Abstract
Menaquinone is an essential component of the electron transport chain in many pathogens and consequently enzymes in the menaquinone biosynthesis pathway are potential drug targets for the development of novel antibacterial agents. In order to identify leads that target MenB, the 1,4-dihydroxy-2-naphthoyl-CoA synthase from Mycobacterium tuberculosis, a high throughput screen was performed. Several 1,4-benzoxazines were identified in this screen and subsequent SAR studies resulted in the discovery of compounds with excellent antibacterial activity against M. tuberculosis H37Rv with MIC values as low as 0.6 µg/ml. The 1,4-benzoxazine scaffold is thus a promising foundation for the development of antitubercular agents.
The emergence of multi-drug resistant (MDR-TB) and extensively-drug resistant (XDR-TB) strains of M. tuberculosis represents a severe threat to human health. Consequently, novel drugs are needed that are active against drug resistant bacteria and that also shorten the course of chemotherapy. In addition, novel agents are also required that are active against latent, non-replicating populations of bacteria since M. tuberculosis can persist in this state for many years.1–2 Since it seems likely that latent M. tuberculosis bacteria must respire in order to remain viable, compounds that target respiration are promising candidates for the development of drugs that are active against both replicating and non-replicating bacterial populations. Currently we are pursuing the hypothesis that menaquinone biosynthesis is essential for bacterial viability in vivo, and are actively engaged in identifying compounds that inhibit enzymes in this pathway.
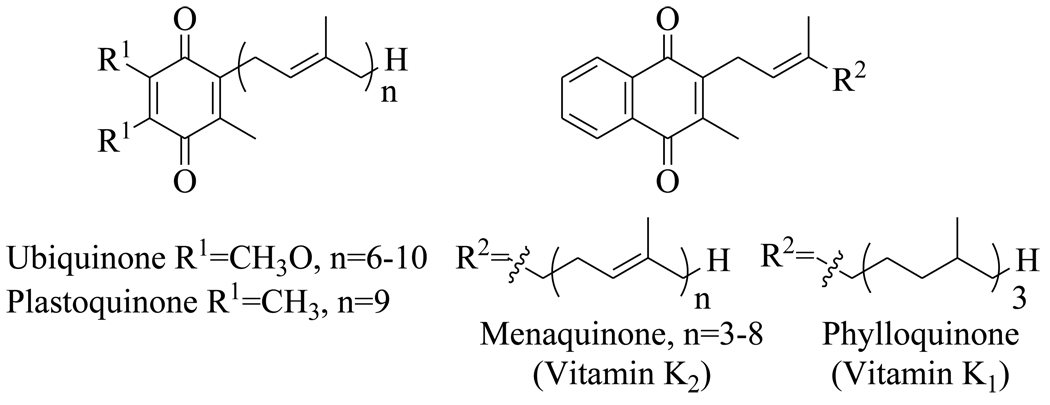
Menaquinone (vitamin K1) (Figure 1) is a polyisoprenylated naphthoquinone that shuttles electrons between membrane-bound protein complexes in the electron transport chain. In mammalian cells this function is performed by ubiquinone (Figure 1), and although menaquinone is required for blood clotting, the biosynthetic pathway for this vitamin is absent in humans. This has resulted in the proposal that enzymes involved in menaquinone biosynthesis may be promising targets for drug discovery.
Figure 1.
Structures of the benzoquinone and naphthoquinone redox cofactors.
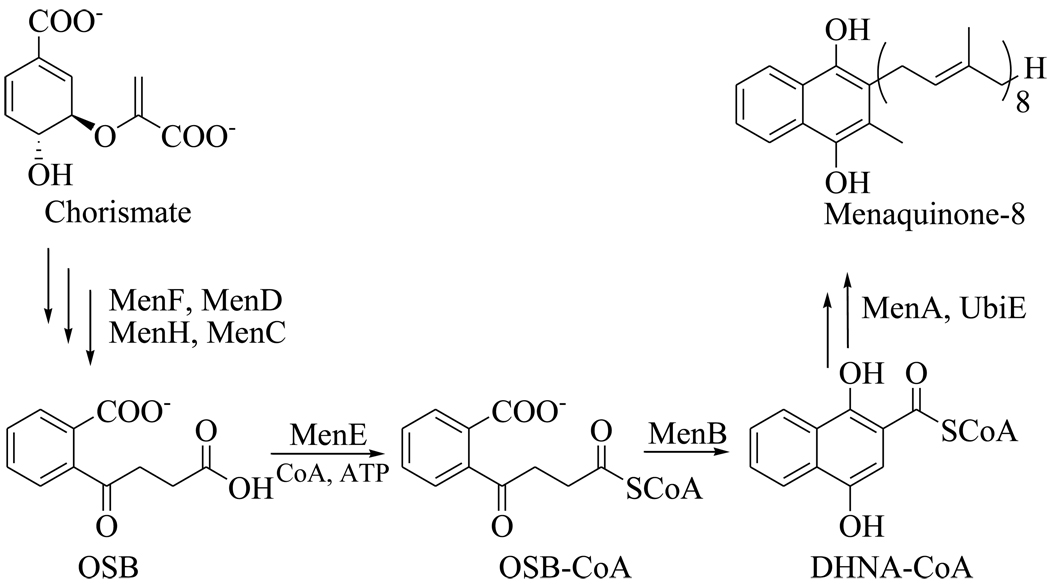
The biosynthesis of menaquinone in M. tuberculosis is thought to mirror the pathway found in organisms such as Escherichia coli, Bacillus subtilis and Mycobacterium phlei (Figure 2),3–5 where it is known that the men genes are essential for survival.6 Previous inhibitor discovery efforts have focused on the o-succinylbenzoate synthase from E. coli and Bacillus anthracis (MenE),7–8 while Crick and coworkers have demonstrated the antibacterial activity of inhibitors of the polyprenyl transferase (MenA) from M. tuberculosis,9 validating this pathway as a target for tuberculosis drug discovery.
Figure 2.
The menaquinone biosynthesis pathway in E. coli showing the reactions catalyzed by MenE and MenB.
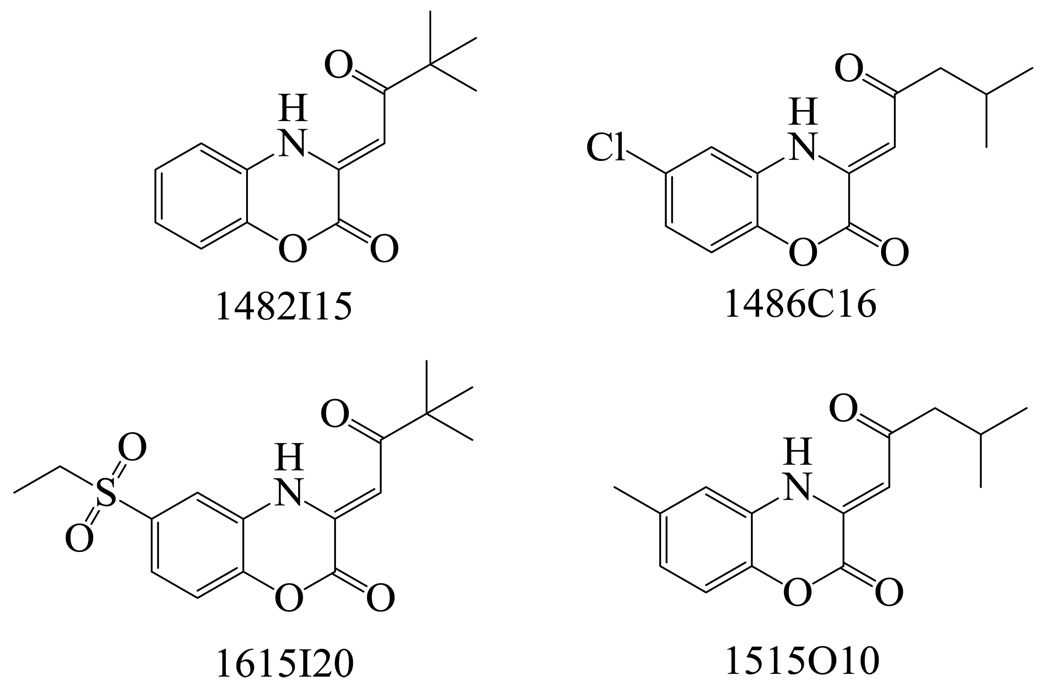
MenB, the 1,4-dihydroxy-2-naphthoyl-CoA (DHNA-CoA) synthase catalyzes an intramolecular Claisen condensation leading to the formation of DHNA from o-succinylbenzoate (Figure 2).5 In order to provide a foundation for the discovery of compounds that target MenB, we used a coupled assay10 to screen 105,091 small drug-like molecules that contained a large variety of chemical structures. Following the primary screen, in which compounds were tested at a single concentration of 125 µg/ml, we obtained 455 hits that had at least 30% enzyme inhibition relative to control (data not shown). Within these hits, we identified 4 compounds based on the 1,4-benzoxazine scaffold (Figure 3, Table 1). Since a coupled assay was used for screening, the ability of each compound to inhibit MenE was also evaluated by directly monitoring the formation of PPi.11 However, none of the cherry-picked compounds affected MenE activity at a concentration of 50 µM.
Figure 3.
Chemical structures of hits from the HTS that have the common 1,4-benzoxazine backbone.
Table 1.
Enzyme inhibition data for 1,4-benzoxazines identified in the high-throughput screen.a
| Compound | % inhibition of MenB at 125 µg/ml |
|---|---|
| 1482I15 | 68.4±7.3 |
| 1486C16 | 98.0±1.2 |
| 1615I20 | 60.3±10.3 |
| 1515O10 | 77.3±5.2 |
MenB concentration was 150 nM and OSB concentration was 30 µM.
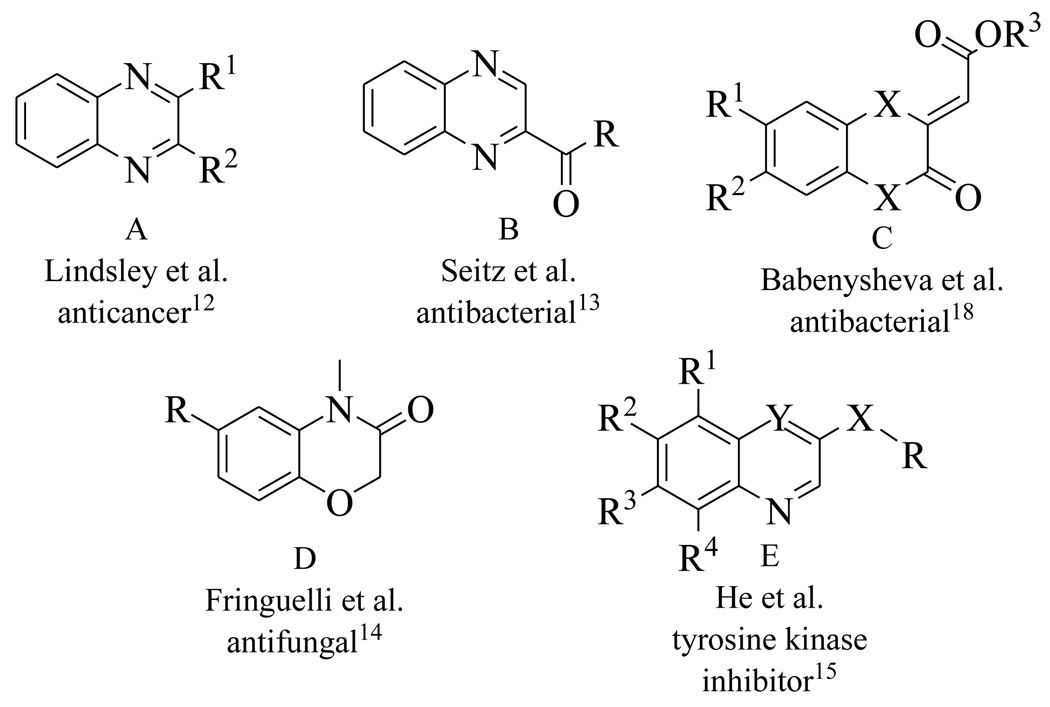
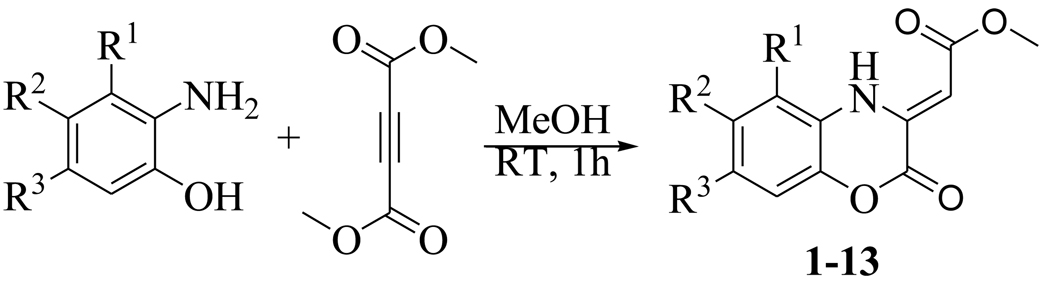
Benzoxazines and quinoxalines (Figure 4) are privileged ring systems that are found in a broad range of biologically active molecules with anticancer,12 antibacterial,13 and antifungal activity,14 and also serve as scaffolds for kinase inhibitors.15–16 In order to explore the utility of this ring system as a starting point for developing MenB inhibitors, we synthesized 13 compounds as shown in Scheme 1. In this procedure, the keto group is replaced by an ester for convenience and the 1,4-benzoxazines are generated by mixing commercially available 2-aminophenols with dimethyl but-2-ynedioate. After 1 h at room temperature the product precipitates and is recrystallized from MeOH/EtOAc.
Figure 4.
Representative structures of existing benzoxazines and quinoxalines.
Scheme 1.
Synthetic route for the (Z)-methyl 2-(2-oxo-2H-benzo[b][1,4]oxazin-3(4H)ylidene)acetates.
The synthesized compounds were evaluated for their ability to inhibit the reaction catalyzed by MenB as well as the growth of M. tuberculosis H37Rv (Table 2). Several compounds had MIC values of less than 1 µg/ml in which there was either no substituent on the benzoxazine core (1), or in which R2 = F (4) or R3 = F or Cl (9, 10). However introduction of a methyl group (2, 3, 8) resulted in a greater than 100-fold reduction in antibacterial activity. In addition, for substituents at R2 and R3, introduction of larger electron withdrawing groups (R2 = Cl (5), R2 = NO2 (6), R2 = EtSO2 (7), R3 = NO2 (11)) also caused a dramatic reduction in antibacterial activity. Since the methyl group is electron donating, we speculate that the effect on activity results primarily from the size of the substituent and not from electronic effects. Interestingly, a series of benzoxazines bearing an ethylsulfonyl group (similar to compound 7), had poor antibacterial activity against E. coli and S. aureus,17 raising the possibility that a reduction in substituent size in the latter case might lead to an increase in compound activity. The data in Table 2 also demonstrate that the benzoxazine core is essential for antibacterial activity as well as enzyme inhibition, since compounds with quinoxaline (12) or benzothiozine (13) cores had greatly reduced MIC and IC50 values compared to the parent compounds. This result is in agreement with studies by Reynolds and coworkers who observed moderate MIC values (6.25 µg/ml) against H37Rv for a series of quinoxalines. These compounds also had poor activity against M. tuberculosis H37Ra and M. avium (> 64 µg/ml),13 while in separate studies it was also shown that quinoxalines had low activity against both E. coli and S. aureus.18
Table 2.
In vitro activity of the (Z)-methyl 2-(2-oxo-2H-benzo[b][1,4]oxazin-3(4H)ylidene)acetatesa
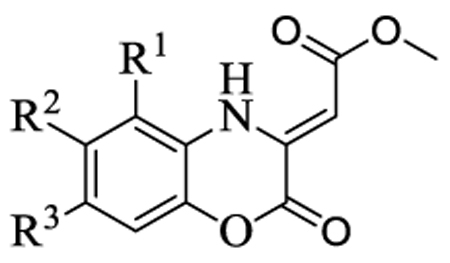 | |||||||
|---|---|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | IC50b (µM) |
Ki (µM) |
Ki’ (µM) |
MICc (µg/ml) |
| 1 | H | H | H | 10.0±1.0 | 9.1±1.2 | 67.0±7.9 | 0.64 |
| 2 | Me | H | H | 24.1±1.8 | 25 | ||
| 3 | H | Me | H | 23.1±1.0 | >100 | ||
| 4 | H | F | H | 27.0±3.0 | 11.5±1.5 | 10.1±0.9 | 0.63 |
| 5 | H | Cl | H | 46.3±3.5 | 22.5±1.1 | 18.5±2.6 | 5 |
| 6 | H | NO2 | H | 28.2±4.4 | 50 | ||
| 7 | H | EtSO2 | H | 17.9±3.0 | >100 | ||
| 8 | H | H | Me | 18.2±2.8 | 100 | ||
| 9 | H | H | F | 30.0±3.7 | 0.63 | ||
| 10 | H | H | Cl | 35.7±4.8 | 0.63 | ||
| 11 | H | H | NO2 | 20.3±1.8 | >100 | ||
| 12 | 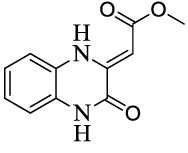 |
>122 | >100 | ||||
| 13 | 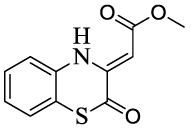 |
>140 | >100 | ||||
MenB concentration was 150 nM.
Compound concentration giving 50% inhibition of enzyme activity.
Compound concentration giving 90% inhibition of M. tuberculosis H37Rv growth.
Although substitution of the benzoxazine core generally had a dramatic effect on the MIC values, the impact of the inhibition of MenB was much less pronounced. Although it can be seen from Table 2 that the benzoxazine core is essential for MenB inhibition, the IC50 values for the benzoxazine analogues only varied by 4–5 fold. These data suggest that other factors such as altered cell permeability and/or evasion of detoxification strategies may also modulate antibacterial activity, together with the possibility that MenB might not be the only target in the cell. To gain further insight into the mechanism of enzyme inhibition, detailed kinetic studies revealed that compounds 1, 4 and 5 are non-competitive inhibitors of MenB with Ki and Ki’ values that were similar to the IC50 values. These compounds can thus bind to the enzyme both before and after the varied substrate (OSB-CoA). Since MenB is a hexamer, it is possible that binding of inhibitor to one subunit affects the ability of substrate to bind to an adjacent monomer. Alternatively, there must be a distinct binding site for the benzoxazines in the MenB monomer separate from the active site, occupation of which perturbs catalysis. X-ray crystallography is currently being used to distinguish between these possibilities.
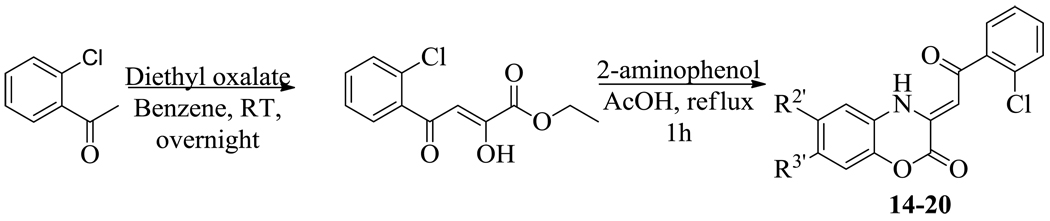
To further explore the antibacterial properties of the 1,4-benzoxazines, we therefore synthesized a second series of compounds in which the methyl ester was replaced with a substituted phenyl ring (Scheme 2). In this procedure the commercially available 2’-chloroacetophenone was converted to the corresponding (Z)-ethyl 2-hydroxy-4-oxo-4-(2’chlorophenyl)-but-2-enoate by treating with diethyl oxalate at room temperature overnight. The but-2-enoate was then refluxed with a selection of substituted 2-aminophenols in acetic acid for 2 h in order to generate the 1,4-benzoxazine. Interestingly, none of these compounds (14–20) were able to inhibit MenB up to concentrations of 100 µM. Thus, introduction of a bulky group into the side chain significantly perturbs the ability of the 1,4-benzoxazines to inhibit MenB. In addition, the MIC values increased 5-fold for 14 and 16, and 2.5-fold for 17, (Table 3), compared to the analogous compounds in Table 2, suggesting that while a portion of their antibacterial activity could stem from an ability to inhibit MenB, there must be additional targets for these compounds in the cell. Many antibacterial benzoxazines previously reported also have bulky side chains and relatively poor activity against M. tuberculosis19 as well as other bacteria.17, 20 Thus, again we would suggest that activity might be improved in the latter cases by reducing the size of the side chain.
Scheme 2.
Synthetic route for the (Z)-3-(2-aryl-2-oxoethylidene)-3,4-dihydro-2H-benzo[b][1,4]oxazin-2-ones.
Table 3.
In vitro activity of the (Z)-3-(2-aryl-2-oxoethylidene)-3,4-dihydro-2H-benzo[b][1,4]oxazin-2-ones
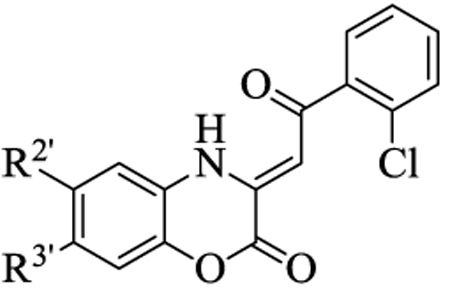 | |||
|---|---|---|---|
| Compound | R2’ | R3’ | MICa (µg/ml) |
| 14 | H | H | 3.13 |
| 15 | Me | H | 100 |
| 16 | F | H | 3.13 |
| 17 | Cl | H | 12.5 |
| 18 | H | Me | >100 |
| 19 | H | F | 1.56 |
| 20 | H | Cl | 3.13 |
Compound concentration giving 90% inhibition of M. tuberculosis H37Rv growth.
The compounds in Table 3 also inhibit the growth of E. coli M17 and S. aureus P-209,20 and the SAR data presented here follows a similar trend to that reported previously in which halogen substituents lead to an increase in antibacterial activity. In addition, the MIC data also show that antibacterial activity is abolished by the introduction of a methyl group into the benzoxazine ring (15 and 18), as was observed for the methyl esters (Table 2, compounds 3 and 8). Thus, while the introduction of a bulky side chain has reduced antibacterial activity, both series of compounds in Tables 2 and 3 likely have a common target(s) in the cell, inhibition of which is very sensitive to methylation at R2 (R2’). Current studies are focused on elucidating the mode of action of the antibacterial benzoxazines and identifying the proteins in bacteria to which they bind.
In summary, we have identified a group of 1,4-benzoxazines with promising in vitro antibacterial activity toward M. tuberculosis H37Rv. These compounds were identified by screening a compound library against the 1,4-dihydroxy-2-naphthoyl-CoA (DHNA-CoA) synthase (MenB) in the M. tuberculosis menaquinone biosynthesis pathway. However the current SAR data suggest that these compounds act by binding to additional targets within the cell. These molecules provide a foundation for the development of novel antimycobacterial agents that may ultimately lead to the discovery of new therapeutics for treating patients with tuberculosis.
Supplementary Material
Acknowledgement
This work was supported in part by National Institutes of Health grants AI044639, AI070383, and AI058785 to PJT. High throughput screening was performed at The National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Diseases (NSRB) with the support of National Institutes of Health grant U54AI057159. We thank members of the NSRB and ICCB-Longwood for their help with compound screening.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data, including procedures for compound synthesis, enzyme assays, and antibacterial activity together with H-NMR data for the synthesized compounds, can be found in the online version at www.sciencedirect.com.
References
- 1.Kochi A. Tubercle. 1991;72:1. doi: 10.1016/0041-3879(91)90017-m. [DOI] [PubMed] [Google Scholar]
- 2.Bloom BR, Murray CJL. Science. 1992;257:1055. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 3.Begley TP, Kinsland C, Taylor S, Tandon M, Nicewonger R, Wu M, Chiu HJ, Kelleher N, Campobasso N, Zhang Y. In: Biosynthesis - Polyketides and Vitamins. Topics in Current Chemistry. Leeper FJ, Vederas JC, editors. Vol. 195. Berlin: Springer-Verlag; 1998. p. 93. [Google Scholar]
- 4.Meganathan R. Vitam. Horm. 2001;61:173. doi: 10.1016/s0083-6729(01)61006-9. [DOI] [PubMed] [Google Scholar]
- 5.Truglio JJ, Theis K, Feng Y, Gajda R, Machutta C, Tonge PJ, Kisker C. J. Biol. Chem. 2003;278:42352. doi: 10.1074/jbc.M307399200. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers J, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4678. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu XQ, Zhang HN, Tonge PJ, Tan DS. Bioorg. Med. Chem. Lett. 2008;18:5963. doi: 10.1016/j.bmcl.2008.07.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Suk DH, Cai F, Crich D, Mesecar AD. Biochemistry. 2008;47:12434. doi: 10.1021/bi801311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurosu M, Narayanasamy P, Biswas K, Dhiman R, Crick DC. J. Med. Chem. 2007;50:3973. doi: 10.1021/jm070638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truglio JJ, Theis K, Feng YG, Gajda R, Machutta C, Tonge PJ, Kisker C. J. Biol. Chem. 2003;278:42352. doi: 10.1074/jbc.M307399200. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien WE. Anal. Biochem. 1976;76:423. doi: 10.1016/0003-2697(76)90337-7. [DOI] [PubMed] [Google Scholar]
- 12.Lindsley CW, Zhao ZJ, Leister WH, Robinson RG, Barnett SF, Defeo-Jones D, Jones RE, Hartman GD, Huff JR, Huber HE, Duggan ME. Bioorg. Med. Chem. Lett. 2005;15:761. doi: 10.1016/j.bmcl.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Seitz LE, Suling WJ, Reynolds RC. J. Med. Chem. 2002;45:5604. doi: 10.1021/jm020310n. [DOI] [PubMed] [Google Scholar]
- 14.Fringuelli R, Giacche N, Milanese L, Cenci E, Macchiarulo A, Vecchiarelli A, Costantino G, Schiaffella F. Bioorg. Med. Chem. 2009;17:3838. doi: 10.1016/j.bmc.2009.04.051. [DOI] [PubMed] [Google Scholar]
- 15.He W, Myers MR, Hanney B, Spada AP, Bilder G, Galzcinski H, Amin D, Needle S, Page K, Jayyosi Z, Perrone MH. Bioorg. Med. Chem. Lett. 2003;13:3097. doi: 10.1016/s0960-894x(03)00655-3. [DOI] [PubMed] [Google Scholar]
- 16.La DS, Belzile J, Bready JV, Coxon A, DeMelfi T, Doerr N, Estrada J, Flynn JC, Flynn SR, Graceffa RF, Harriman SP, Larrow JF, Long AM, Martin MW, Morrison MJ, Patel VF, Roveto PM, Wang L, Weiss MM, Whittington DA, Teffera Y, Zhao ZY, Polverino AJ, Harmanget JC. J. Med. Chem. 2008;51:1695. doi: 10.1021/jm701129j. [DOI] [PubMed] [Google Scholar]
- 17.Gein V, Rassudikhina N, Voronina E. Pharm. Chem. J. 2006;40:554. [Google Scholar]
- 18.Babenysheva A, Lisovskaya N, Belevich I, Lisovenko N. Pharm. Chem. J. 2006;40:611. [Google Scholar]
- 19.Gein V, Rassudikhina N, Shepelina N, Vakhrin M, Babushkina E, Voronina E. Pharm. Chem. J. 2008;42:529. [Google Scholar]
- 20.Mashevskaya I, Tolmacheva I, Voronova É, Odegova T, Aleksandrova G, Goleneva A, Kol'tsova S, Maslivets A. Pharm. Chem. J. 2002;36:32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.