Abstract
Recent discoveries have revealed previously unappreciated complexity with which retroviruses interact with their hosts. In particular, we have become aware that many mammals, including humans, are equipped with genes encoding so-called “restriction factors,” that provide considerable resistance to retroviral infection. Such antiretroviral genes are sometimes constitutively expressed, and sometimes interferon-induced. Thus they can be viewed as comprising an intrinsic immune system that provides a pre-mobilized defense against retroviral infection or, alternatively, as a specialized extension of conventional innate immunity. Antiretroviral restriction factors have evolved at an unusually rapid pace, particularly in primates, and some startling examples of evolutionary change are present in genes encoding restriction factors. Our understanding of the mechanisms by which restriction factors interfere with retroviral replication, and how their effects are avoided by certain retroviruses, is accruing, but far from complete. Such knowledge could allow for novel forms of therapeutic intervention in pathogenic retroviral infections, as well as the development of animal models of human disease.
Introduction
Recent discoveries have revealed a previously unappreciated complexity with which HIV-1 and related retroviruses interact with their hosts. In particular, we have become aware that many mammals, including humans, are equipped with genes encoding so-called “restriction factors,” that provide considerable resistance to infection by HIV-1 and other retroviruses. Because antiretroviral genes are sometimes constitutively expressed, they can provide an intrinsic pre-mobilized system of defense against retroviral infection (Bieniasz 2004; Wolf and Goff 2008). In addition, in some cell types and tissues, the expression of antiretroviral genes can be up-regulated by type I interferons, suggesting they form part of the inducible innate immune system. The importance of restriction factors in the replication and pathogenesis of retroviruses is underscored by the mechanisms that some have evolved in order to evade the antiviral activity of these proteins. The array of restriction factors that are known to be active against HIV-1 have evolved at an unusually rapid pace, particularly in primates, and some startling examples of evolutionary change are present in genes encoding them. Our understanding of the mechanisms by which restriction factors interfere with retroviral replication, and how their effects are avoided by HIV-1 in human cells, is accruing, but far from complete. Such knowledge could allow for novel forms of therapeutic intervention in HIV/AIDS, as well as the development of animal models of the disease. Here, we briefly review current progress in studies of antiretroviral genes that can inhibit HIV-1, emphasizing recent advances, questions that are yet to be resolved, and their relationship to the wider innate immune system.
The Concept of Retroviral “Restriction” and Historical Perspectives
Until the last decade, the species-specific and tissue tropism of retroviruses was thought to depend largely on whether or not a given cell type provided factors and conditions that are required for optimal replication completion of the viral life cycle (reviewed in Goff 2007). For example, a failure to replicate in a given cell type could often be ascribed to failed viral entry due to the absence of a specific cell surface receptor, or species-specific differences in receptor molecules that render them unable to support infection. Alternatively, once inside the cell, a retrovirus might encounter conditions, such as low concentrations of deoxyribonucleotides, which are not conducive for reverse transcription of the viral genome. In some cases, mitosis is required for entry into the nucleus, and the viral genome cannot traverse the nuclear membrane in nondividing cell types. At the level of gene expression, retroviral replication might then be halted because the correct transcriptional machinery is not in place to mediate synthesis of retroviral mRNAs. Finally, essential cellular factors required for the building and release of new viral particles again might be expressed in a tissue-specific manner or vary in species-specific ways. Investigations of all aspects of restricted retroviral tropism have resulted in the identification of a range of cellular factors that are required for the replication of retroviruses. Indeed, throughout the quarter century of HIV-1 research, the notion that the presence or absence of required factors governs HIV-1 tropism has been particularly informative in unraveling HIV-1–host cell interactions. However, more recent studies have indicated that the lack of expression essential factors did not explain at least some facets of HIV-1 tropism.
The concept that there might be innate and/or intrinsic cellular inhibitors of retroviral replication originated with studies showing that it was possible to isolate inbred mouse strains that were resistant to leukemia induced by specific variants of Friend murine leukemia virus (MLV). These studies demonstrated that there were several “Friend virus susceptibility” (Fv) loci that could give rise to inherited “immunity” to retrovirus-induced disease (Lilly and Pincus 1973). Crucially, at least 2 of these loci, Fv1 and Fv4, were dominant and could inhibit MLV replication in cell culture. While Fv4 encodes an endogenous MLV-like virus whose envelope protein directly interferes with exogenous viral entry (Ikeda and others 1985), Fv1 was more interesting in that was shown to encode a cellular protein with homology to an endogenous retroviral structural protein, Gag (Best and others 1996). Fv1 blocks the replication of sensitive MLV isolates by targeting incoming viral capsids (DesGroseillers and Jolicoeur 1983), inhibiting a step after reverse transcription, but prior to provirus integration. However, its precise mechanism of action is yet to be determined.
For some years, Fv1 remained the sole identified example of an antiretroviral restriction factor. However, beginning in the late 1990s, evidence that specific restriction factors could inhibit HIV-1 replication began to accumulate. These findings were based on investigations of restricted tropism that could not be explained by the absence of necessary factors for HIV-1 replication. A key experimental tool was the use of heterokaryons formed between “permissive” cells (ie, cells where HIV-1 replication proceeded normally) and “nonpermissive” cells (where replication was restricted at a specified step). In a number of cases, the heterokaryons were nonpermissive, implying the existence of dominant factor therein that inhibited HIV-1 replication (Simon and others 1998a; Cowan and others 2002; Münk and others 2002; Varthakavi and others 2003). This type of heterokaryon experiment was used to infer the existence of capsid-specific restriction factors in monkey cells that blocked early events of HIV-1 in replication (Cowan and others 2002; Münk and others 2002), as well as restriction factors that imposed a requirement for particular HIV-1 accessory genes (Madani and Kabat 1998; Simon and others 1998a; Varthakavi and others 2003). Specifically, all lentiviruses, including HIV-1, encode accessory genes (Vif, Vpr/Vpx, Vpu, and Nef for primate immunodeficiency viruses) that are dispensable for virus replication in many “permissive” cultured cell lines. However, in some “nonpermissive” cell types, particularly in the primary cell targets of HIV/SIV, the absence of accessory genes can cause a marked defect in the production of infectious virion particles. In general, HIV-1 and SIVs lacking one or more of these accessory proteins are attenuated in vivo (Kestler and others 1991; Hirsch and others 1998). Heterokaryon experiments were instrumental in demonstrating that the HIV-1 Vif and Vpu genes facilitated replication by antagonizing the effects of antiviral restriction factors (Simon and others 1998a; Varthakavi and others 2003).
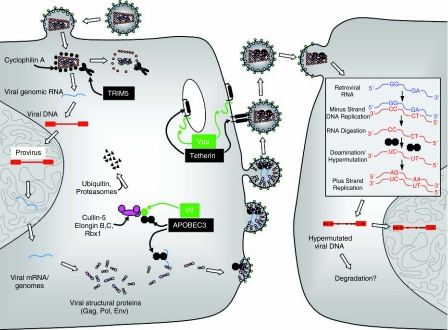
To date, 3 major types of antiretroviral restriction factor that are potentially capable of inhibiting HIV-1 replication have been identified (Sheehy and others 2002; Stremlau and others 2004; Neil and others 2008) (Fig. 1). Moreover, there are strong suspicions that other lentivirus accessory genes are also antagonists of restriction factors that are yet to be uncovered. Of particular interest, these restriction factors are constitutively expressed in some relevant cell types but can be induced by type I interferon in others.
FIG. 1.
Major host restriction factors capable of inhibiting HIV-1 replication, and their viral antagonists. TRIM5 recognizes incoming HIV-1 capsids and inactivates them; accelerated capsid disassembly accompanies the inactivating lesion. Cyclophilin A also binds to incoming capsids and can affect recognition by TRIM5. APOBEC3 proteins are cytidine deaminases, can be incorporated into assembling HIV-1 particles, and catalyze the deamination of nascent minus strand DNA during the subsequent round of infection. The HIV-1 Vif protein binds to APOBEC3 proteins, and also to a Cullin5-based ubiquitin ligase complex. This results in APOBEC3 protein ubiquitination and degradation in proteasomes. Tetherin is a cell surface protein that causes entrapment of nascent virions on the infected cell surface. Vpu antagonizes tetherin by sequestering tetherin from virions and by reducing the levels of tetherin on the cell surface.
The APOBEC3 Family of Cytidine Deaminases
The first host gene to be identified as an inhibitor of HIV-1 replication was found to encode a cytidine deaminase, whose expression was shown to define cells that were nonpermissive for replication of HIV-1 mutants lacking a functional Vif gene (Sheehy and others 2002). The genomes of primates contain an unusually large number of genes encoding cytidine deaminases, named after the prototype member of this family of enzymes—apolipoprotein B editing complex, catalytic subunit (APOBEC) (Harris and Liddament 2004). Subsequently, several members of the APOBEC3 family have been shown to inhibit the replication of HIV-1 and other retroviruses (Bishop and others 2004; Harris and Liddament 2004; Wiegand and others 2004; Yu and others 2004; OhAinle and others 2006), and an intense research effort has illuminated significant aspects of the mechanism by which these enzymes inhibit HIV-1 replication (Fig. 1). Specifically, APOBEC3 proteins can be incorporated into HIV-1 and other retrovirus particles, at least in part through interactions with RNA that is inevitably packaged therein (Schäfer and others 2004; Svarovskaia and others 2004; Zennou and others 2004; Wang and others 2007; Xu and others 2007), and carried by the virion to a new target cell. Thereafter, upon infection and generation of nascent minus strand retroviral DNA, APOBEC3 enzymes catalyze the deamination of deoxycytidines in a sequence context-based manner, generating minus strand DNA containing many deoxyuracil nucleotides whose replication results in plus strand G to A mutations (Harris and others 2003; Lecossier and others 2003; Mangeat and others 2003; Zhang and others 2003).
The various APOBEC3 enzymes exhibit varying degrees of antiretroviral activity, depending on which retrovirus is targeted and from which primate species the APOBEC3 protein is derived, with APOBEC3G being the most prominent and consistently active against retroviruses (Bishop and others 2004; Zennou and Bieniasz 2006; Virgen and Hatziioannou 2007). In general, APOBEC3-induced hypermutation of retroviral DNA can itself be lethal to a retrovirus through the deposition of many inactivating missense and nonsense mutations in protein-coding sequences. However, recent finding suggest that the mutator activity of APOBEC3 enzymes appears to constitute only a part of the mechanism by which antiviral activity is achieved (Newman and others 2005). From the outset, it was clear that the amount of viral DNA that accumulates during HIV-1 infection is reduced when incoming virions carry APOBEC3G, or its close relative APOBEC3F, and while this was reasonably postulated to be due to destruction of uracil-containing retroviral DNA (Schröfelbauer and others 2005), repair enzymes that are known to recognize uracil-containing DNA appear to be dispensable for the APOBEC3G antiviral activity (Kaiser and Emerman 2006). Moreover, the propensity of APOBEC3G and its close relative, APOBEC3F, to reduce viral DNA accumulation appears to be at least partly genetically separable from mutator activity, and mutants that lack cytidine deaminase activity possess residual antiretroviral activity (Newman and others 2005; Holmes and others 2007). Several studies have further suggested that APOBEC3 proteins can inhibit the replication of retroelements without causing their mutation (Bogerd and others 2006; Chen and others 2006b; Chiu and others 2006; Stenglein and Harris 2006; Hulme and others 2007), and that APOBEC3G can directly inhibit specific steps of HIV-1 reverse transcription or integration (Luo and others 2007; Mbisa and others 2007; Bishop and others 2008). However, no consensus has yet emerged as to what is dominant process by which APOBEC proteins inhibit viral or retroelement DNA accumulation, and how intimately connected is this effect cytidine deaminase activity. Nonetheless, in the case of APOBEC3G, enzymatic activity clearly increases the magnitude with which viral DNA accumulation is impaired and it is not completely clear that some of the cytidine deaminase-independent antiretroviral activities ascribed to APOBEC3 proteins occur at physiological levels of protein expression (Miyagi and others 2007; Schumacher and others 2008). Other studies have further elaborated the range of activities reported for APOBEC3G and suggest that it can function in target cells to block HIV-1 infection and that this underlies the resistance of certain cell types or cells in particular physiological states to HIV-1 infection (Chiu and others 2005; Chiu and others 2006). The molecular details of this mode of inhibition are completely unclear at present and the finding itself is controversial (Kamata and others 2009). Finally, in a fascinating re-evaluation of classical genetic resistance to Friend MLV-induced disease in mice, the recovery from Friend virus 3 (RFν3) gene has been identified as murine APOBEC3 (Santiago and others 2008; Takeda and others 2008). RFν3 controls the ability of mice to produce neutralizing antibodies to F-MLV and clear plasma viremia (Hasenkrug and others 1995). The recessive allele of RFν3 encodes a less active APOBEC3 whose mRNA may be aberrantly spliced, and RFν3-sensitive mice progress to erythroleukemia. Similarly APOBEC3-deficient mice exhibit an RFν3-sensitive phenotype (Santiago and others 2008), and are also more susceptible to the betaretrovirus, mouse mammary tumor virus (MMTV) (Okeoma and others 2007). MLV has been shown to be at least partly resistant to murine APOBEC3 through exclusion from assembling virions (Doehle and others 2005), so how the antiretroviral activity of murine APOBEC3 relates to the RFν3 phenotype in F-MLV pathogenesis remains to be determined. However, these data suggest the possibility of linkage between APOBEC3 and downstream mobilization of adaptive immunity against a retrovirus. If this proves to be the case, then these findings clearly have wider implications for the role of these genes in HIV/AIDS.
TRIM5, a Capsid-Targeting Restriction Factor
TRIM5α was originally discovered as the gene product responsible for a series of dominant retrovirus restriction phenotypes that can be observed in human and nonhuman primate cells (Towers and others 2000; Besnier and others 2002; Cowan and others 2002; Münk and others 2002; Hatziioannou and others 2003; Owens and others 2003; Stremlau and others 2004). In particular, TRIM5α is responsible for the inability of HIV-1 to infect many old world monkey cells (Stremlau and others 2004), as well as the resistance of human and other primate cells to certain strains of MLV, and equine infectious anemia virus, a lentivirus (Hatziioannou and others 2004; Keckesova and others 2004; Perron and others 2004; Yap and others 2004). More recent studies have expanded the range of retroviruses that are potentially targeted by TRIM5 proteins to include betaretroviruses and foamy viruses (Diehl and others 2008; Yap and others 2008). Even before TRIM5α was discovered, it was clear that it represented an activity that targeted a range of incoming retroviral capsids, blocking infection before the establishment of a provirus in the target cell (Towers and others 2000; Besnier and others 2002; Cowan and others 2002; Münk and others 2002; Hatziioannou and others 2003) (Fig. 1). The activity was saturable by high amounts of challenge virus, and thus shared several characteristics with the aforementioned Fv1-induced resistance to MLV. Indeed, Fv1 and TRIM5 target overlapping determinants on the MLV capsid, although Fv1 appears to allow reverse transcription to proceed, while TRIM5α prevents retroviral DNA synthesis, in most cases.
The identification of TRIM5α as the protein responsible for the resistance of certain cells to HIV-1 (Stremlau and others 2004) enabled many detailed studies of its evolution and mechanism of action. However, because of the inherent technical difficulty in studying the particular step of the viral lifecycle targeted by TRIM5α, several aspects of how TRIM5α functions remain poorly understood. Nonetheless, we have a rudimentary understanding of the molecular details by which TRIM5 proteins block retroviral infection. Specifically, a C-terminal domain of the TRIM5α protein directly recognizes the incoming viral capsids (Sebastian and Luban 2005; Stremlau and others 2006a) and thereby governs antiretroviral specificity (Perez-Caballero and others 2005a; Stremlau and others 2005; Yap and others 2005; Perron and others 2006), while a central coiled-coil drives TRIM5α multimerization, which is essential for inhibition. An N-terminal “effector” domain markedly increases the potency with which TRIM5α inhibits infection, through unknown mechanisms (Javanbakht and others 2005; Perez-Caballero and others 2005a). The molecular events that lead to virus inactivation are not well understood, but it is apparent that an irreversible, lethal lesion in the incoming viral capsid occurs within minutes of entry of the viral capsid into the target cell cytoplasm (Perez-Caballero and others 2005b). The timing of this lesion significantly precedes the completion of viral DNA synthesis, and in most (but not all) cases, TRIM5α proteins inhibit the accumulation of retroviral DNA (Stremlau and others 2004; Ylinen and others 2005). One clear and highly suggestive observation is that TRIM5 proteins induce the premature disassembly of incoming retrovirus capsids (Stremlau and others 2006a). Curiously, however, the application of proteasome inhibitors prevents TRIM5-induced capsid disassembly and rescues TRIM5α-inhibited synthesis of viral DNA but does not reverse TRIM5α-inhibited infection (Wu and others 2006; Diaz-Griffero and others 2007). Thus, neither proteasome activity itself, inhibition of reverse transcription, nor accelerated capsid disassembly appears essential for TRIM5 to block retroviral infection. These observations leave significant mechanistic questions; unfortunately, probing the mechanism by which TRIM5α proteins work is not straightforward—biochemical and microscopic analysis of the events that follow retrovirus entry into the target cytoplasm is notoriously difficult. Nonetheless, the urge to understand how TRIM5 proteins work is providing additional impetus for the development of new biochemical and imaging assays to probe this heretofore comparatively impenetrable area of retrovirus biology (Stremlau and others 2006a; Campbell and others 2008).
Tetherin, an Inhibitor of HIV-1 Release
Recent work has identified an antiviral protein that works by a novel mechanism in that it can inhibit the release of HIV-1 particles from infected cells (Neil and others 2008; Van Damme and others 2008) (Fig. 1). This protein, termed tetherin (otherwise known as BST-2, CD317, or HM1.24) is a membrane protein, with unusual topology (Kupzig and others 2003). Specifically, it harbors a transmembrane anchor near its N-terminus, and a putative glycophosphatidyl-inositol lipid anchor at its C-terminus, which is essential for its antiviral function. Tetherin appears to induce the formation, and may be a component, of protein-based tethers that cause the retention of HIV-1 particles on infected cell surfaces (Neil and others 2006; Neil and others 2008) (Fig. 1). Interestingly, the retained particles are fully formed and mature and have independent lipid bilayers that are discontinuous with cell membranes (Neil and others 2006). Thus, tetherin seems to act after the formation of virus particles and prevents their dissemination to uninfected cells, perhaps simply by cross-linking the virion and cell membranes. Subsequently, tetherin-retained virions can be reinternalized into the infected cell and targeted to late endosomes, where they may be retained or destroyed by lysosomal enzymes (Neil and others 2006). Ordinarily, tetherin is expressed on a limited subset of cell types, but upon exposure to type I interferon, tetherin is broadly expressed (Blasius and others 2006), suggesting that it is part of a broader innate immune defense that limits the replication of, perhaps, many enveloped viruses. In fact recent data has shown that virus-like particles derived from a wide range of retroviruses, as well as filoviruses and arenaviruses, are also similarly sensitive to tetherin (Neil and others 2007; Neil and others 2008; Jouvenet and others 2009; Kaletsky and others 2009; Sakuma and others 2009).
Antagonism of, and Adaptation to, Antiretroviral Genes by HIV-1
While the aforementioned restriction factors can impart very strong blocks at various stages of retrovirus replication, HIV-1 has evolved a number of strategies to avoid and antagonize these host defenses (Fig. 1). In the case of capsid recognition by TRIM5α proteins, it is sometimes the case that minor sequence variation in capsid proteins can ablate or confer recognition by TRIM5α proteins (Ylinen and others 2005). Additionally, however, a variety of lentivirus capsids have the ability to bind cyclophilin A (CypA), a very abundant cytoplasmic host protein. The reasons for this are not entirely clear, but in some instances it appears that CypA binding can inhibit restriction by TRIM5α proteins (Towers and others 2003; Zhang and others 2006), perhaps by coating the incoming capsid, or by isomerizing peptidy–prolyl bonds (Fig. 1). In the case of HIV-1, its capsid sequence and cyclophilin-binding activity both appear to contribute to resistance to human TRIM5α. However, in at least some instances, CypA binding by capsids appears to have the opposite effect and enhance TRIM5α recognition (Berthoux and others 2005; Keckesova and others 2006; Stremlau and others 2006b). Thus, while CypA binding by retroviral capsids can clearly modify the effect of TRIM5α proteins, whether lentiviruses evolved to acquire this interaction in response to restriction by TRIM5α is not clear at present.
Some entirely new biological activities have been uncovered as a consequence of studies of antiretroviral genes and their antagonists. Indeed, the APOBEC3 and tetherin proteins were discovered based on the ability of their antagonists (the HIV-1 Vif and Vpu proteins) to selectively enhance HIV-1 replication in the so-called “nonpermissive” cells that express them (Sheehy and others 2002; Neil and others 2008). For example, the HIV-1 Vif protein is essential for primate lentivirus replication in cells that express certain APOBEC3 proteins, including primary T cells, and functions by simultaneously binding to a Cullin5-based ubiquitin ligase complex and to APOBEC3 proteins, resulting in their polyubiquitination and degradation (Conticello and others 2003; Marin and others 2003; Sheehy and others 2003; Yu and others 2003) (Fig. 1). Similarly, tetherin was discovered based on the finding that the HIV-1 Vpu protein was required for efficient particle release from certain human cell types, and that type I interferon induced a requirement for Vpu in others (Neil and others 2007). The mechanism by which Vpu antagonizes tetherin is not well established, although it appears that Vpu sequesters tetherin from sites of HIV-1 particle assembly, thereby preventing it for encountering nascent virions (Neil and others 2008; Jouvenet and others 2009) (Fig. 1). This may be linked to the ability of Vpu to reduce the level of tetherin at the cell surface (Van Damme and others 2008) and to trigger its proteasome-dependent degradation (Goffinet and others 2009). It is likely that many other viruses encode mechanisms to antagonize tetherin (Bartee and others 2006). Indeed, the Ebola virus surface glycoprotein (GP) associates with tetherin and antagonizes its ability to inhibit virion release (Kaletsky and others 2009), and a similar activity may be conferred by the envelope glycoprotein of HIV-2, which, like most primate immunodeficiency viruses, lacks a Vpu protein.
Evolution of APOBEC3, TRIM5, and Tetherin Genes
The genomic loci harboring APOBEC3, TRIM5, and tetherin genes show striking evidence of positively selected evolutionary change during primate speciation (Sawyer and others 2004; Sawyer and others 2005; Song and others 2005; McNatt and others 2009). Specifically, the number of nonsynonymous nucleotide substitutions uncovered during interspecies, and in some cases of intraspecies, sequence comparisons of orthologous genes are unexpectedly high. In the case of antiviral genes, positive, or diversifying, selection could be driven by the need to inhibit infection by new viruses, or viral variants. Alternatively, it could arise as a consequence of the need for the antiviral gene to avoid antagonism by viral countermeasures. In the case of TRIM5, positively selected sequences are clearly concentrated in the 3′ exons—precisely those that encode the protein domains responsible for retroviral capsid recognition (Sawyer and others 2005; Song and others 2005). Indeed, individual residues that govern capsid recognition show evidence of positive selection. In contrast, the 5′ TRIM5 exons that are required for antiviral activity but do not appear to govern specificity show the more usual signature of negative, or purifying, selection (Song and others 2005), suggesting that they have been selected to conserve, rather than diversify, function. The distribution of positively selected sites in APOBEC3 genes varies according to which particular APOBEC3 gene is analyzed (Sawyer and others 2004; OhAinle and others 2006), while in tetherin, positively selected sites are enriched toward the 5′ end of the coding sequence that encode the N-terminal cytoplasmic tail and transmembrane segments (McNatt and others 2009). Notably, these positively selected sequence coincide with the protein domains that are known to be targeted by viral antagonists, including the HIV-1 Vpu protein.
In addition to high rates of nonsynonymous nucleotide substitution in primate APOBEC3, tetherin, and TRIM5 genes, there are other quite remarkable evolutionary signatures in these genes that suggest potent selective pressures have been applied to them. For example, so-called “balancing” selection, whereby multiple alleles are maintained in a single species, is evident in old world monkey TRIM5 genes (Newman and others 2006). Another striking phenomenon, particularly at the primate APOBEC3 locus, is gene proliferation; mice possess a single APOBEC3 gene, while humans have no less than 7 (Harris and Liddament 2004). Gene proliferation, albeit less dramatic, is also present at the tetherin locus of bovines, where the gene has been duplicated, with significant divergence in the duplicated copy (PDB, unpublished observation). Again, this gene duplication would likely permit a broader spectrum of viruses to be inhibited, or facilitate the avoidance of viral antagonism. Additionally, the ability of certain retroviral capsids to bind CypA likely provided the impetus for a remarkable example of convergent evolution in primate TRIM5 genes that has recently been observed. In 2 separate species, owl monkeys (a new world monkey) and macaques (an old world monkey) that are separated by 35 million years of divergence, a retrotransposition event has placed a cyclophilin cDNA into the TRIM5 locus (Sayah and others 2004; Nisole and others 2004; Liao and others 2007; Brennan and others 2008; Newman and others 2008; Virgen and others 2008; Wilson and others 2008). While the site of CypA insertion into the locus is clearly distinct in the 2 species, both events lead to the expression of TRIM5–CypA fusion proteins in which the C-terminal portion of TRIM5α that ordinarily determines restriction specificity is replaced by CypA. This obviously changes, in a profound way, the spectrum of retroviruses that can be inhibited by TRIM5-encoded proteins. In macaques, a single retransposition event occurred in an ancestral macaque, and this lesion was maintained at different frequencies in the various descendent macaque species (Liao and others 2007; Brennan and others 2008; Newman and others 2008; Virgen and others 2008; Wilson and others 2008). For example, the CypA insertion in TRIM5 is present in ∼10% of rhesus macaque alleles, but appears to be present in 100% of pigtailed macaque alleles. Remarkably, the retrotransposed CypA cDNA acquired a single amino acid substitution that affects the specificity which retroviral capsids are recognized, and abolishes binding to the HIV-1 capsid (Virgen and others 2008). Thus, pigtailed macaques, unlike most old world monkeys, lack a TRIM5 protein that is capable of inhibiting HIV-1 infection.
The rapid evolution and consequent wide divergence in other primate restriction factor genes also have profound biological consequences. In both APOBEC3 and tetherin proteins, diversity that results from positive selection means that the primate lentivirus Vif and Vpu proteins antagonize their respective host target proteins in a highly species-specific manner (Simon and others 1998b; Mariani and others 2003; Gaddis and others 2004; Virgen and Hatziioannou 2007; McNatt and others 2009). In particular, the Vif genes that are found in most SIVs have specifically adapted to antagonize the particular variants of APOBEC3 proteins that are encountered therein. Therefore, many SIV Vif proteins simply do not function effectively in human cells because they cannot bind to and remove human APOBEC3G, and perhaps other inhibitory human APOBEC3 proteins. Thus, the transmission of potentially pathogenic relatives of HIV-1 from primates to humans has likely been limited by restriction factor divergence. Similarly, the HIV-1 Vif protein fails to function effectively in many nonhuman primate cells, because of a failure to recognize and eliminate several monkey APOBEC3 proteins (Mariani and others 2003; Virgen and Hatziioannou 2007). Recently, it has also become clear that the HIV-1 Vpu protein, while an affective antagonist of human tetherin, is inactive against at least 2 monkey tetherin proteins (McNatt and others 2009). In this case, residues in the tetherin transmembrane domain that have evolved under positive selection appear to be responsible for sensitivity/resistance to Vpu, and it is possible that selective pressures driving the evident positive selection was by ancient SIV Vpu proteins, or similar viral antagonists.
A corollary of the blocks to zoonoses imposed by these antiretroviral proteins is that HIV-1 is not able to replicate nonhuman primate species. Consequently, an authentic animal model of HIV-1 infection is lacking and this has proven to be a significant impediment to AIDS research. Nonetheless, understanding the basis for species tropism of HIV-1 and its simian relatives has allowed the engineering of minimally modified HIV-1 strains that can replicate in macaque cells (Hatziioannou and others 2006; Kamada and others 2006). Recently it has been shown that replacing HIV-1 Vif sequences with those of SIV allows high-level replication and an persistence in pigtailed macaques, whose TRIM5 protein is, unusually, ineffective against HIV-1 (Hatziioannou and others 2009). Thus the development of an authentic animal model of HIV-1 infection appears feasible.
HIV-1, Interferon, and Restriction Factors
Recent evidence indicates that HIV-1 particles are capable of stimulating release of IFN-α from dendritic cells through engagement with Toll-like receptors, particularly recognition of viral genomic RNA by TLR7 from endocytosed virions (Beignon and others 2005). Moreover HIV-1-infected individuals often have high levels of plasma type I IFN (von Sydow and others 1991), and this is rapidly induced during the acute phase of HIV-1 infection, as early as a week after exposure (Stacey and others 2009). Coupled with the multiple and potentially cell type-specific effects of type I IFN on viral replication in vitro (Stetson and Medzhitov 2006), this suggests that the IFN system could potentially present a powerful barrier to HIV-1 systemic spread, and drive the virus to acquire mechanisms for its evasion. These findings, along with the current interest in the roles of retroviral restriction factors as effectors of innate antiviral immunity, have sparked renewed interest in how type I interferons (type I IFN) could modulate the replication of HIV-1. Historically, recombinant IFN-α was investigated early in the HIV epidemic as a potential therapeutic for AIDS-associated Kaposi's sarcoma (Volberding and others 1984) (now known to be caused by the human herpesvirus 8 (Chang and others 1994)), and administration in some patients was shown to reduce HIV plasma viremia (Lane and others 1988; Lane and others 1990). Additionally, various studies demonstrated that replication of HIV-1 in culture could be inhibited by IFN-α, but the nature of the block to virus replication was somewhat nebulous as it often differed between investigators using different cell types or viral strains (Poli and others 1989; Shirazi and Pitha 1992; Agy and others 1995; Korth and others 1998; Taylor and others 1998). Nonetheless, a common observation in primary cell types was that IFN-α-induced block to virus release and concomitant accumulation of cell-associated virions, particularly in macrophages and primary CD4+ T cells (Kornbluth and others 1989; Poli and others 1989; Göttlinger and others 1991; Smith and others 1991; Biswas and others 1992). We now know that this effect was due to the induction of tetherin (Neil and others 2007; Neil and others 2008), and that early studies of HIV-1 used highly passaged laboratory adapted strains that sometimes had lesions in 1 or more of the viral accessory genes, including Vpu.
Indeed, tetherin constitutes the clearest link between the antiretroviral activity of type I IFN and retroviral restriction factors. In cell types that do not normally express tetherin, type I IFN potently induces its expression. In primary HIV-1 target cells such as CD4+ T cells and macrophages, tetherin expression is up-regulated by cellular activation and maturation, but further enhanced by IFN-α (Neil and others 2008; Miyagi and others 2009). Thus, in vitro replication of HIV-1 strains carrying a lesion in the Vpu gene in primary human T cells is effectively inhibited by IFN-α (Neil and others 2007). Wild-type HIV-1 strains are substantially, but not completely, resistant to inhibition by type I IFN—virus spread through the culture occurs, but is significantly delayed (Neil and others 2007). It remains to be determined whether the residual anti-HIV-1 activity of type I interferon is due to incomplete antagonism of high levels of tetherin by inadequate expression of Vpu, or other mechanisms. Notably, in vitro studies indicate that overexpression of tetherin can result in residual inhibition of particle release, even when the viral genome encodes Vpu (Van Damme and others 2008; McNatt and others 2009). Determining the magnitude of the tetherin antiviral effect and the effectiveness of Vpu antagonism in vivo is essential to understand the importance of tetherin as an antiviral factor in HIV/AIDS, and for providing rationale to target Vpu with novel therapeutics. Interestingly, examination of the tetherin promoter reveals likely binding sites for IFN-regulated transcription factors, and also a variety of single-nucleotide polymorphisms in the same area (http://www.ensembl.org/index.html). This raises the possibility that tetherin promoter polymorphisms might influence the course of in HIV infection, and perhaps other viral infections.
Expression of both APOBEC3G and TRIM5α can also be enhanced by IFN-α. APOBEC3G induction by type I IFN has been reported in T cells, macrophages, dendritic cells, and brain microvascular endothelial cells, and suggested to contribute to a block in HIV-1 replication in these cells upon IFN treatment (Chen and others 2006a; Peng and others 2006; Argyris and others 2007; Wang and others 2008). These effects appear to be primarily related to the less well-characterized block to HIV-1 infection when APOBEC3G is present in target cells in low-molecular-weight complexes (Chiu and others 2005), rather than the conventional mechanism by which APOBEC3G is thought to act. Additional, TRIM5 induction by IFN-α has been reported to enhance the ability of human cells to restrict infection by human TRIM5-sensitive MLV strains, and the ability of monkey cells to restrict infection by HIV-1 (Asaoka and others 2005; Sakuma and others 2007; Carthagena and others 2008). In each of these cases, TRIM5α was expressed, and able to inhibit HIV-1 infection in the absence of IFN-α, but IFN-α treatment increased TRIM5α mRNA levels and intensified the infection block. However, since human TRIM5α only marginally inhibits HIV-1 infection (Stremlau and others 2004), even at high expression levels, it is unclear at present whether IFN-α induction would result in sufficiently high TRIM5α protein levels to impact HIV-1 replication in human primary cells. Nonetheless, a significant fraction of TRIM family genes have been reported to be regulated by type I IFN and thus may harbor antiviral activities (Carthagena and others 2009). One recent report suggests that TRIM22, another type I IFN-induced TRIM protein, is capable of restricting the assembly and release of HIV-1 in some human cells (Barr and others 2008) and another study indicated that a number of TRIM proteins have activity against MLV or HIV-1 (Uchil and others 2008). Finally, TRIM28, a nuclear TRIM associated with transcriptional repression, has been found to be recruited to the primer-binding site of integrated MLV proviruses in embryonic stems cells (Wolf and Goff 2007) by the DNA-binding protein ZFP809 (Wolf and Goff 2009). This interaction mediates a silencing of MLV transcription, and may be an antiviral strategy to limit retrovirus replication/reactivation during development. Thus, it may be the case that antiretroviral activity is common among TRIM proteins, although their viral targets and mechanisms of action remain largely unexplored.
Little is known about whether the “classical” IFN-induced genes have any direct effects on HIV-1 replication. The ubiquitin homolog ISG15, which is essential in mice for innate immune resistance to RNA and DNA viruses (Lenschow and others 2007; Lai and others 2009), has been suggested to interfere with HIV-1 assembly. Specifically, conjugation of ISG15 to HIV-1 Gag has been reported to inhibit association with TSG101, an essential cellular factor for HIV-1 budding (Okumura and others 2006). A caveat with these findings is that they were generated by overexpression of both ISG15 as well as its cognate E2-conjugating enzyme. Moreover, inhibiting TSG101 engagement by Gag induces a highly characteristic “late-budding” block, whereby the lipid envelope of nascent particles remains continuous with the host cell membrane, a phenotype that is not typically observed when HIV-1-infected cells are treated with type I IFN (Smith and others 1991; Göttlinger and others 1993; Neil and others 2006; Neil and others 2007). Hints that other “classical” IFN-induced genes might have anti-HIV-1 activity derive from studies of other retroviruses and retroelements, For example, 2′6′ oligoadenylate synthetase and RNAseL have been shown to be able to inhibit the replication of a MLV strain (XMRV) (Dong and others 2007) that was recently found in human prostate cancer tissue (Urisman and others 2006), suggesting the possibility that they may be able to inhibit other retroviruses. Moreover, an interesting recent study identified the ER-associated Trex1 DNA 3′ exonuclease as being required for the degradation of cytoplasmic dsDNA (Stetson and others 2008). In its absence, dsDNA triggers type I IFN release, and mice and human with lesions in Trex1 develop lethal autoimmunity that is dependent on type I IFN. Trex1-deficient cells accumulate cytoplasmic DNAs derived from endogenous retroelements, and Trex1 overexpression suppresses retrotransposition (Stetson and others 2008). These findings raise the intriguing possibility that exogenous retroviral reverse transcripts might be removed by Trex1 or similar factors. However, 1 recent report indicates that the SET-1 complex, of which TREX is a component, actually facilitates HIV-1 infection by inhibiting autointegration (Yan and others 2009).
Possible Roles for HIV-1 Accessory Genes in Antagonizing Novel Restriction Factors or Attenuating the IFN Response
TRIM5, APOBEC3, and tetherin proteins are currently the most intensively studied of the restriction factors, primarily because there is clear evidence that specific adaptations have occurred in HIV-1 and its relatives to evade or antagonize these host defenses. However, overexpression or underexpression of a number of cellular genes can inhibit and enhance retrovirus replication, respectively (Goff 2007). Moreover, there are strong hints that other primate lentivirus accessory genes might have evolved activities to counteract additional, as yet unidentified, restriction factors or to attenuate the type I IFN response.
Inhibition of interferon and/or Toll-like receptor signaling through IRF, STAT, or NF-κB activation is a common mechanism by which mammalian viruses attempt to evade innate immune responses (Haller and others 2006). Interestingly, proteasomal degradation of STAT1 or 2 by the V proteins of mammalian paramyxoviruses requires their recruitment to a Cullin4 E3–ubiquitin ligase complex via DDB1 (Barry and Früh 2006). This same complex is engaged by HIV-1 Vpr and SIVmac Vpx through their binding to DCAF1 (VprBP) (Hrecka and others 2007; Le Rouzic and others 2007; Schröfelbauer and others 2007; Tan and others 2007; Wen and others 2007). Intriguingly, Vpx, carried in virion particles, is able to suppress the resistance of human dendritic cells or macrophages to HIV-1 and SIVmac/HIV-2 infection and this property requires engagement of the ubiqutin ligase complex (Goujon and others 2007; Goujon and others 2008; Sharova and others 2008). We as yet do not know what “substrates” are targeted to the Cul4 ubiqutin ligase complex by Vpr/Vpx, but it is likely to be a cellular factor that has, or that mobilizes, an antiretroviral activity. It is also tempting to speculate that this accessory gene might modulate IFN-induced innate immunity through this mechanism, although this has not been demonstrated. Additionally, another HIV-1 accessory protein, Vpu binds the ubiquitin ligase adaptor β-TRCP, and in so doing, has been reported to inhibit degradation of phosphorylated IκB-α, preventing NF-κB liberation (Bour and others 2001). Thus, it too, could have a role in blunting the effects of IFNs on infected cells.
Antiretroviral Genes and the Treatment of Human Disease
Retrovirus restriction factors and their antagonists are intriguing in their own right, both because they function in biologically novel and unique ways, and because they illuminate how hosts and viruses have coevolved. Additionally, our understanding of some restriction factors, particularly the APOBEC3 and TRIM5 proteins, has progressed to the point that knowledge of them might be usefully applied to translational problems. One could envisage, for example that small molecules that disrupt the Vif:APOBEC3G interaction, or the putative Vpu:tetherin interaction, could be clinically useful inhibitors of HIV-1 replication. Indeed in a recent study, a compound that can block Vif-induced APOBEC3G degradation has been identified and shows antiviral activity against Vif+ HIV-1 replication in T-cell lines (Nathans and others 2008). Similarly, it might be possible to derive small molecules that promote TRIM5α–capsid interactions, or molecules that directly bind to capsid and mimic the apparent effects of TRIM5 on capsid (in)stability.
In addition, the rapid evolution and consequent wide divergence in primate restriction factor genes have impacted human disease and its investigation—on the one hand, the transmission of potentially pathogenic retroviruses from primates to humans has likely been limited by restriction factor divergence. On the other hand, the inability of HIV-1 to infect nonhuman primate species as a consequence of restriction factor activity has proved a significant impediment to AIDS research. Nonetheless, understanding the basis for species tropism of HIV-1 and its simian relatives has allowed the engineering of minimally modified HIV-1 strains that can replicate in macaque cells and, thus, an animal model of HIV-1 infection appears feasible. Thus, the discovery and characterization of natural antiretroviral factors have led to fascinating science, as well as possible translation applications.
Acknowledgments
S.N. is supported by a Wellcome Trust Research Career Development Fellowship WT082274MA. P.D.B. is supported by NIH Grants R01AI064003 and R01AI050111 and is an investigator of the Howard Hughes Medical Institute.
References
- Agy MB. Acker RL. Sherbert CH. Katze MG. Interferon treatment inhibits virus replication in HIV-1- and SIV-infected CD4+ T-cell lines by distinct mechanisms: evidence for decreased stability and aberrant processing of HIV-1 proteins. Virology. 1995;214(2):379–386. doi: 10.1006/viro.1995.0047. [DOI] [PubMed] [Google Scholar]
- Argyris EG. Acheampong E. Wang F. Huang J. Chen K. Mukhtar M. Zhang H. The interferon-induced expression of APOBEC3G in human blood-brain barrier exerts a potent intrinsic immunity to block HIV-1 entry to central nervous system. Virology. 2007;367(2):440–451. doi: 10.1016/j.virol.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka K. Ikeda K. Hishinuma T. Horie-Inoue K. Takeda S. Inoue S. A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem Biophys Res Commun. 2005;338(4):1950–1956. doi: 10.1016/j.bbrc.2005.10.173. [DOI] [PubMed] [Google Scholar]
- Barr SD. Smiley JR. Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4(2):e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M. Früh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE. 2006;2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- Bartee E. McCormack A. Früh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2(10):e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beignon AS. McKenna K. Skoberne M. Manches O. DaSilva I. Kavanagh DG. Larsson M. Gorelick RJ. Lifson JD. Bhardwaj N. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest. 2005;115(11):3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L. Sebastian S. Sokolskaja E. Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc Natl Acad Sci USA. 2005;102(41):14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnier C. Takeuchi Y. Towers G. Restriction of lentivirus in monkeys. Proc Natl Acad Sci USA. 2002;99(18):11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S. Le Tissier P. Towers G. Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382(6594):826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5(11):1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Bishop KN. Holmes RK. Sheehy AM. Davidson NO. Cho SJ. Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14(15):1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bishop KN. Verma M. Kim EY. Wolinsky SM. Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas P. Poli G. Kinter AL. Justement JS. Stanley SK. Maury WJ. Bressler P. Orenstein JM. Fauci AS. Interferon gamma induces the expression of human immunodeficiency virus in persistently infected promonocytic cells (U1) and redirects the production of virions to intracytoplasmic vacuoles in phorbol myristate acetate-differentiated U1 cells. J Exp Med. 1992;176(3):739–750. doi: 10.1084/jem.176.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL. Giurisato E. Cella M. Schreiber RD. Shaw AS. Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177(5):3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- Bogerd HP. Wiegand HL. Hulme AE. Garcia-Perez JL. O'Shea KS. Moran JV. Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA. 2006;103(23):8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S. Perrin C. Akari H. Strebel K. The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP-mediated degradation of Ikappa B. J Biol Chem. 2001;276(19):15920–15928. doi: 10.1074/jbc.M010533200. [DOI] [PubMed] [Google Scholar]
- Brennan G. Kozyrev Y. Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105(9):3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM. Perez O. Anderson JL. Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J Cell Biol. 2008;180(3):549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthagena L. Bergamaschi A. Luna JM. David A. Uchil PD. Margottin-Goguet F. Mothes W. Hazan U. Transy C. Pancino G. Nisole S. Human TRIM gene expression in response to interferons. PLoS ONE. 2009;4(3):e4894. doi: 10.1371/journal.pone.0004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthagena L. Parise MC. Ringeard M. Chelbi-Alix MK. Hazan U. Nisole S. Implication of TRIM alpha and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology. 2008;5:59. doi: 10.1186/1742-4690-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. Cesarman E. Pessin MS. Lee F. Culpepper J. Knowles DM. Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chen K. Huang J. Zhang C. Huang S. Nunnari G. Wang FX. Tong X. Gao L. Nikisher K. Zhang H. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006a;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Lilley CE. Yu Q. Lee DV. Chou J. Narvaiza I. Landau NR. Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006b;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chiu YL. Soros VB. Kreisberg JF. Stopak K. Yonemoto W. Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435(7038):108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Chiu YL. Witkowska HE. Hall SC. Santiago M. Soros VB. Esnault C. Heidmann T. Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci USA. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG. Harris RS. Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13(22):2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Cowan S. Hatziioannou T. Cunningham T. Muesing MA. Gottlinger HG. Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci USA. 2002;99(18):11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L. Jolicoeur P. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J Virol. 1983;48(3):685–696. doi: 10.1128/jvi.48.3.685-696.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F. Kar A. Lee M. Stremlau M. Poeschla E. Sodroski J. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007;369(2):400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl WE. Stansell E. Kaiser SM. Emerman M. Hunter E. Identification of postentry restrictions to Mason-Pfizer monkey virus infection in New World monkey cells. J Virol. 2008;82(22):11140–11151. doi: 10.1128/JVI.00269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle BP. Schäfer A. Wiegand HL. Bogerd HP. Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J Virol. 2005;79(13):8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B. Kim S. Hong S. Das Gupta J. Malathi K. Klein EA. Ganem D. Derisi JL. Chow SA. Silverman RH. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci USA. 2007;104(5):1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis NC. Sheehy AM. Ahmad KM. Swanson CM. Bishop KN. Beer BE. Marx PA. Gao F. Bibollet-Ruche F. Hahn BH. Malim MH. Further investigation of simian immunodeficiency virus Vif function in human cells. J Virol. 2004;78(21):12041–12046. doi: 10.1128/JVI.78.21.12041-12046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5(4):253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Goffinet C. Allespach I. Homann S. Tervo HM. Habermann A. Rupp D. Oberbremer L. Kern C. Tibroni N. Welsch S. Krijnse-Locker J. Banting G. Kräusslich HG. Fackler OT. Keppler OT. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5(3):285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Göttlinger HG. Dorfman T. Cohen EA. Haseltine WA. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci USA. 1993;90(15):7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlinger HG. Dorfman T. Sodroski JG. Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88(8):3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C. Arfi V. Pertel T. Luban J. Lienard J. Rigal D. Darlix JL. Cimarelli A. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J Virol. 2008;82(24):12335–12345. doi: 10.1128/JVI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C. Rivière L. Jarrosson-Wuilleme L. Bernaud J. Rigal D. Darlix JL. Cimarelli A. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology. 2007;4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O. Kochs G. Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344(1):119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. Bishop KN. Sheehy AM. Craig HM. Petersen-Mahrt SK. Watt IN. Neuberger MS. Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Harris RS. Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4(11):868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Hasenkrug KJ. Valenzuela A. Letts VA. Nishio J. Chesebro B. Frankel WN. Chromosome mapping of Rfv3, a host resistance gene to Friend murine retrovirus. J Virol. 1995;69(4):2617–2620. doi: 10.1128/jvi.69.4.2617-2620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T. Ambrose Z. Chung NP. Piatak M. Yuan F. Trubey CM. Coalter V. Kiser R. Schneider D. Smedley J. Pung R. Gathuka M. Estes JD. Veazey RS. KewalRamani VN. Lifson JD. Bieniasz PD. A macaque model of HIV-1 infection. Proc Natl Acad Sci USA. 2009;106(11):4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T. Cowan S. Goff SP. Bieniasz PD. Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22(3):385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T. Perez-Caballero D. Yang A. Cowan S. Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci USA. 2004;101(29):10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T. Princiotta M. Piatak M. Yuan F. Zhang F. Lifson JD. Bieniasz PD. Generation of simian-tropic HIV-1 by restriction factor evasion. Science. 2006;314(5796):95. doi: 10.1126/science.1130994. [DOI] [PubMed] [Google Scholar]
- Hirsch VM. Sharkey ME. Brown CR. Brichacek B. Goldstein S. Wakefield J. Byrum R. Elkins WR. Hahn BH. Lifson JD. Stevenson M. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4(12):1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RK. Koning FA. Bishop KN. Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282(4):2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- Hrecka K. Gierszewska M. Srivastava S. Kozaczkiewicz L. Swanson SK. Florens L. Washburn MP. Skowronski J. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc Natl Acad Sci USA. 2007;104(28):11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE. Bogerd HP. Cullen BR. Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390(1–2):199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H. Laigret F. Martin MA. Repaske R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol. 1985;55(3):768–777. doi: 10.1128/jvi.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H. Diaz-Griffero F. Stremlau M. Si Z. Sodroski J. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;280(29):26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- Jouvenet N. Neil SJ. Zhadina M. Zang T. Kratovac Z. Lee Y. McNatt M. Hatziioannou T. Bieniasz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83(4):1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SM. Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80(2):875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL. Francica JR. Agrawal-Gamse C. Bates P. Tetherinmediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci USA. 2009;106(8):2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K. Igarashi T. Martin MA. Khamsri B. Hatcho K. Yamashita T. Fujita M. Uchiyama T. Adachi A. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc Natl Acad Sci USA. 2006;103(45):16959–16964. doi: 10.1073/pnas.0608289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata M. Nagaoka Y. Chen IS. Reassessing the role of APOBEC3G in human immunodeficiency virus type 1 infection of quiescent CD4+ T-cells. PLoS Pathog. 2009;5(3):e1000342. doi: 10.1371/journal.ppat.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z. Ylinen LM. Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci USA. 2004;101(29):10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z. Ylinen LM. Towers GJ. Cyclophilin A renders human immunodeficiency virus type 1 sensitive to Old World monkey but not human TRIM5 alpha antiviral activity. J Virol. 2006;80(10):4683–4690. doi: 10.1128/JVI.80.10.4683-4690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW. Ringler DJ. Mori K. Panicali DL. Sehgal PK. Daniel MD. Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Kornbluth RS. Oh PS. Munis JR. Cleveland PH. Richman DD. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med. 1989;169(3):1137–1151. doi: 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth MJ. Taylor MD. Katze MG. Interferon inhibits the replication of HIV-1, SIV, and SHIV chimeric viruses by distinct mechanisms. Virology. 1998;247(2):265–273. doi: 10.1006/viro.1998.9249. [DOI] [PubMed] [Google Scholar]
- Kupzig S. Korolchuk V. Rollason R. Sugden A. Wilde A. Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4(10):694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- Lai C. Struckhoff JJ. Schneider J. Martinez-Sobrido L. Wolff T. García-Sastre A. Zhang DE. Lenschow DJ. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol. 2009;83(2):1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HC. Davey V. Kovacs JA. Feinberg J. Metcalf JA. Herpin B. Walker R. Deyton L. Davey RT. Falloon J. Interferon-alpha in patients with asymptomatic human immunodeficiency virus (HIV) infection. A randomized, placebo-controlled trial. Ann Intern Med. 1990;112(11):805–811. doi: 10.7326/0003-4819-112-11-805. [DOI] [PubMed] [Google Scholar]
- Lane HC. Feinberg J. Davey V. Deyton L. Baseler M. Manischewitz J. Masur H. Kovacs JA. Herpin B. Walker R. Metcalf JA. Salzman N. Quinnan G. Faucia AS. Anti-retroviral effects of interferon-alpha in AIDS-associated Kaposi's sarcoma. Lancet. 1988;2(8622):1218–1222. doi: 10.1016/s0140-6736(88)90811-2. [DOI] [PubMed] [Google Scholar]
- Le Rouzic E. Belaïdouni N. Estrabaud E. Morel M. Rain JC. Transy C. Margottin-Goguet F. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. 2007;6(2):182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- Lecossier D. Bouchonnet F. Clavel F. Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300(5622):1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lenschow DJ. Lai C. Frias-Staheli N. Giannakopoulos NV. Lutz A. Wolff T. Osiak A. Levine B. Schmidt RE. García-Sastre A. Leib DA. Pekosz A. Knobeloch KP. Horak I. Virgin HW. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104(4):1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CH. Kuang YQ. Liu HL. Zheng YT. Su B. A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS. 2007;21(Suppl. 8):S19–S26. doi: 10.1097/01.aids.0000304692.09143.1b. [DOI] [PubMed] [Google Scholar]
- Lilly F. Pincus T. Genetic control of murine viral leukemogenesis. Adv Cancer Res. 1973;17:231–277. [Google Scholar]
- Luo K. Wang T. Liu B. Tian C. Xiao Z. Kappes J. Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol. 2007;81(13):7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N. Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72(12):10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B. Turelli P. Caron G. Friedli M. Perrin L. Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mariani R. Chen D. Schröfelbauer B. Navarro F. König R. Bollman B. Münk C. Nymark-McMahon H. Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114(1):21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Marin M. Rose KM. Kozak SL. Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9(11):1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mbisa JL. Barr R. Thomas JA. Vandegraaff N. Dorweiler IJ. Svarovskaia ES. Brown WL. Mansky LM. Gorelick RJ. Harris RS. Engelman A. Pathak VK. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81(13):7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW. Zang T. Hatziioannou T. Bartlett M. Fofana IB. Johnson WE. Neil SJ. Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5(2):e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E. Andrew AJ. Kao S. Strebel K. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc Natl Acad Sci USA. 2009;106(8):2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E. Opi S. Takeuchi H. Khan M. Goila-Gaur R. Kao S. Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81(24):13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C. Brandt SM. Lucero G. Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci USA. 2002;99(21):13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R. Cao H. Sharova N. Ali A. Sharkey M. Stranska R. Stevenson M. Rana TM. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26(10):1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ. Eastman SW. Jouvenet N. Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2(5):e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ. Sandrin V. Sundquist WI. Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2(3):193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ. Zang T. Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Newman RM. Hall L. Connole M. Chen GL. Sato S. Yuste E. Diehl W. Hunter E. Kaur A. Miller GM. Johnson WE. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci USA. 2006;103(50):19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RM. Hall L. Kirmaier A. Pozzi LA. Pery E. Farzan M. O'Neil SP. Johnson W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4(2):e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EN. Holmes RK. Craig HM. Klein KC. Lingappa JR. Malim MH. Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15(2):166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nisole S. Lynch C. Stoye JP. Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101(36):13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M. Kerns JA. Malik HS. Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80(8):3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM. Lovsin N. Peterlin BM. Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445(7130):927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- Okumura A. Lu G. Pitha-Rowe I. Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103(5):1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CM. Yang PC. Göttlinger H. Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J Virol. 2003;77(1):726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G. Lei KJ. Jin W. Greenwell-Wild T. Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203(1):41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D. Hatziioannou T. Yang A. Cowan S. Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005a;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D. Hatziioannou T. Zhang F. Cowan S. Bieniasz PD. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol. 2005b;79(24):15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ. Stremlau M. Sodroski J. Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J Virol. 2006;80(11):5631–5636. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ. Stremlau M. Song B. Ulm W. Mulligan RC. Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101(32):11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G. Orenstein JM. Kinter A. Folks TM. Fauci AS. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science. 1989;244(4904):575–577. doi: 10.1126/science.2470148. [DOI] [PubMed] [Google Scholar]
- Sakuma R. Mael AA. Ikeda Y. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J Virol. 2007;81(18):10201–10206. doi: 10.1128/JVI.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T. Noda T. Urata S. Kawaoka Y. Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J Virol. 2009;83(5):2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML. Montano M. Benitez R. Messer RJ. Yonemoto W. Chesebro B. Hasenkrug KJ. Greene WC. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321(5894):1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL. Emerman M. Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2(9):E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL. Wu LI. Emerman M. Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102(8):2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM. Sokolskaja E. Berthoux L. Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Schäfer A. Bogerd HP. Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328(2):163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schröfelbauer B. Hakata Y. Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc Natl Acad Sci USA. 2007;104(10):4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröfelbauer B. Yu Q. Zeitlin SG. Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol. 2005;79(17):10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ. Haché G. Macduff DA. Brown WL. Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82(6):2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S. Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova N. Wu Y. Zhu X. Stranska R. Kaushik R. Sharkey M. Stevenson M. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 2008;4(5):e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM. Gaddis NC. Choi JD. Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM. Gaddis NC. Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9(11):1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shirazi Y. Pitha PM. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J Virol. 1992;66(3):1321–1328. doi: 10.1128/jvi.66.3.1321-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JH. Gaddis NC. Fouchier RA. Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998a;4(12):1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- Simon JH. Miller DL. Fouchier RA. Soares MA. Peden KW. Malim MH. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 1998b;17(5):1259–1267. doi: 10.1093/emboj/17.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MS. Thresher RJ. Pagano JS. Inhibition of human immunodeficiency virus type 1 morphogenesis in T cells by alpha interferon. Antimicrob Agents Chemother. 1991;35(1):62–67. doi: 10.1128/aac.35.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. Gold B. O'Huigin C. Javanbakht H. Li X. Stremlau M. Winkler C. Dean M. Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005;79(10):6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey AR. Norris PJ. Qin L. Haygreen EA. Taylor E. Heitman J. Lebedeva M. DeCamp A. Li D. Grove D. Self SG. Borrow P. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83(8):3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD. Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stetson DB. Ko JS. Heidmann T. Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB. Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stremlau M. Owens CM. Perron MJ. Kiessling M. Autissier P. Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M. Perron M. Lee M. Li Y. Song B. Javanbakht H. Diaz-Griffero F. Anderson DJ. Sundquist WI. Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006a;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M. Perron M. Welikala S. Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79(5):3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M. Song B. Javanbakht H. Perron M. Sodroski J. Cyclophilin A: an auxiliary but not necessary cofactor for TRIM5alpha restriction of HIV-1. Virology. 2006b;351(1):112–120. doi: 10.1016/j.virol.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Svarovskaia ES. Xu H. Mbisa JL. Barr R. Gorelick RJ. Ono A. Freed EO. Hu WS. Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279(34):35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Takeda E. Tsuji-Kawahara S. Sakamoto M. Langlois MA. Neuberger MS. Rada C. Miyazawa M. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J Virol. 2008;82(22):10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. Ehrlich E. Yu XF. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J Virol. 2007;81(19):10822–10830. doi: 10.1128/JVI.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD. Korth MJ. Katze MG. Interferon treatment inhibits the replication of simian immunodeficiency virus at an early stage: evidence for a block between attachment and reverse transcription. Virology. 1998;241(1):156–162. doi: 10.1006/viro.1997.8964. [DOI] [PubMed] [Google Scholar]
- Towers G. Bock M. Martin S. Takeuchi Y. Stoye JP. Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci USA. 2000;97(22):12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ. Hatziioannou T. Cowan S. Goff SP. Luban J. Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9(9):1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- Uchil PD. Quinlan BD. Chan WT. Luna JM. Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4(2):e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman A. Molinaro RJ. Fischer N. Plummer SJ. Casey G. Klein EA. Malathi K. Magi-Galluzzi C. Tubbs RR. Ganem D. Silverman RH. DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2(3):e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Van Damme N. Goff D. Katsura C. Jorgenson RL. Mitchell R. Johnson MC. Stephens EB. Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V. Smith RM. Bour SP. Strebel K. Spearman P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc Natl Acad Sci USA. 2003;100(25):15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA. Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J Virol. 2007;81(24):13932–13937. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA. Kratovac Z. Bieniasz PD. Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105(9):3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volberding P. Valero R. Rothman J. Gee G. Alpha interferon therapy of Kaposi's sarcoma in AIDS. Ann NY Acad Sci. 1984;437:439–446. doi: 10.1111/j.1749-6632.1984.tb37165.x. [DOI] [PubMed] [Google Scholar]
- von Sydow M. Sönnerborg A. Gaines H. Strannegård O. Interferon-alpha and tumor necrosis factor-alpha in serum of patients in various stages of HIV-1 infection. AIDS Res Hum Retroviruses. 1991;7(4):375–380. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- Wang FX. Huang J. Zhang H. Ma X. Zhang H. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008;89(Pt 3):722–730. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- Wang T. Tian C. Zhang W. Luo K. Sarkis PT. Yu L. Liu B. Yu Y. Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81(23):13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X. Duus KM. Friedrich TD. de Noronha CM. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J Biol Chem. 2007;282(37):27046–27057. doi: 10.1074/jbc.M703955200. [DOI] [PubMed] [Google Scholar]
- Wiegand HL. Doehle BP. Bogerd HP. Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23(12):2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SJ. Webb BL. Ylinen LM. Verschoor E. Heeney JL. Towers GJ. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105(9):3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D. Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131(1):46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Wolf D. Goff SP. Host restriction factors blocking retroviral replication. Annu Rev Genet. 2008;42:143–163. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D. Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Anderson JL. Campbell EM. Joseph AM. Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103(19):7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. Chertova E. Chen J. Ott DE. Roser JD. Hu WS. Pathak VK. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360(2):247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Yan N. Cherepanov P. Daigle JE. Engelman A. Lieberman J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 2009;5(3):e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW. Lindemann D. Stanke N. Reh J. Westphal D. Hanenberg H. Ohkura S. Stoye JP. Restriction of foamy viruses by primate Trim5alpha. J Virol. 2008;82(11):5429–5439. doi: 10.1128/JVI.02462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW. Nisole S. Lynch C. Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101(29):10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW. Nisole S. Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Ylinen LM. Keckesova Z. Wilson SJ. Ranasinghe S. Towers GJ. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J Virol. 2005;79(18):11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q. Chen D. König R. Mariani R. Unutmaz D. Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279(51):53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Yu X. Yu Y. Liu B. Luo K. Kong W. Mao P. Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302(5647):1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zennou V. Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349(1):31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zennou V. Perez-Caballero D. Göttlinger H. Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78(21):12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Hatziioannou T. Perez-Caballero D. Derse D. Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353(2):396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Zhang H. Yang B. Pomerantz RJ. Zhang C. Arunachalam SC. Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424(6944):94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]