the Gene Ontology Consortium (www.geneontology.org).14 This public domain database is divided into three major categories: molecular function, biological processes, and cellular component. “Biological processes” was selected to better stratify the role of a particular protein. Each protein was assigned a weighed value based on the number of stents from which it was identified. Category percentages were calculated by totaling the weighed value per group divided by the total weighted number of all proteins.
FIG. 1.
Proteins identified from encrusted (A) and nonencrusted (B) stents grouped by function. Defense/injury and adhesion/motility proteins were more commonly identified from encrusted stents (*p < 0.05 and p < 0.1, respectively), while plasma and transport proteins were identified more commonly from nonencrusted stents (p < 0.1).
Inflammatory and defense proteins were the most commonly identified functional group in both encrusted and nonencrusted stent types (49% vs. 36% respectively, p < 0.05). Encrusted stents tended to have higher frequencies of adhesion proteins (15% vs. 6%, p < 0.1), whereas nonencrusted stents had higher frequencies of plasma (19% vs. 5%, p < 0.1) and transport (17% vs. 5%, p < 0.1) proteins. Any protein identified in four or more stents was further evaluated for association with encrustation variable. Of the 312 identified proteins, alpha-1 anti-trypsin, Ig kappa, IgH G1, and histone H2B and H3A were found to be highly associated with stent encrustation (p < 0.05), whereas Tamm-Horsfall protein (THP) and histone H2A were found to have a marginal association (p < 0.1, Table 3).
Table 3.
Stent Proteins Associated with Encrustation
| Protein | Nonencrustationa | Encrustationa | Accession | Isoelectric point | kDa | Coverage | Peptidesb | p-Value |
|---|---|---|---|---|---|---|---|---|
| IgH G1 | 2 | 5 | 15779222 | 8.6 | 52.5 | 9 | 7 | <0.01 |
| Ig kappa light chain | 0 | 5 | 21669427 | 6.2 | 25.5 | 41 | 4 | <0.01 |
| Histone H3a | 0 | 5 | 559902 | 11.1 | 15.4 | 86 | 3 | <0.01 |
| α1 anti-trypsin | 4 | 5 | 28966 | 5.4 | 46.7 | 13 | 4 | 0.04 |
| Histone H2b | 4 | 5 | 9863669 | 10.3 | 13.9 | 42 | 3 | 0.04 |
| Tamm-Horsfall protein | 6 | 4 | 137116 | 5.1 | 69.8 | 21 | 14 | 0.06 |
| Histone H2a | 2 | 2 | 74752099 | 10.9 | 14.2 | 57 | 3 | 0.06 |
Boldface signifies statistically significant association.
Frequency of protein detection between groups.
Number of peptides identified in given protein.
Discussion
Polyurethane, a material with a variety of medical applications, is the most common polymeric biomaterial used in modern ureteral stents.15 The Bard Inlay ureteral stent, utilized in our study, is a polyurethane-based stent coated with polyvinyl pyrrolidone, a proprietary, hydrophilic polymer that absorbs water, increases lubricity and elasticity, and minimizes patient discomfort.16 Much like a ureteral stent coated with a hydrogel, this polymer is attracted to water along with the small, weakly charged molecules that are soluble within water. Recall that exposed amino acid residues of proteins interact with water and their environment. The net charge of a protein surface depends on pH and the number and identity of its charged amino acids. For most proteins, no net charge (neutral) occurs at a pH called the pI, usually somewhere between 5.5 and 8. A soluble protein most likely to interact with water and/or a hydrophilic stent would be one that is far away from its pI (charged) and small in size (less than 50 kDa).
Table 3 summarizes the seven proteins most commonly identified within early conditioning films. As predicted, several have calculated pI (charged) that differ from normal urinary pH range, and all but THP are approximately 50 kDa or smaller. One of the more commonly identified proteins was histone. Histones, nuclear DNA-condensing proteins, are the chief protein components of chromatin and are normally present in small amounts in human urine as breakdown products from sloughed transitional epithelial cells. Of the six classes of human histones, H2b, H3, and H4 have the greatest net positive charges because of their unique α-helical configuration.17,18 This large net positive charge makes them particularly water soluble and likely contributes to their affinity to early stent accumulation. Additionally, they may be more abundant after ureteroscopy because of urothelial cell disruption and trauma. In our cohorts, both proteins H3a and H2b were highly associated with early conditioning films and encrustation.
Compared with nonencrusted, encrusted stents had higher numbers of inflammatory (p < 0.05, Fig. 1) and adhesion/motility proteins (p < 0.1, Fig. 1) identified. The cause and effect of both of these is difficult to determine. It is possible that conditioning film adhesive proteins promoted crystal binding along the stent surface, which led to a more robust immune response, or encrustation patients could have developed a greater host response to the stents, leading to binding of adhesive proteins, further inflammation, and encrustation. In addition to pattern profiling, two inflammatory proteins (Ig kappa and alpha-1 antitrypsin) were individually associated with greater encrustation. Immunoglobulin kappa and associated light chain is a plasma protein involved in antigen processing and presentation by major histocompatibility complex 1. It is a ligand of the glycoprotein receptor for cubilin and megalin in the proximal renal tubule and for THP in the distal tubule and is seen as a low abundance protein within human urine.19,20 These light chains were exclusively seen from stents in our encrustation cohort and may be a reflection of increased host cell response to ureteroscopic manipulation or stent trauma. Alpha-1 antitrypsin is a 52-kDa plasma glycoprotein important for the protection of tissues from proteolytic enzymes. Its higher presence in the encrusted stent group may be in response to enzymes released from inflammatory cells.
THP is a highly abundant, excretory urinary protein that inhibits calcium crystallization and uropathogenic bacterial attachment to epithelial cells by nonspecific surface binding.21 It is notorious for its ability to self-aggregate and to activate human neutrophils through a single class of sialic acid–specific cell surface receptors.22 This protein is likely present along stents because of its high urine abundance and ability to adhere to almost any charged surface. Once bound, THP could activate human polymorphonuclear leukocytes within the urine and result in an inflammatory cascade of crystal, protein, or even bacterial binding.22
Only one group, Santin et al,23 has previously attempted to identify the proteins responsible for urinary conditioning films. This group studied stents retrieved from two stone formers (encrusted) and two patients with ureteral neoplasia (nonencrusted). In vitro dynamic modeling of cut stent portions demonstrated marked precipitation of material in the encrusted group, while protein analysis by 1D gel electrophoresis and immunoblotting with three antibodies demonstrated only one common protein, human serum albumin, on all four ureteral stent pieces. THP and α-1 microglobulin were positively identified within the nonencrusted controls. In our analysis, no difference in encrustation patterns were seen with the protein albumin (p = 0.58, data not shown), and THP had only a marginal association.
This study has several limitations. First, multiple comparisons and small cohort enrollment limit the power of our study. Protein loss or degradation during stent storage or during the extraction and clean-up process may have occurred. Protein may also have been lost because of urinary proteases, have precipitated during sample preparation, or have been below the detection limit of our mass spectrometer. As protein absorption is a dynamic effect, biofilm composition may change proportional to the amount of indwelling stent time. Additionally, only one stent type, the Bard Inlay, was evaluated during the trial. As this stent has a proprietary coating, the findings may not be generalized to other stent types or materials.
In conclusion, the stent proteome we describe is an accurate but limited first step in exploring stent conditioning films. Validation of our findings in a larger population is certainly warranted, and future studies may include inflammatory urinary cytokines in addition to pattern protein profiling of ureteral stents. Overall, an understanding of conditioning biofilm proteins, in particular, Tamm-Horsfall and protein classes of immunoglobulins and histones, may aid in the design of stents that can resist encrustation and biofilm formation.
Abbreviations Used
- ACN
acetonitrile
- MS
mass spectrometry
- pI
isoelectric points
- THP
Tamm-Horsfall protein
Acknowledgments
B.K.C. is supported by a Foundation Research Scholar Grant from the American Urological Association, a Biomedical Genomics Center Seed Grant from the University of Minnesota, and an NIH RO1 DK065658-03S1. MS/MS data were acquired at the University of Minnesota Center for Mass Spectrometry and Proteomics, Minneapolis, and St. Paul Campus.
Disclosure Statement
No competing financial interests exist.
References
- 1.Riedl CR. Plas E. Hubner WA. Zimmerl H. Ulrich W. Pfluger H. Bacterial colonization of ureteral stents. Eur Urol. 1999;36:53–59. doi: 10.1159/000019927. [DOI] [PubMed] [Google Scholar]
- 2.Singh I. Gupta NP. Hemal AK. Aron M. Seth A. Dogra PN. Severely encrusted polyurethane ureteral stents: Management and analysis of potential risk factors. Urology. 2001;58:526–531. doi: 10.1016/s0090-4295(01)01317-6. [DOI] [PubMed] [Google Scholar]
- 3.Mouzakis DE. Bouropoulos N. Bithelis G. Liatsikos E. Aging assessment by dynamic mechanical analysis of in vivo encrusted polymeric urinary stents. J Endourol. 2006;20:64–68. doi: 10.1089/end.2006.20.64. [DOI] [PubMed] [Google Scholar]
- 4.Keane PF. Bonner MC. Johnston SR. Zafar A. Gorman SP. Characterization of biofilm and encrustation on ureteric stents in vivo. Br J Urol. 1994;73:687–691. doi: 10.1111/j.1464-410x.1994.tb07557.x. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y. Simonovsky FI. Ratner BD. Horbett TA. The role of adsorbed fibrinogen in platelet adhesion to polyurethane surfaces: A comparison of surface hydrophobicity, protein adsorption, monoclonal antibody binding, and platelet adhesion. J Biomed Mater Res A. 2005;74:722–738. doi: 10.1002/jbm.a.30381. [DOI] [PubMed] [Google Scholar]
- 6.Hu WJ. Eaton JW. Ugarova TP. Tang L. Molecular basis of biomaterial-mediated foreign body reactions. Blood. 2001;98:1231–1238. doi: 10.1182/blood.v98.4.1231. [DOI] [PubMed] [Google Scholar]
- 7.Choong S. Whitfield H. Biofilms and their role in infections in urology. BJU Int. 2000;86:935–941. doi: 10.1046/j.1464-410x.2000.00949.x. [DOI] [PubMed] [Google Scholar]
- 8.Gristina AG. Biomaterial-centered infection: Microbial adhesion versus tissue integration. Science. 1987;237:1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- 9.Jung SY. Lim SM. Albertorio F, et al. The Vroman effect: A molecular level description of fibrinogen displacement. J Am Chem Soc. 2003;125:12782–12786. doi: 10.1021/ja037263o. [DOI] [PubMed] [Google Scholar]
- 10.Holmberg M. Stibius KB. Larsen NB. Hou X. Competitive protein adsorption to polymer surfaces from human serum. J Mater Sci Mater Med. 2008;19:2179–2185. doi: 10.1007/s10856-007-3318-9. [DOI] [PubMed] [Google Scholar]
- 11.Yates JR., III Mass spectrometry as an emerging tool for systems biology. Biotechniques. 2004;36:917–919. doi: 10.2144/04366TE01. [DOI] [PubMed] [Google Scholar]
- 12.Kapphahn RJ. Ethen CM. Peters EA. Higgins L. Ferrington DA. Modified alpha A crystallin in the retina: Altered expression and truncation with aging. Biochemistry. 2003;42:15310–15325. doi: 10.1021/bi034774e. [DOI] [PubMed] [Google Scholar]
- 13.Canales BK. Anderson L. Higgins L, et al. Second prize: Comprehensive proteomic analysis of human calcium oxalate monohydrate kidney stone matrix. J Endourol. 2008;22:1161–1167. doi: 10.1089/end.2007.0440. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M. Ball CA. Blake JA, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew BH. Denstedt JD. Technology insight: Novel ureteral stent materials and designs. Nat Clin Pract Urol. 2004;1:44–48. doi: 10.1038/ncpuro0014. [DOI] [PubMed] [Google Scholar]
- 16.Tunney MM. Gorman SP. Evaluation of a poly(vinyl pyrollidone)-coated biomaterial for urological use. Biomaterials. 2002;23:4601–4608. doi: 10.1016/s0142-9612(02)00206-5. [DOI] [PubMed] [Google Scholar]
- 17.Eisenberg D. The discovery of the alpha-helix and beta-sheet, the principal structural features of proteins. Proc Natl Acad Sci USA. 2003;100:11207–11210. doi: 10.1073/pnas.2034522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulton CS. Seirafi A. Hinton JC, et al. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 19.Klassen RB. Allen PL. Batuman V. Crenshaw K. Hammond TG. Light chains are a ligand for megalin. J Appl Physiol. 2005;98:257–263. doi: 10.1152/japplphysiol.01090.2003. [DOI] [PubMed] [Google Scholar]
- 20.Huang ZQ. Sanders PW. Localization of a single binding site for immunoglobulin light chains on human Tamm-Horsfall glycoprotein. J Clin Invest. 1997;99:732–736. doi: 10.1172/JCI119218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hession C. Decker JM. Sherblom AP, et al. Uromodulin (Tamm-Horsfall glycoprotein): a renal ligand for lymphokines. Science. 1987;237:1479–1484. doi: 10.1126/science.3498215. [DOI] [PubMed] [Google Scholar]
- 22.Thomas DB. Davies M. Peters JR. Williams JD. Tamm Horsfall protein binds to a single class of carbohydrate specific receptors on human neutrophils. Kidney Int. 1993;44:423–429. doi: 10.1038/ki.1993.260. [DOI] [PubMed] [Google Scholar]
- 23.Santin M. Motta A. Denyer SP. Cannas M. Effect of the urine conditioning film on ureteral stent encrustation and characterization of its protein composition. Biomaterials. 1999;20:1245–1251. doi: 10.1016/s0142-9612(99)00026-5. [DOI] [PubMed] [Google Scholar]