Natural outbreaks of multidrug-resistant microorganisms can cause widespread devastation, and several can be used or engineered as agents of bioterrorism. A key challenge is the ability to detect and then efficiently control, within hours, the spread and the potential effects of an emergent outbreak, without recourse to either antibiotics or vaccines. The authors describe phage engineering as a potentially highly flexible and effective means to both detect and eradicate threats from emergent bacterial strains.
Abstract
Natural outbreaks of multidrug-resistant microorganisms can cause widespread devastation, and several can be used or engineered as agents of bioterrorism. From a biosecurity standpoint, the capacity to detect and then efficiently control, within hours, the spread and the potential pathological effects of an emergent outbreak, for which there may be no effective antibiotics or vaccines, become key challenges that must be met. We turned to phage engineering as a potentially highly flexible and effective means to both detect and eradicate threats originating from emergent (uncharacterized) bacterial strains. To this end, we developed technologies allowing us to (1) concurrently modify multiple regions within the coding sequence of a gene while conserving intact the remainder of the gene, (2) reversibly interrupt the lytic cycle of an obligate virulent phage (T4) within its host, (3) carry out efficient insertion, by homologous recombination, of any number of engineered genes into the deactivated genomes of a T4 wild-type phage population, and (4) reactivate the lytic cycle, leading to the production of engineered infective virulent recombinant progeny. This allows the production of very large, genetically engineered lytic phage banks containing, in an E. coli host, a very wide spectrum of variants for any chosen phage-associated function, including phage host-range. Screening of such a bank should allow the rapid isolation of recombinant T4 particles capable of detecting (ie, diagnosing), infecting, and destroying hosts belonging to gram-negative bacterial species far removed from the original E. coli host.
The therapeutic potential of obligate lytic (virulent) phages in the treatment of pathological bacterial infections has long been known and successfully used. However, in western societies, the development of highly effective antibiotics that are easy to produce and administer and that can quickly and efficiently control many life-threatening bacterial infections has rapidly made obsolete the more cumbersome phage therapies. With the advent of mass-resistance to most antibiotics in a plethora of pathogenic bacterial strains, phage therapy is now regaining attention.1
T4, the archetype of virulent phages, has evolved very efficient strategies to subvert host functions and impose the expression of its genome.2 T4 has a myriad of relatives in nature that differ significantly in their host range.3 These phages detect and bind to the outer membrane proteins of their hosts, such as the Omp transporters, via tail fibers host specificity determinants (adhesins) encoded by the genes gp37 and gp38.4 The amino acid sequences of the receptor recognition proteins encoded by these genes is critical for phage-host interactions, largely defining the host-range of a phage. Thus, mutations in these genes restrict or increase the range of bacterial hosts a phage can detect and infect.5,6 These genes appear as mosaics with parts derived from a common gene pool, and different segments within each gene constitute host-range cassettes consisting of 2 constant domains and 1 contiguous array of 4 hypervariable regions.5,7 Exchanges involving these constant domains can replace the endogenous specificity determinants, thus mediating acquisition of determinants allowing a phage to cross species boundaries and infect taxonomically distant hosts.8 Mutations affecting the host range of tailed phages map to these regions, and specific mutagenesis within the hypervariable domains has, in the past, been used on T4 phages active against E. coli to generate host range mutants redirected toward Yersinia pseudotuberculosis.9
As a result of their continuous co-evolution with phages, bacteria have evolved numerous strategies allowing them to efficiently resist phage predation, even when several different phages are used concurrently.10 Among these strategies, the accumulation of mutations affecting the structure and the expression levels of outer membrane transporters figures prominently.11 Furthermore, the majority of bacterial pathogens contain prophages or phage remnants integrated into the bacterial DNA, many of which encode virulence factors that contribute to protecting the cell from phage predation.12
Thus, just as with antibiotics, the rapid emergence of bacterial resistance in response to recurrent predation by the same group of phages remains a major biosecurity issue.13 The countermeasures most frequently adopted have attempted to isolate, from natural environments, new phages capable of infecting and destroying resistant bacterial populations.10 But this takes considerable time and effort (generally on the order of at least 1 year). Other avenues envisaged have been the use of filamentous as well as obligate lytic phages engineered to express enzymes that (1) repress the host's DNA repair machinery (SOS DNA-repair system), thus inhibiting the development of resistance to antibiotics while enhancing the lethal effects of these drugs,14 or that (2) degrade biofilms, thus locally enhancing bacterial lysis.15
However, although these approaches present undeniable merits, they perpetuate a situation where countermeasures always lag well behind the appearance of resistance, since these approaches can be implemented only when the occurrence of the undesirable phenomenon, known to be inevitable, has become evident. Indeed, in each case, the bacteria targeted must have been characterized at both structural and molecular levels. Filamentous phages can only address flagellate cells, and bacteria possess numerous mechanisms that rapidly counteract SOS inhibition to maintain highly efficient DNA-repair and recombination systems accommodating horizontal genetic transfers,16 while the option using lytic phages must be initiated from a virus that can adsorb to and replicate in the target cells. This not only leaves the emergence of bacterial resistance to phage predation unaffected, but recent studies demonstrate that a high adsorption rate is detrimental to phage effectiveness in biofilm environments,17 hence favoring passive effects (relying on self-replication to bring the phage concentration above the bacterial “inundation threshold” at the site of infection), which can be effective only if both target bacteria and infective phage concentrations are low.18
An alternative would be the production of large, genetically engineered, virulent phage populations from which particles capable of defeating any bacterial resistance strategy could be rapidly isolated. Moreover, provided such phage banks can be produced, they need not be limited to addressing gram-negative bacteria only. Phages targeting gram-positive cells also could be used. Thus, after thorough screening against currently known bacterial pathogens (be they natural or engineered), the availability of such phage banks could provide the means to both rapidly detect the presence and treat topical infections by any of the known pathogens as well as by previously uncharacterized emergent strains, provided a sample can be obtained for rapid screening against the banks.
We used systems biology to model both the mechanisms indispensable for virulent phage infection and replication concurrently with those the host needs to implement to resist phage predation. The models demonstrated that 3 different novel technological approaches would be required for the production of genetically engineered virulent phage banks.
First, a technology (TAPE) allowing us to modify concurrently and stochastically multiple coding domains within any gene, such as those critical for phage-host interactions, while preserving intact multiple coding regions within the same gene would be needed to generate a multitude of domain-targeted variants.
Then, another technology (AB-Accus) allowing us to efficiently and stochastically recombine, at a population level, the multitude of variants generated for at least 2 genes into the genomes of the members of a wild-type phage population in replacement of the native sequences would be necessary.
Finally, a third technology (RIPh) allowing us to reversibly interrupt the lytic cycle of an obligate virulent phage within its host would be indispensable to allow the above homologous recombinations to be performed at high efficiency so that, upon reinstatement and completion of the lytic cycle, each recombinant parental particle would produce a progeny different from that of most other recombinant particles.
Results
Host-Range Engineering
To produce the widest possible range of functional host-range variants, large-scale stochastic modifications will have to be introduced into the variable regions that constitute the host-range cassettes in the gp37 and gp38 genes. This implies that, in each gene, X regions must be simultaneously targeted and stochastically modified, at Y different sites, in Z different manners, while conserving intact N different domains—all of this independently and concurrently. To efficiently achieve this, we developed a nested PCR-based technology, named TAPE (for Targeted Accelerated Protein Evolution), that uses an interplay between error-prone and high-fidelity reactions to introduce random point mutations into a sequence while preserving the identity of predefined inner segments. The modifications were introduced by (1) decreasing the fidelity of the Taq polymerase during DNA synthesis without significantly decreasing the level of amplification achieved in the PCR reaction, and (2) the random incorporation of nucleotide analogues. Different regions can be subjected to different mutation rates depending on application. This is followed by a series of selective high-fidelity reactions that reconstruct a wide spectrum of variant forms of the genetic region of interest while automatically eliminating the products containing mutations within the domains selected to remain constant. The T4 genes gp37 and gp38 were used to develop and validate the method described in Figure 1.
Figure 1.
Schematic representation of the “TAPE” procedure. The gene to be stochastically modified is first isolated by PCR amplification. Comparative sequence analysis with other isoforms is used to identify the positions of conserved domains and/or variable regions. PCR primers corresponding to entire conserved domains or the extremities thereof are then used in nested, error-prone PCR reactions, generating fragments embodying 2 inner variable regions and 2 outer conserved domains. This approach allows one to compensate for the fact that stochastic mutagenesis is not very efficient over very short regions. With each successive PCR cycle, mutations will be stochastically and incrementally introduced anywhere over the entire length of each fragment (Step 1). It is now necessary to eliminate the products carrying mutations in the conserved domains while starting reconstruction, in the correct order, of the entire coding sequence. This is achieved in 2 steps. A high-fidelity PCR reaction, using the outer conserved domains as primers, is carried out to selectively amplify, in each of the mutagenesis PCR reaction products, the fragments where the outer priming sites have been largely preserved mutation-free (Step 2). This high-stringency reaction uses at least 25 PCR cycles, leading to the elimination by dilution of most undesirable fragments generated by each mutagenesis reaction. The amplified products are recovered, and overlapping fragments, in which one outer conserved domain in one fragment corresponds to the inner conserved domain in its overlapping counterpart (Step 3), are then used in high-fidelity PCR reactions where each fragment serves as the primer for its overlapping counterpart. In this reaction, where the participating fragments all have largely intact outer conserved domains, only fragments where the inner conserved domain has been preserved largely mutation-free will allow effective priming and subsequent exponential amplification of the merged products. This constitutes a negative selection step, in which undesirable fragments gradually and automatically exclude themselves from inclusion into partly reconstructed and amplified gene segments containing 4 conserved domains interspersed among 3 variable regions. This step is then sequentially repeated, using partly overlapping reconstructed fragments, for the final reconstruction of the entire coding sequence, generating a large variety of constructs (Step 4), all presenting identical structures and successions of conserved domains interspersed among a large spectrum of stochastically modified variable regions. Color images available online at www.liebertonline.com/bsp.
The DNA sequence to be modified is first analyzed to define the domains that must be conserved (Blast and CD searches: http://www.ncbi.nlm.nih.gov/sutils/static/blinkhelp.html and http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). These are then used to construct overlapping PCR primers. The gene or genetic region to be modified is then isolated from the corresponding genome using high-fidelity PCR amplification. This PCR product is then used as a template in a series of error-prone PCR reactions (see Figure 1, Step 1) addressing different variable domains bracketed by primers corresponding to constant domains or parts thereof. Each PCR reaction can be independently adjusted to different theoretical mutation rates (1.3% to 15%; Tables 1a and 1b). The resulting PCR products are then recovered and individually reamplified using high-fidelity PCR reactions (Figure 1, Step 2, and Figure 2A, Ca). The purposes of this process are to “lock-in” the mutations introduced while ensuring that the fragments' ends (priming sites) have been conserved intact. Only the templates in which the priming sites have been conserved mutation-free will be efficiently amplified.
Table 1a.
Comparative efficiencies (%) of the two mutagenesis methods used
| Methods | P1 (60 bp) | P2 (75 bp) | P3 (27 bp) | P4 (258 bp) | P5 (33 bp) |
|---|---|---|---|---|---|
| 1 | 1.7 ± 2.3 | 1.3 ± 0.0 | 1.9 ± 2.6 | 1.6 ± 0.5 | 1.6 ± 2.2 |
| 2 | 7.5 ± 1.1 | 5.3 ± 0.0 | 9.3 ± 2.6 | 8.8 ± 3 | 7.6 ± 6.4 |
Table 1b.
Influence of PCR cycle numbers upon the rates (%) of mutagenesis using the nucleotides analogues method
| Number of cycle | P1 (225 bp) | P2 (255 bp) |
|---|---|---|
| 5 | 3.8 ± 0.3 | 3.7 ± 0.3 |
| 10 | 7.8 ± 0.4 | 7.7 ± 1.3 |
| 15 | 8.2 ± 0.3 | 7.8 ± 0.5 |
| 20 | 10.3 ± 1.9 | 9.0 ± 0.6 |
| 25 | 13.8 ± 1.3 | 11.8 ± 2.8 |
| 30 | 16.9 ± 0.6 | 13.2 ± 1.3 |
The mutagenesis efficiencies arising from the random incorporation of nucleotide analogues (0.5 mM dPTP; method 2) were compared to those achieved by decreasing the fidelity of the Taq polymerase (method 1). For this comparison, the 5 overlapping hypervariable domains of gp38 fragments (P1 to P5) encoding the C terminus receptor-recognition domain of this tail fiber protein (Table 1a) and the first 2 (P1 and P2) of the 15 overlapping fragments constituting the gp37 gene (Table 1b) were independently subjected to a PCR reaction made error-prone by either of the 2 methods. The resulting products were then sequenced (n = 4) and the frequencies of mutated sites recorded in terms of percentages. Using 15 error-prone PCR cycles, the random incorporation of nucleotide analogues produces much higher levels of mutagenesis than the destabilization of Taq fidelity (averages of 7.7% ± 2.6% versus 1.6% ± 1.5%, respectively, Table 1a). However, the results obtained with the latter approach appeared considerably more stochastic (standard error as large as the mean) and thus difficult to control. While neither method appeared particularly dependent on the lengths of DNA fragments (Table 1a), mutagenesis by random incorporation of nucleotide analogues is highly sensitive to the number of error-prone PCR cycles (Table 1b), thus allowing control over the levels of mutagenesis through the modulation of both the concentrations of nucleotide analogues in the reaction and the number of error-prone PCR cycles.
Figure 2.
Mutagenesis and reconstruction of gp37 and gp38. The 2 genes encoding the host recognition receptor functions in the T4 genome were subjected to the TAPE procedure. The gp37 gene (2897 bp) was organized into 13 conserved domains interspersed among 12 variable regions (designated P1 to P12), and the coding region corresponding to the C-terminus of the gp38 protein (583 bp) was organized into 6 conserved domains interspersed among 5 variable regions (designated P1 to P5) and subjected to PCR mutagenesis by random incorporation of nucleotide analogues (0.5 mM dPTP and 20 PCR-cycles [10% nominal mutation rate; see Table 2]). The resulting randomly mutated fragments were then subjected to the entire high-fidelity reconstruction procedure described in Figure 1. The products generated at each step were visualized by gel electrophoresis. Figure 2A presents the results obtained following high-fidelity amplification of each post-mutagenesis fragment (Step 2) generated for gp37 (P1 to P12), each lane (1 to 12) showing the corresponding PCR fragment (P1 = lane 1, expected size of 275 bp; P2 = lane 2, expected size of 300 bp; P3 = lane 3, expected size of 111 bp; P4 = lane 4, expected size of 102 bp; P5 = lane 5, expected size of 326 bp; P6 = lane 6, expected size of 690 bp; P7 = lane 7, expected size of 302 bp; P8 = lane 8, expected size of 130 bp; P9 = lane 9, expected size of 254 bp; P10 = lane 10, expected size of 224 bp; P11 = lane 11, expected size of 132 bp; and P12 = lane 12, expected size of 341 bp; M = molecular size marker). Figure 2B shows the results obtained at completion of sequential reconstruction of gp37 variants (lane 1 = construct of P1 to P12, expected size of 2897 bp) containing 13 conserved domains and 12 variable regions (Step 4), while Figure 2C shows the results obtained during partial (Fig. 2Ca, lane 1 = P1 + P2, expected size of 203 bp; lane 2 = P3 + P4, expected size of 345 bp [Step 3]) and complete sequential reconstruction of the C-terminus-encoding gp38 region (Fig. 2Cb, lane 1 = construct of P1 to P5, expected size of 583 bp [Step 4]) from randomly mutated PCR products representing 6 conserved domains and 5 variable regions. Color images available online at www.liebertonline.com/bsp.
The PCR products are then purified, and fragments with overlapping extremities are used for selective reconstruction of the entire genetic region consisting of randomly variable domains interspersed between constant domains (Figure 1, Step 3, and Figure 2B, Cb). In this high-fidelity PCR reaction, each fragment serves as the primer for its partly overlapping counterpart, and dual extension (partial reconstruction) will be possible only if the overlapping segments (conserved domains or parts thereof ) have nearly intact complementary sequences. Different reconstruction schemes (Figure 1, Step 4) were tested. For both gp37 (2897 bp) and gp38 (C-terminus, 583 bp) genes, batch reconstructions were successfully achieved using up to 6 fragments simultaneously (data not shown). However, we did not encounter limits using sequential schemes, and versions of the gp37 gene made to contain up to 13 conserved domains interspersed between variable regions were successfully obtained (Figure 2B).
Reconstructed genes were then analyzed by batch sequencing to define the frequencies, positions, and types of mutations introduced in the variable regions and to verify the levels of sequence conservation within the constant domains. Peak superpositions in sequencing chromatograms, indicating the presence of a mixture of different sequences, revealed the positions and frequencies of alternative nucleotides stochastically introduced in variable domains (Figure 3B, C). Peak superpositions were rarely observed in the conserved domains, their sequences remaining essentially identical to those of the corresponding regions in the wild-type gp37 (Figure 3A, B) and gp38 T4 genes, indicating that all had been largely preserved, irrespective of their numbers and lengths (Figure 3C).
Figure 3.
Sequencing analyses of stochastically mutated and partly reconstructed gp37. Fragments of the wild-type gp37 gene corresponding to the P1 and P2 segments were generated by PCR and subjected to the PCR-mediated reconstruction procedure without prior mutagenesis. Both the wild-type (Figure 3A) and mutated (Figure 3B, nominal mutation rate of 6%) P1 + P2 reconstructed fragments were then subjected to batch DNA sequencing analyses (n = 4; 200 ng of PCR products each). This approach allows the chromatograms to reveal the positions and densities of mutations generated (peaks superpositions marked*). In addition, comparative sequence analyses (Figure 3C) were performed on individual P1 + P2 fragments reconstructed from material subjected to different nominal mutation rates (from 3%, line 1, to 18%, line 4). These analyses demonstrate that (a) the reconstruction protocol does not introduce sequence shuffling, and (b) the TAPE procedure efficiently performs selective stochastic mutagenesis of multiple domains while concurrently preserving the sequence integrity of multiple selected regions within the same gene, even when very high nominal mutation rates are being implemented (line 4 = 18%, 5 = 13%, and 6 = 9%). These analyses were independently repeated 3 times and gave highly comparable results. Color images available online at www.liebertonline.com/bsp.
Hence, TAPE appears to be a robust, trustworthy technology that is easy to implement and to manipulate and that can be applied for the large-scale stochastic diversification of multiple targeted domains within any known gene and protein coding sequence (enzymes, receptors, antibodies, etc).
Production of a Genetically Engineered T4 Phage Bank with Vastly Increased Host-Range
The very large diversification of T4 coding sequences obtained through TAPE implies that homologous recombinations into the phage's genome need to be carried out at population levels if this diversity is to be preserved and harnessed. Plasmid-encoded molecular systems allowing very efficient in vivo homologous recombination reactions using electroporated PCR products19,20 can easily be introduced into bacteria permissive to T4 replication, such as the E. coli strain DK8. However, the T4 genome is very rapidly activated upon its entry into a host, resulting in the quasi-immediate appropriation of the bacterial transcriptional machinery and the conditional inhibition of host-dependent gene activation,21,22 including the expression of plasmid-encoded functions. This phenomenon effectively abolishes the efficacy of plasmid-borne recombination systems. But our models indicated how the mechanisms used by the phage to impose, step by step, its genetic program on the host cell could be used to reversibly inhibit the production of the essential T4 early proteins, which, through mechanisms such as ADP-ribosylation of RNA polymerase and of other host proteins, allow initial phage-directed mRNA synthesis reactions to escape from host control.23 Thus, reversibly blocking T4 transcriptional takeover together with the entire phage replication program would preserve the efficacy of plasmid-encoded recombination systems while maintaining, at least for a few hours, the integrity of the phage's genome within the host, thereby allowing homologous recombination to be efficiently performed.
Reversible Inhibition of the T4 Lytic Cycle Within the Bacterial Host
The T4 early genes essential for transcriptional takeover (such as motA, asiA, alt, and mod22-24) are apparently transcribed as concatenated, run-through RNAs, and the host-encoded homo-hexameric transcription termination factor Rho25 should be required for the production of the corresponding very early phage proteins. Our models clearly suggested that conditional over-expression of a mutated, nonfunctional Rho protein within the host would interfere with the formation of a functional Rho complex, thereby reversibly inhibiting production of the essential T4 early proteins while minimally affecting host viability under controlled laboratory conditions. We used TAPE to produce nonfunctional variants of the E. coli rho gene (Rho*), which were then cloned into IPTG-inducible expression vectors and introduced into DK8 cells harboring the heat-shock-inducible Red recombinase system.26 The cells were then infected with wild-type T4 phages with and without prior induction of Rho* over-expression. Induction of Rho* practically coincident with phage infection had no effects on phage replication, and massive host lysis ensued within 30 minutes postinfection.
However, induction of Rho* over-expression 5 to 10 minutes prior to exposure to T4 particles, while without effects on the efficacy of host infection, entirely blocked phage replication without causing the loss or affecting the integrity of the phage's genome within the host, even after several days of continuous Rho* over-expression (Figure 4A). Following 30 minutes of strong induction, the levels of Rho* expression can be reduced to minimal levels and maintained so for several days without restoring the T4 replication cycle (Figure 4A). Interruption of Rho* expression through the removal of all traces of ITPG from bacterial cultures results in the systematic reinstatement of the phage lytic cycle and the production of an infective progeny (Figure 5A), irrespective of the duration of prior inhibition, thus demonstrating both the efficacy and the reversibility of the process.
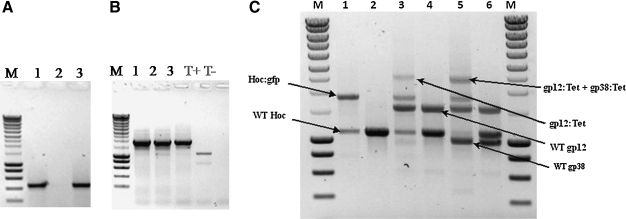
Figure 4.
Rho* expression inhibits phage replication, preserves the phage genome within the host, and allows high-efficiency, stable homologous recombination of phage's genes. DK8 E. coli host cells overexpressing a nonfunctional variant of the rho gene (Rho*) were infected with wild-type T4 phages. Host cells then were collected 3 hours and 96 hours following phage infections, lysed, and the total cellular DNA subjected to PCR analysis directed toward the T4-specific gp38 gene followed by gel electrophoresis. Figure 4A shows that, in spite of the near complete absence of host lysis, PCR analyses revealed the presence at high frequency of T4 gp38 genes in host cells 3 hours (lane 1) and 96 hours (lane 3) following infection, while failing to give a product from noninfected host cells (lane 2). A hoc-GFP construct was electroporated into Rho* overexpressing DK8 cells harboring the cloned Red-recombinase system together with a “deactivated” wild-type T4 genome. Following the induction of Red-mediated homologous recombination, parts of the cells were used for PCR analysis directed toward the hoc-GFP construct (Figure 4B, lane 1). The remainder of the cells were left to grow for 12 hours and the T4 lytic cycle restored (see text). An aliquot of the ensuing phage progeny was used for PCR analysis of the hoc genetic locus (Figure 4B, lane 2), and the remainder were used to infect standard DK8 host cells. The second-generation progeny were analyzed by PCR as above (Figure 4B, lane 3). PCR analysis of the hoc locus in the first-generation progeny (lane 2) revealed a remarkably high apparent recombination rate. The strong PCR band corresponding to the hoc-GFP construct with the concurrent presence of a weak wild-type hoc signal (cf: lane T+) indicated that a very significant proportion of the first-generation progeny appeared to carry the recombinant construct instead of the wild-type gene. This was confirmed by the analytical results obtained from the second-generation progeny (lane 3), in which the recombinant construct was overwhelmingly present and the wild-type hoc sequence hardly detectable. (T+: wild-type PCR hoc signal from DK8 hosts harboring a “deactivated T4 genome; T-: noninfected control DK8 cells). Simultaneous recombinations at multiple loci within the phage's genome were efficiently achieved (Figure 4C, lanes 1, 3, and 5) following concurrent electroporation of hoc-GFP and constructs consisting of the gp12 and gp38 genes fused to a tetracycline resistance encoding insert (gp12:Tet and gp38:Tet, respectively), as demonstrated by postrecombination PCR analyses of the ensuing T4 progeny shown in Figure 4C (M: Molecular weights marker; lane 1: Hoc:GFP insert into the T4 genome; lane 2: WT hoc control; lane 3: Hoc:GFP + gp12:Tet inserts into the T4 genome; lane 4: WT hoc + WT gp12 control; lane 5: Hoc:GFP + gp12:Tet + gp38:Tet inserts into the T4 genome; lane 6: WT hoc + WT gp12 + WT gp38 control).
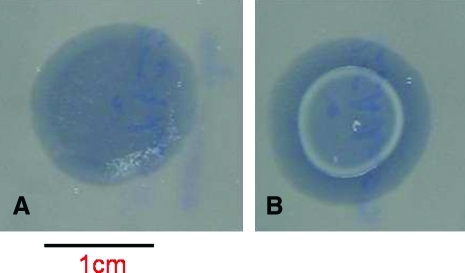
Figure 5.
The Rho*-mediated inhibition of the T4 lytic cycle is reversible, and cells harboring a “deactivated” T4 genome are immune to subsequent T4 superinfection. Overexpression of the nonfunctional variant of the rho gene (Rho*) in DK8 E. coli hosts harboring a wild-type T4 phage genome was inhibited (see text), and, following a latent period of 10 min, the hosts were mixed with noninfected cells and plated onto either standard indicator DK8 cells (panel A) or onto Rho* overexpressing DK8 cells harboring a wild-type T4 phage genome, aliquoted on absorbant discs overlaid onto standard indicator DK8 cells (panel B). In both cases, inhibition of Rho* overexpression prior to plating resulted in reactivation of the phage's lytic cycle and the production of a T4 progeny capable of infecting and lysing standard indicator DK8 cells (dark discs in panels A and B). However, this treatment could not cause the lysis of DK8 cells harboring a “deactivated” T4 genome (white halo in panel B), thus demonstrating that these cells are resistant to T4 superinfection and present the behavior of lysogenic cells. Color images available online at www.liebertonline.com/bsp.
Furthermore, Rho*-mediated inhibition of phage replication (a technology we labeled “RIPh”) is very stable and robust. Only a few cells in large populations (109 cells/ml) exhibit spontaneous reinduction of the T4 lytic cycle, and, most importantly, electroporation of PCR fragments (0.6 to 3 kb) into bacterial cells over-expressing Rho* and harboring a “deactivated” T4 genome is without significant effects on the inhibition of phage replication (Figure 5B).
Large-Scale Recombinations into the Genomes of an Infective Wild-type T4 Population
The T4 hoc gene was used as a target for the insertion, via Red-driven homologous recombination,19 of a fused Hoc-GFP synthetic construct into Rho* “deactivated” T4 genomes. To this effect, the PCR synthetic constructs were electroporated into Rho*-expressing DK8 cells harboring both the heat-shock-inducible Red recombinase system together with a T4 genome, and, following a 30-minute recovery period, the cells were placed at 34°C for 10 minutes to induce the recombinase system and subsequently returned to 25°C for 60 minutes. An aliquot of the cells was then lysed, and the total cellular DNA used for PCR verifications while Rho* expression was suppressed and the T4 lytic cycle restored in the remaining cells. Following lysis, the phage progeny were recovered and an aliquot used for PCR and immunoassay verifications, while the remaining particles were used to infect standard DK8 cells. The subsequent second-generation progeny were recovered and used for PCR and immunoassay verifications.
The results (Figure 4) indicated remarkable levels of homologous recombination efficiency (mean of 10−4 ± 1 × 103, n = 4), as ascertained by both PCR and immunoassays, together with stable transmission of the fused Hoc-GFP gene within the descent of recombinant T4 particles (Figure 4B). Experiments where up to 3 T4 genes were simultaneously targeted for independent homologous recombination gave very similar results (Figure 4C).
Construction of a T4 Bank of Host-Range Variants
To produce the widest possible range of functional host-range variants, we used TAPE to introduce large-scale stochastic modifications into the variable regions that constitute the host-range cassettes in the gp37 and gp38 genes. The pools of TAPE-generated gp37 and gp38 variants were then sequentially recombined as above into the genomes of a wild-type T4 population. The lytic cycle was restored and allowed to proceed for one round of lysis only (25-30 minutes) in order to preserve the recombinant progeny produced in this first lytic cycle that would otherwise be rapidly swamped by nonrecombinant, wild-type progeny with each successive round of host infection and lysis.
The ensuing bank of T4 recombinant progeny was screened against bacterial strains evolutionarily close to (Yersinia ruckeri ATCC 29908) as well as far removed from (Pseudomonas aeruginosa ATCC 47053) the original E. coli K12 strain used for production of the bank. On the first screening pass, recombinant particles capable of infecting and destroying both bacterial species tested were isolated (Figure 6).
Figure 6.
Recombinant T4 banks engineered for extended host-range contain particles capable of infecting and killing bacteria very different from the original DK8 host. In the T4 phage bank used for these assays, generated from a T4 strain known to be restricted to E. coli hosts (ATCC # 110303-B4), both gp38 and gp37 were modified using TAPE. For the assays using Yersinia ruckeri as target cells (panels A, B, C), the bank (100 ml in TE) was lyophilised to 1 ml, which was then used to produce 10-fold dilution series (100 μl into 1 ml final volume, lines starting with “10−2” in panel C), each of which was in turn further diluted in 10 successive steps (1/2 = 100 μl into 200 μl final volume; 1/3 = 100 μl into 300 μl, etc), corresponding to the positions labeled “1/2 to 1/7” above each column in Figures C and F. For the assays using Pseudomonas aeruginosa as target cells (panels D, E, F), banks constructed using different mutation levels (0.6% to 2%) introduced in both gp38 and gp37 were diluted as above. For each dilution, 100 μl were aliquoted onto absorbant discs that were arrayed onto agar plates. The discs were then overlaid with soft agar containing, as indicator cells, bacteria belonging to species far removed from the original E. coli K12 host strain (DK8). With both Yersinia and Pseudomonas indicator cells, multiple lysis plaques (LPs), under the form of dark halos over a clearer background (panels A, E, the part of panel C above the black horizontal bar and that of panel F below the bar), were observed, indicating the presence, within the bank, of numerous phage particles capable of initiating and maintaining their lytic replication cycles in the new host species. Areas of unaffected indicator cells (no lysis) appearing as an absence of distinction with the general background (lower half of panel C, panel B [shown in negative], top of panel F, and panel D) were associated with higher dilution levels (panel C) or low mutation rates (panel F), thus providing evidence that the approach described here allows us to produce, within a single bank, a spectrum of phage particles capable of controlling bacterial cells belonging to multiples and very different species. With both Yersinia and Pseudomonas indicator cells, samples of the parental T4 phage strain were arrayed onto each agar plate as a control. No lysis plaques were observed, confirming that the wild-type T4 strain used here did not infect the indicator bacterial species (data not shown).
For the assays using Yersinia ruckeri as target cells, the 10-fold dilution series from 10−2 to 10−5 are shown. Lysis of Yersinia ruckeri cells remained observable up to an effective 103-fold dilution of the bank. For the assays using Pseudomonas aeruginosa as target cells, only the higher mutation rates for both gp37 and gp38 produced banks containing T4 variants capable of infecting and lysing the target cells. Lysis remained observable up to an 80-fold (100 μl in 8 ml final volume) effective dilution of these banks. Color images available online at www.liebertonline.com/bsp.
Discussion
The mutability of bacteriophages offers particular advantages in the treatment of bacterial infections not afforded by other antimicrobial therapies. Indeed, antibiotics are static molecules, incapable of selectively responding to resistant bacteria in vivo. However, while mutations at genome level can generate potentially beneficial variants, they also produce deleterious genetic loads. Natural phage types with a historical beneficial-to-deleterious mutation ratio of approximately 0.1 per genome per generation have been reported to offer a reasonable balance between beneficial diversity and deleterious mutational load.27 Recent studies, addressing situations where phages and hosts co-evolve, indicate that for phage populations with minimal genetic loads, stochastically engineered moderate increases in mutation rate beyond the optimal value of mutation-selection balance (10 deleterious mutation per genome per generation) may provide even greater protection against emergent bacterial types, albeit only with very weak selective coefficients for de novo genome-wide deleterious mutations (below −0.01).28 However, in such studies, it is the entire viral genome that is being considered and not hyper-variable domains within genes encoding tail fiber proteins exclusively. Here, different tailed phages that are structurally very similar and can infect the same hosts—such as T4, RB69, K3, and T5—carry tail fiber genes that have very few sequence similarities to each other.29 Indeed, between K3 and T4, 2 closely homologous phages, the global long tail fiber proteins (gp37) sequence similarity of 76% falls to 21.7% over the last 231 C-terminus amino acids. The same can be said of a comparison between adhesin protein sequences in T4 (gp37) and T2 (gp38), yielding 50% homology over the last 98 amino acids (blast analyses carried out for the design of constant domains within the T4 gp37 C-terminus; data not shown).
Furthermore, previous studies have demonstrated that rearrangements, fusions, and insertions can be made to gp37 without disrupting the functional integrity of the mature protein structure.30,31 Hence, there is considerable scope for mutagenesis beyond the 0.6-2% levels conservatively used in this study, let alone beyond the levels advocated by mutation-selection balance studies. All the more so since, in the present study, desirable variants are selected by direct exposure to various bacterial strains, thus de facto bypassing all nonfunctional variants in the phage bank, irrespective of their actual frequencies. Thus, once isolated from the phage bank, the desirable novel variants form a population with a genetic load practically identical to that of any wild type T4 population, since, apart from the tail fiber coding sequences, their genome is that of the T4 of origin, all mutations with deleterious effects on the tail fiber proteins having been largely weeded out by the selection process.
Although active phage therapies could be very effective at suppressing susceptible bacteria, phage-resistant cells are readily found both in vitro and in vivo and are a particular problem when the bacterial population is large or growing. Here, single-strain active phage therapies are unlikely to avoid problems of resistance. Single-step mutants can be highly or totally resistant to infection by the phage, and combinations of multiple phages for which cross-resistance does not arise will probably be the only option for closing the mutant selection window.10,32 Combinations of phages that are used in an active mode (the concentration of phages exceeds the rate at which bacterial growth is equaled by the rate of phage infection) must be closely matched for their life cycle properties in order to avoid the “faster” phage dominating the treatment and making the combination little more effective than this faster phage would have been alone, thereby favoring the appearance of resistant bacterial mutants. Furthermore, as the differences between phage strains increase, there is a rapid transition from a relatively low probability of multistrain-resistant cells emerging to a very high probability.10,32
Our approach provides an elegant solution to this problem. The technological approaches (named TAPE, RIPh, and AB-Accus) we have developed allow the efficient production of very large, stochastically engineered, virulent phage banks containing a very wide spectrum of variants that differ for their host-targeting specificities but are all identical for their life cycle properties and from which multiple particles capable of controling a pathogenic, uncharacterized gram-negative bacterial population can be easily and rapidly isolated, as demonstrated by our results. The large lysis plaques that were obtained with multiple dilution series of the phage bank on both Yersinia and Pseudomonas cells strongly suggest the concurrent presence, within the bank, of multiple variants capable of targeting each bacterial species. Indeed, within such a bank, the various constituents of the multitude of variants produced are highly unlikely to be represented with quantitative homogeneity, and any given variant cannot be expected to quantitatively dominate all others. Hence, for lysis to remain highly effective across several dilution levels of the bank, multiple variants represented with quantitative heterogeneity must be present. This amounts to producing complex therapeutic cocktails consisting of phages perfectly matched in terms of life cycle properties. From a technological standpoint, our novel methodologies appear to be robust, trustworthy, and easy to implement and manipulate, and they could be directly applicable to a wide-spectrum of obligate lytic phages.
Nevertheless, any given form of phage-based treatment cannot be expected to be exempt from all types of resistance problems, and this must be accounted for if the full potential of phage therapy is to be realized. Activation of existing restriction-modification or abortive infection mechanisms, such as the ToxN and Abi systems,33,34 and stationary, or other phases of growth that confer temporary resistance to phages,33 figure prominently among the arsenal of very effective escape strategies developed by bacteria. The approach we described here most certainly cannot allow, in its present form, to bypass or counteract these effective defense mechanisms. However, some of the means whereby phages can counteract the induction of Abi-mediated “altruistic suicide” of infected cells, active across multiple genera of gram-negative bacteria and against a spectrum of phages, have been described35,36 and can be relatively easily engineered into the genome of a lytic phage, using some of the techniques described here, prior to modifying its host-targeting mechanism. From a fundamental standpoint, this work constitutes the first direct experimental demonstration explaining the mechanisms governing the expression of T4 very early proteins in E. coli; their entire dependence on the immediate availability of functional, host-encoded transcription termination factors, such as the Rho homo-hexameric complex; and the effects of their functional absence on the T4 replication program.
During the course of this work, several entirely unexpected phenomena associated with Rho*-mediated inhibition of phage replication became apparent. In DK8 cells, the “deactivated” T4 genome behaves as a phagemid and is replicated concurrently with the host cell's genome, resulting in its propagation within the growing, Rho*-expressing bacterial population (Figure 4A). Rho*-expressing bacteria harboring a “deactivated” T4 genome are resistant to subsequent T4 super-infection (pseudo-lysogeny), irrespective of the duration of Rho* expression (Figure 5B), hence protecting the bacterial population from the effects of spontaneous reinduction of the T4 lytic cycle in some of the cells. Finally, Rho*-expressing cells harboring a “deactivated” T4 genome can be glycerolated prior to or after induction of homologous recombination into the T4 genome, stored at −80°C for weeks, and reused without liberating the “deactivated” phage (data not shown). Nevertheless, the duration of storage has adverse effects, since, with increasing time beyond 4-5 weeks, we observed rapidly increasing levels of spontaneous reinduction of the T4 lytic cycle in the revived bacterial cultures (data not shown).
In conclusion, the methods we have developed allow the efficient production of very large, stochastically engineered, virulent phage banks containing a very wide spectrum of variants for any chosen phage-associated function. The banks that can be produced using these approaches can naturally address a wide variety of virulent phage, the genomes of which have been sequenced and annotated. A collection of such banks, comprising gram-negative and gram-positive phages, could conceivably provide the means to largely precede and counteract the escape strategy that may be implemented by any given bacterial population, without recourse to natural phage pools and the lengthy, labor-intensive isolation processes necessarily attached. In addition, these approaches also open the way to an entirely novel use of virulent phages.
Within such a bank, a significant number of phages will be capable of adsorbing to a variety of molecules never encountered on bacterial surfaces. Since the capsids of these phages may be made to contain a variety of molecules other than the usual genomic DNA,34 these particles, in effect, constitute targetable nano-devices that could be used to deliver cargos of small molecules onto materials bearing the surfactants appropriate for efficient phage adsorption.
Thus, besides providing a broadly applicable solution for the long-term maintenance of effective phage therapies (T4 being merely one such example) against emergent, multiresistant bacterial pathogens, these engineered phage banks also provide a source of widely diversified targetable delivery nano-devices. To our knowledge, this is the first time that such an approach to broadly improved biosecurity has been successfully developed and implemented.
Methods
All standard laboratory procedures used in this work have been extensively described by Sambrook and Russell.36
High-fidelity PCR
T4 gp37 (NC_000866.4, GeneID:1258629) and gp38 (NC_000866.4, GeneID:1258706) genes DNA served as a template. The primers used in this study are listed in Table 2. PCRs were performed with 1 unit of Taq polymerase (Roche) in the supplier's buffer. Reaction mixes contained 10 μM each primer and 1 mM dNTPs. The PCR program involved 1 step at 94°C for 2 min, followed by 30 cycles of amplification of 3 steps at (a) 94°C for 30 s, (b) 55°C for 30 s, and (c) 72°C for 1-3 min, depending on the fragment length. PCR products were maintained at 72°C for 5 min.
Table 2.
Primers Used for PCR Amplification
| Primer Pairs | Target Sequence | Variable Domain Size (bp) | Nucleotide Sequences |
|---|---|---|---|
| 37 P1-F | P1 | 225 | 5′-GATTCAGGAAATATCATCGATCTAGG-3′ |
| 37 P1-R | 5′-GGGCGAGCATATATAACGCCACGTTC-3′ | ||
| 37 P2-F | P2 | 255 | 5′-GAACGTGGCGTTATATATGCTCGCCC-3′ |
| 37 P2-R | 5′-GCGAACTTTACGAAGATAGTTTACA-3′ | ||
| 37 P3-F | P3 | 57 | 5′-GGTGTAAACTATCTTCGTAAAGTTCGC-3′ |
| 37 P3-R | 5′-TACACCAGACCACCAAGAAACTTCATC-3′ | ||
| 37 P4-F | P4 | 54 | 5′-GATGAAGTTTCTTGGTGGTCTGGTGAT-3′ |
| 37 P4-R | 5′-TGTACCTAATGCAAGGCTATTACG-3′ | ||
| 37 P5-F | P5 | 267 | 5′-CGTAATAGCCTTGCATTAGGTACA-3′ |
| 37 P5-R | 5′-ATATGGGTTTGTCCGTCTCCAGCATTGTTATTATC-3′ | ||
| 37 P6-F | P6 | 624 | 5′-GCTGGAGACGGACAAACCCATATCGGGTAC-3′ |
| 37 P6-R | 5′-TTCCGAACGACGGAAAATAGCACCATATTCAGCGT-3′ | ||
| 37 P7-F | P7 | 246 | 5′-AACGCTGAATATGGTGCTATTTTCCGTCGTTCGGA-3′ |
| 37 P7-R | 5′-GCAAATGAACCTGCACCCG-3′ | ||
| 37 P8-F | P8 | 75 | 5′-CCCGGCGGGTGCAGGTTCAT-3′ |
| 37 P8-R | 5′-ATTGCCTTGAACATAACGTTGTTTCAAAATAGG-3′ | ||
| 37 P9-F | P9 | 180 | 5′-CCTATTTTGAAACAACGTTATGTTCAAGGCAA-3′ |
| 37 P9-R | 5′-TTTAAGTTAGCAAAATTACCAGAACCACCAGTGATATTAC-3′ | ||
| 37 P10-F | P10 | 135 | 5′-GGTAATTTTGCTAACTTAAACAGTACAATTGAATCACTTAAAACTGATATC-3′ |
| 37 P10-R | 5′-GGTTTACCCTTGATAGTTTGCCCGCGCATATCTGGAAT-3′ | ||
| 37 P11-F | P11 | 51 | 5′-GGGCAAACTATCAAGGGTAAACCAAGTGGTCGTGCTGTTTTGAGC-3′ |
| 37 P11-R | 5′-GTCAAAGCTTGATGTGGTTTTAGTACCTAAGTCAGT-3′ | ||
| 37 P12-F | P12 | 260 | 5′-GGTACTAAAACCACATCAAGCTTTGACTATGGTACGAAG-3′ |
| 37 P12-R | 5′-CCTGTACTATTTACAGTGATAGTATGACCATGTGATCCAATT-3′ | ||
| 38 P1-F | P1 | 60 | 5′-CGGCCCTTCTAAATATGAAAATATATCAT-3′ |
| 38 P1-R | 5′-TGTTGAATGAGCAGGAAGACCG-3′ | ||
| 38 P2-F | P2 | 75 | 5′-CGGTCTTCCTGCTCATTCAACA-3′ |
| 38 P2-R | 5′-TTTTCCGCGATGGTCTTCT-3′ | ||
| 38 P3-F | P3 | 27 | 5′-GAAGACCATCGCGGAAAACG-3′ |
| 38 P3-R | 5′-TTCCAATGTCACTTATAAAAATT-3′ | ||
| 38 P4-F | P4 | 258 | 5′-TTTTTATAAGTGACATTGGA-3′ |
| 38 P4-R | 5′-CTTCTGTTCTTTTTCAGGCCA-3′ | ||
| 38 P5-F |
P5 |
33 |
5′-TGGCCTGAAAAAGAACAGAAG-3′ |
| 38 P5-R | 5′-CCCTCCTTTTAAGATATTTA-3′ |
Error-Prone PCR
The JBS Error-Prone Kit (Jena Bioscience) was used to introduce stochastic mutations by decreasing the fidelity of Taq polymerase (method 1). These reactions were carried out using the supplier's buffer (1×) mixed with 1 μl Taq polymerase (1U), 50 ng DNA template, 2 μl dNTP Error-prone Mix, 1× Error-prone Solution, and 10 mM of a primer pair amplifying an internal portion of gp37 or gp38. Each amplification reaction involved 1 step at 94°C for 2 min, followed by 30 cycles of amplification of 3 steps at (a) 94°C for 30 s, (b) 55°C for 30 s, and (c) 72°C for 1-3 min, depending on the fragment length. PCR products were maintained at 72°C for 5 min. The error-prone solution enhances mutational rate by modifying parameters of PCR-reaction such as a higher Mg2+ concentration of up to 7 mM, a partial substitution of Mg2+ by Mn2+, and unbalanced rates of dNTPs, resulting in a rate of mutagenesis of 0.6% to 2%.
Stochastic insertion of nucleotide analogues (method 2) was performed using the JBS dNTP-Mutagenesis Kit (Jena Bioscience). These reactions were carried out using the supplier's buffer (1×) mixed with 1 μl Taq polymerase (1U), 50 ng DNA template, 1 mM dNTPs, 0.5 mM 8-oxo-dGTP, a range of 0.1 to 0.5 mM dPTP, and 10 mM of a primer pair amplifying an internal portion of gp37 or gp38. Each amplification reaction involved 1 step at 94°C for 2 min, followed by 5 to 30 amplification cycles of 3 steps at (a) 94°C for 30 s, (b) 55°C for 30 s, and (c) 72°C for 1-3 min, depending on the fragment length. PCR products were maintained at 72°C for 5 min. 8-oxo-dGTP is incorporated opposite a template adenine, yielding 2 transition mutations: A → C and T → G. dPTP has been reported37,38 to be approximately 10-fold more mutagenic than 8-oxo-dGTP and yields 4 types of transition mutations: A → G, T → C, G → A, C → T. The resulting rate of mutagenesis is between 3% and 17%, depending on dPTP concentration and number of PCR cycles. For elimination of the mutagenic dNTPs and to conserve the mutations introduced, an aliquot of 1 μl of this first PCR reaction was used in a second high-fidelity PCR amplification.
Selective High-Fidelity Amplification of Desired Fragments
In order to select fragments with priming sites preserved, fragments produced by the error-prone reactions were reamplified using a 1:1 mixture of Taq (Roche) and Isis (MP, proofreading) polymerases and the appropriate primer pairs (Table 2). The reaction mixes and the cycling profiles were as described above (high-fidelity PCR).
Reconstruction of Sequence via PCR
Following error-prone and high-fidelity PCR amplifications, selected fragments in partial overlap were used as templates in equal proportion, and PCR amplifications were performed with 1 unit of Taq polymerase (Roche) in the supplier's buffer and 1 mM dNTPs. After 15 amplification cycles, 1 μM each of the primers corresponding to the termini of the reconstructed fragments were added, and the reaction resumed for another 15 cycles. The cycling protocol was (a) 94°C for 30 s, (b) 55°C for 30 s, and (c) 72°C for 1-3 min, depending on the fragment length.
DNA Sequencing and Analysis
Reconstructed fragments were batch-sequenced by BIOFIDAL (http://www.biofidal.com/), and the ApE program was used to analyze the sequencing chromatograms (http://www.biology.utah.edu/jorgensen/wayned/ape/).
Production and Expression of Nonfunctional E. coli rho Genes
The E. coli genome of DK8 cells (F-δ (srl-recA)306lacZya536(lacZam)rpsL39) was used to PCR amplify and isolate the rho coding sequence (NC_000913.2, GeneID: 948297). The primers used were 5′-TACTTAGAATGGCTTAATTTCTTATGC (ERf) and 5′-TTATGAGCGTTTCATCATTTCGA (ERr). The PCR product was then subjected to error-prone mutagenesis as above. Following sequencing, a mutant bearing an in-frame stop codon at position 634 (Rho*) was cloned into the IPTG inducible expression vector pHSG299 (Takara) and electroporated21 into DK8 cells that were then plated onto LB X-gal/IPTG plates containing 30 μg/ml kanamycin. Following isolation of a white colony and PCR verification for the presence of Rho*, expression of the truncated gene was obtained by exposure of the cells to a final concentration of 0.1 mM IPTG. To inhibit Rho* expression, the cells (15 ml) are centrifuged (5000 × g) and thoroughly resuspended 3 consecutive times in 40 ml minimal media21 with a final resuspension in 20 ml LB.
Construction and Expression of the Heat-Inducible Red-Recombinase System
Genomic DNA from the phage λcI857 was used as a template in a PCR reaction amplifying the attP-cro region that contains the exo, bet, and gam under the control of the heat-sensitive cI repressor. The PCR primers used were 5′-GTATGCATGCTGGGTGTGG (MλRf ) and 5′-CGCACTCTCGATTCGTAGAGCCTCG (MλRr). The PCR product was then cloned into the low-copy number, constitutive expression vector pFN476 (ATCC 86962) and electroporated as above into DK8 cells. The cells were then grown in LB at 26°C where the cI repressor is expressed and actively prevents exo, bet, and gam induction. Upon transfer to 34°C, the cI repressor becomes inactivated, allowing implementation of the exo, bet, gam–mediated recombinase system.
Acknowledgments
We thank Thierry Reynaud and Manuel Géa for their support throughout this work.
Conflicts of Interest
The authors disclose that the funders of this work (Pherecydes Pharma and Bio-Modeling Systems) have submitted international patent applications for 3 technologies described in the manuscript: TAPE and AB-Accus were filed on behalf of Pherecydes Pharma (sole owner) on December 20, 2006 (Ref: BIF117147FR and BIF117138FR, respectively). The patent application for RIPh was filed on behalf of Pherecydes Pharma (sole owner) on March 25, 2009 (ref: B090081FRA).
The authors also disclose that, as legal entities, the funders of this work were not involved in the study design, nor the collection, analysis, and interpretation of data, nor the writing of the paper. However, both entities are fully aware of the contents of this paper, and both agreed to the decision to submit for publication. The authors state that they fully adhere to all journal policies on sharing data and materials.
References
- 1.Summers WC. Bacteriophage therapy. Annu Rev Microbiol. 2001;55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 2.Uzan M. RNA processing and decay in bacteriophage T4. Prog Mol Biol Transl Sci. 2009;85:43–89. doi: 10.1016/S0079-6603(08)00802-7. [DOI] [PubMed] [Google Scholar]
- 3.Desplats C. Krisch HM. The diversity and evolution of the T4-type bacteriophages. Res Microbiol. 2003;154:259–267. doi: 10.1016/S0923-2508(03)00069-X. [DOI] [PubMed] [Google Scholar]
- 4.Riede I. Degen M. Henning U. The receptor specificity of bacteriophages can be determined by a tail fiber modifying protein. EMBO J. 1985;4:2343–2346. doi: 10.1002/j.1460-2075.1985.tb03936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drexler K. Dannull J. Hindennach I. Mutschler B. Henning U. Single mutations in a gene for a tail fiber component of an Escherichia coli phage can cause an extension from a protein to a carbohydrate as a receptor. J Mol Biol. 1991;219:655–663. doi: 10.1016/0022-2836(91)90662-p. [DOI] [PubMed] [Google Scholar]
- 6.Morita M. Fischer CR. Mizoguchi K, et al. Amino acid alterations in Gp38 of host range mutants of PP01 and evidence for their infection of an ompC null mutant of Escherichia coli O157:H7. FEMS Microbiol Lett. 2002;216:243–248. doi: 10.1111/j.1574-6968.2002.tb11442.x. [DOI] [PubMed] [Google Scholar]
- 7.Snyder M. Wood WB. Genetic definition of two functional elements in a bacteriophage T4 host-range “cassette.”. Genetics. 1989;122:471–479. doi: 10.1093/genetics/122.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tetart F. Desplats C. Krisch HM. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J Mol Biol. 1998;282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- 9.Tétart F. Repoila F. Monod C. Krisch HM. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol. 1996;258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- 10.Cairns BJ. Payne RJ. Bacteriophage therapy and the mutant selection window. Antimicrob Agents Chemother. 2008;52:4344–4350. doi: 10.1128/AAC.00574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanji Y. Hattori K. Suzuki K. Miyanaga K. Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl Environ Microbiol. 2008;74:4256–4263. doi: 10.1128/AEM.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzaki S. Rashel M. Uchiyama J, et al. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother. 2005;11:211–219. doi: 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 13.Donlan RM. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009;17:66–72. doi: 10.1016/j.tim.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Lu TK. Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu TK. Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labrie SJ. Moineau S. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J Bacteriol. 2007;189:1482–1487. doi: 10.1128/JB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallet R. Shao Y. Wang IN. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evol Biol. 2009;9:241–253. doi: 10.1186/1471-2148-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cairns BJ. Timms AR. Jansen VA. Connerton IF. Payne RJ. Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog. 2009;5:e1000253. doi: 10.1371/journal.ppat.1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesic B. Rahme LG. Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol Biol. 2008;9:20. doi: 10.1186/1471-2199-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko KA. Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma UK. Chatterji D. Differential mechanisms of binding of anti-sigma factors Escherichia coli Rsd and bacteriophage T4 AsiA to E. coli RNA polymerase lead to diverse physiological consequences. J Bacteriol. 2008;190:3434–3443. doi: 10.1128/JB.01792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinton DM. Pande S. Wais N, et al. Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology. 2005;151:1729–1740. doi: 10.1099/mic.0.27972-0. [DOI] [PubMed] [Google Scholar]
- 23.Tiemann B. Depping R. Gineikiene E. Kaliniene L. Nivinskas R. Rüger W. ModA and ModB, two ADP-ribosyltransferases encoded by bacteriophage T4: catalytic properties and mutation analysis. J Bacteriol. 2004;186:7262–7272. doi: 10.1128/JB.186.21.7262-7272.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkens K. Tiemann B. Bazan F. Ruger W. ADP-ribosylation and early transcription regulation by bacteriophage T4. Adv Exp Med Biol. 1997;419:71–82. doi: 10.1007/978-1-4419-8632-0_8. [DOI] [PubMed] [Google Scholar]
- 25.Chalissery J. Banerjee S. Bandey I. Sen R. Transcription termination defective mutants of Rho: role of different functions of Rho in releasing RNA from the elongation complex. J Mol Biol. 2007;371:855–872. doi: 10.1016/j.jmb.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto S. Izumiya H. Morita M. Arakawa E. Watanabe H. Application of lambda Red recombination system to Vibrio cholerae genetics: simple methods for inactivation and modification of chromosomal genes. Gene. 2009;438:57–64. doi: 10.1016/j.gene.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 27.de la Pena M. Elena SF. Moya A. Effect of deleterious mutation-accumulation on the fitness of RNA bacteriophage MS2. Evolution. 2000;54:686–691. doi: 10.1554/0014-3820(2000)054[0686:eodmao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Kysela DT. Turner PE. Optimal bacteriophage mutation rates for phage therapy. J Theor Biol. 2007;249:411–421. doi: 10.1016/j.jtbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Casjens SR. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res Microbiol. 2008;159:340–348. doi: 10.1016/j.resmic.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyman P. Valluzzi R. Goldberg E. Design of protein struts for self-assembling nanoconstructs. Proc Natl Acad Sci U S A. 2002;99:8488–8493. doi: 10.1073/pnas.132544299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abedon ST. Kinetics of phage-mediated biocontrol of bacteria. Foodborne Pathog Dis. 2009;6:807–815. doi: 10.1089/fpd.2008.0242. [DOI] [PubMed] [Google Scholar]
- 32.Blower TR. Fineran PC. Johnson MJ. Toth IK. Humphreys DP. Salmond GP. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol. 2009;191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bidnenko E. Chopin A. Ehrlich SD. Chopin MC. Activation of mRNA translation by phage protein and low temperature: the case of Lactococcus lactis abortive infection system AbiD1. BMC Mol Biol. 2009;10:4–20. doi: 10.1186/1471-2199-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller TE. Molineux IJ. Bull JJ. Viral resistance evolution fully escapes a rationally designed lethal inhibitor. Mol Biol Evol. 2009;26:2041–2046. doi: 10.1093/molbev/msp111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanguino A. Lopez-Berestein G. Sood AK. Strategies for in vivo siRNA delivery in cancer. Mini Rev Med Chem. 2008;8:248–255. doi: 10.2174/138955708783744074. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J. Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Vol. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 37.Zaccolo M. Williams DM. Brown DM. Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 38.Pritchard L. Corne D. Kell D. Rowland J. Winson M. A general model of error-prone PCR. J Theor Biol. 2005;234:497–509. doi: 10.1016/j.jtbi.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Kurnit DM. Escherichia coli recA deletion strains that are highly competent for transformation and for in vivo phage packaging. Gene. 1989;82:313–315. doi: 10.1016/0378-1119(89)90056-5. [DOI] [PubMed] [Google Scholar]