Standfirst header
Epigenomics provides the functional context of genome sequence, analogous to the functional anatomy of the human body provided by Vesalius a half millennium ago. Much of what appear to be inconclusive genetic data for common disease could therefore become meaningful in an epigenomic context.
New Year's Eve in 2014 will mark the 500th anniversary of the birth of Andreas van Wesel, commonly known as Vesalius, author of De humani corporis fabrica1, a treatise almost as influential in its time as was Origin of Species over three centuries later. Vesalius pioneered the rigorous study of human anatomy, and introduced experimental observation into medical education, as a rigorous substitute to hearsay). The late Victor McKusick, who helped to create the genome project and mapped the first human autosomal gene, called gene mapping “neo-Vesalian,”2 as it represented foundational mapping of the genome in order to exploit this information for finding genes. Vesalius was more than a mapper, though, as he debunked dogma of both Galen and Aristotle on the anatomy and physiology of blood circulation, showing how the anatomical map meant that blood must flow through the lungs and return to the heart, not just directly between the ventricles. Similarly, the particular order of genes on chromosomes and arrangement of the chromosomes themselves have only recently been found to be intrinsically meaningful, not just as a map. I suggest here that epigenomics, i.e. the genome science of epigenetics, has transformed genome science, by showing us that the organization of the genome is as important for gene function, as Vesalius showed us how the organization of anatomic structures allowed the function of organs. Moreover, the combination of new epigenomic tools with conventional genetics and a new mathematical language for their interface may have as much impact on understanding human disease as did Vesalius' anatomy a half millennium ago.
Epigenomics Provides a Functional Anatomy of the Genome
Epigenomics has helped to reveal several surprising large-scale functional relationships among the genes themselves and the surrounding nongenic DNA, previously hinted at by the beta-globin cluster. One is the generality of large (10's to 100's of kb) genomic regions regulating gene expression. While the beta-globin gene cluster had been studied for decades3, linking progressive chromatin changes to globin gene switching during development4, the generality and size of multigene chromatin domains only emerged with large-scale epigenomic mapping. As increasing numbers of imprinted genes were found, it was discovered that they were organized in gene clusters, often with common regulatory elements such as CTCF binding sites5. With the advent of genome-scale mapping of histone modifications, many large regions of heterochromatin modifications were found, such as specific modifications associated with the inactive X chromosome6. Moreover, large autosomal regions of heterochromatin modification across HOX gene clusters were found to be more highly conserved across species than the underlying DNA sequence, while not being a simple reflection of exonic boundaries7. Thus, epigenomicstudies revealed that the scope of genome that is apparently functional was at least an order of magnitude greater than that suspected from the sequence alone. Epigenomics provided the functional anatomy of the genome that Vesalius gave gross anatomy a half millennium ago.
Another surprising large-scale genomic relationship is frequent intra-chromosomal and inter-chromosomal interactions mediated by chromatin proteins. These were discovered through chromatin capture methods, described in detail elsewhere in this issue 8, designed to preserve chromatin-mediated interactions over long distances. DNA loop structures, mediated by chromatin, surprisingly common and highly dynamic, were found to be associated with function. For example, multiple interleukin genes in the 200 kb mouse TH2 cytokine locus, when transcriptionally active, are folded into numerous loops, anchored by SATB at their bases9. Remarkably, trans-interactions between chromosomes involve some of the same sequences that epigenetically regulate imprinted gene domains, for example the H19 differentially methylated region, and may act through transvection to regulate genes in trans10.
A recent example of large-scale genomic organization mediated by chromatin is the link between long RNAs, heterochromatin modification and gene activity. At the Cold Spring Harbor Genome Biology meeting in 2005, Tom Gingeras asked for a wager on the number of genes that will ultimately be agreed upon, arguing that the nearly 50% of the genome that may be untranslated RNA will be proved functional11. Growing evidence indicates that much of this RNA mediates chromatin structure. For example, antisense RNAs appears to establish heterochromatin in mammalian genes, independent of dicer and the posttranslational miRNA machinery12. These regions may be >100 kb12, affect multiple genes, and involve Argonaut family proteins13. An exciting recent discovery is the role of long intergenic noncoding RNAs (lincRNAs) in establishing heterochromatin. For example, HOTAIR is a lincRNA that retargets PRC2 over HOX domains with profound changes in gene expression relevant to cancer progression14.
Finally, large organized chromatin K(lysine) modifications (or LOCKs) have been shown to organize the genome into very large blocks (100's to 1000's of kb), some of which are differentiation-specific in their location and extent and correspond to lamin-associated domains (LADs)15–17. These very large regions may provide a dynamic mechanism for functional organization of the genome and are altered in cancer15.
Additional clues that many such large-scale epigenetic networks profoundly influence cellular development and genome function come from large-scale mapping studies. For example, CTCF, which mediates H19 imprinting described earlier, appears to play a general role in defining functional gene region boundaries18. Similarly polycomb target genes, thought to be involved in stable gene silencing, may alternate between functionally active and silent states over large gene regions19. That such networks have a general role in organizing the genome functionally is suggested by the identification of chromosome territories with closely approximated gene-rich regions20.
Epigenomics may supersede single-gene epigenetic disease research
Just as epigenomics provides a functional anatomy of the normal genome, genome-scale studies of epigenetic disease are revealing a Pandora’s box of epigenetic pathology. Just as cancer was the vanguard for gene-specific disease epigenetics21, genome-scale epigenetic studies of disease have also focused first on cancer, and these studies have revealed much more genetic pathology than was suggested by candidate gene approaches. For example, methylation changes can affect large genomic regions in colorectal cancer22, and widespread methylation changes are even more striking outside of the usually examined CpG islands, i.e. in shores and gene bodies23. Similarly, it came as a surprise to most when widespread alterations in histone acetylation and methylation were found ubiquitously in cancer24. Stem cells, a hoped for therapeutic target for many diseases, have shown promiscuous methylation differences from somatic cells on a genome-scale, surprisingly involving non-CpG sites25. Remarkably, the sites of differential methylation largely overlap, with strong statistical significance, among physiological states, such as normal vs. cancer, stem cells vs. differentiated cells, and tissues derived from differing germ layers26. Thus, the language of epigenomic organization appears to be common for normal development and for disease, just as the language of anatomy is common for normal and abnormal physiology.
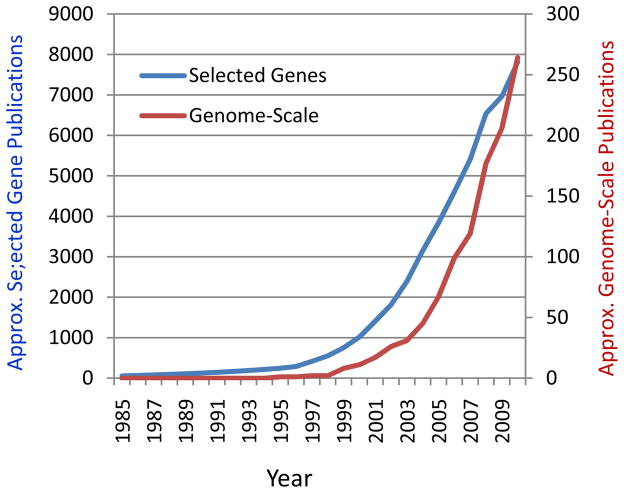
The increasingly appreciated importance of large-scale epigenetic control in regulating gene function has had a profound influence on how disease-based genomic studies are being organized. While published genome-scale studies represent only about 2% of cancer epigenetics, the rate of increase over the last 5 years of cancer epigenomic studies is more than double that of conventional gene-based analyses (Fig. 1). The same relative increase in genome-scale studies also appears to be true for the nascent field of non-cancer human disease epigenetics, such as cardiovascular, immunological and neuropsychiatric disease 27, 28.These differences are of course driven in part by the availability of new technology, but also by the growing realization that variation in both DNA methylation and chromatin are widespread across the genome, and may be organized into large genomic domains.
Figure 1. Greater rate of increase of genome-scale, compared to selected gene-focused publications, addressing cancer epigenetics.
While published genome-scale studies represent only about 2% of cancer epigenetics, the rate of increase over the last 5 years of cancer epigenomic studies appears double that of conventional selected gene-based analyses. Numbers are approximate from PubMed citation analysis; scales are different for gene-based and genome-based plots; 2010 is extrapolated from 2/3 of 2009 plus 2010.
But another important factor driving such “disease epigenomics” is the relatively limited yield to date of conventional SNP-based genetic analysis in explaining most common human disease. As widely described in both scientific29, 30 and lay publications31, the gap between original expectation of genetic analysis and attributable risk of disease is much greater than anticipated a decadeago.
How is epigenomics transforming the search for genetic causes of common human disease? Many have suggested that one contributing factor may be the importance of environmentally driven epigenetic variation in disease risk, particularly as a surrogate for mutational change32–34 (Table 1). But we should also consider another dimension to this epigenetic argument for common disease that has received comparatively less attention. Since the actual “genome anatomy” target is likely much larger than we realized, perhaps involving half or more of the genome, and since the understanding of the normal function of this genome anatomy requires epigenomics, perhaps much of what appears to be negative genetic data could become meaningful in an epigenomic context (Table 1). For example, most GWAS studies identify not genes, but nearby regions or intergenic deserts. Yet these same regions frequently harbor differentially methylated regions (DMRs) that discriminate tissue types, or distinguish cancer from normal cells. They are also the canonical regions for long intergenic noncoding RNAs (LINCs) that help establish chromatin structure and normal gene function. Furthermore, gene deserts may promote trans-associations of chromosomes in epigenetic regulation35. Another way in which disease-associated DNA sequence variants might affect disease risk is through their linkage to DNA sequences regulating DNA methylation or chromatin modification or binding factors. Substantial association of SNPs with DNA methylation has already been found36, 37.
Table 1.
How epigenomics is transforming the search for genetic causes of common human disease
| Epigenome anatomy | Possible disease link | New approach to common disease search |
|---|---|---|
| Environmentally driven epigenetic variation | Epigenome changes in absence of sequence variant | Methylome arrays, capture bisulfite sequencing, ChIPseq |
| Regulatorysite or expression per se | Noncoding RNAs | RNAseq and methods above |
| Key disease sequences unlinked to target genes | Intra-and interchromosomal interactions | Chromatin network mapping; replication timing? |
| Regulatory sequence distant from gene | Co-regulated gene clusters | Genome-scale methylation; chromatin mapping |
| Sequence-defined methylation | Sequence variants controlling epigenome | Linked GWAS and epigenome studies |
| New class of Variably Methylated Regions | Sequence variants controlling epigenomic variance perse | New statistics for reexamining and integrating GWAS |
| Domain disruption, anchoring proteins | LOCKs and LADs | Native chromatin whole-genome analysis |
An intriguing additional possibility we have proposed is that DNA sequence variants might themselves affect the stochastic or environmentally influenced variance in the epigenome. According to this model, complex species would have an evolutionary advantageto include alleles for increased epigenetic variation per se, i.e. genetic alleles that increase epigenetic variance but not the mean38. This would be like an evolutionary “hedging one's bet,” and confer an advantage for genes in pathways for which the environment changes epochally, e.g. the abundance of food and water. Examining inbred mice from the same litter and living in the same cage, we identified hundreds of “variably methylated regions,” or VMRs, that were highly enriched for key genes in development and embryonic pattern formation. Thus development itself, which epigenetics regulates, likely includes a great deal of stochasticism at the epigenetic level. Genetic variants that increase this developmental plasticity at specific targets may confer an evolutionary advantage but might be deleterious to some individuals after a recent epochal change in the environment, such as the recent Western diet38. Intriguingly, several VMRs have recently been linked to body mass index39.
Finally, we are only beginning to understand the role of LOCKs and LADs in functional genome organization, and their assessment in disease will require robust genome-scale approaches to native chromatin measurement, and availability of clinical specimens permitting such analyses (Table 1).
Future technology development that could drive epigenomics
What potential areas for future technology development will fuel growth in this area? Of course, all roads lead to sequencing, including bisulfite genome-scale sequencing for DNA methylation, just as in non-epigenetic genome science. The rollout of inexpensive, comprehensive, high throughput single molecule sequencing has been slower than promised, and second generation sequencing is still impractical for large scale epidemiological studies involving thousands of patients, except for capture-based methods such as padlock probes 40. The conundrum in such studies is that while they offer enormous advantages in throughput, single base resolution, and allele-specific data, they will not reveal regions of differential methylation where we do not already know to look, which may be vast as epigenomics is applied to an ever increasing number of disease states. At the same time, high throughput sequencing is relatively cheap now for examining chromatin modifications, but that is true for modifications representing a relatively small fraction of the genome purified by chromatin immunoprecipitation, for example. For large regional changes such as LOCKs, one faces cost limitations similar to whole-genome bisulfite sequencing.
An important advance will come from reagents that are cheap and amenable to processing by typical university core laboratories, such as Illumina and other arrays. For example, a soon to be released methylation chip will provide ~450,000 targets, including all CpG islands and shores, as well as DNase hypersensitive sites and other regions identified and curated for this purpose by a consortium of laboratories organized by Tom Hudson. While this reagent may not be next or even this year’s most comprehensive approach, last year’s isn’t bad and such cooperative approaches open epigenomic research to any general laboratory, a very exciting development. Other exciting technological initiatives include epigenomic analysis of microdissected samples or even single cells, and enrichment of small chromosomal fragments for biochemical analysis of chromatin 41.
A new epigenetic epidemiology will need to be crafted. We can no longer consider genetic variation in isolation when looking for disease relationships. Samples in ongoing and future large-scale cohorts must be preserved to allow DNA methylation and chromatin analysis. But retrospectively, a great deal can be added to existing cohort studies, since DNA methylation is stable over decades. Much of the existing genetic data might be made clearer by adding epigenomic analysis to those studies. New cohort sampling should include standard sources, such as lymphocytes, but also, as much as possible, target tissues affected by the disease.
Additionally, we need to develop new statistical and epidemiological tools for disease epigenomics, and for its synthesis with conventional genetic analysis. For example, unlike SNPs, epigenetic variation is inherently quantitative, and thus does not lend itself to simple allele designation, e.g. quantitative levels of DNA methylation or polycomb complex members. The quantitative nature of epigenome variation can help explain complex traits with a smaller number of contributing loci, since they do not necessarily require as many of the additive signals originally proposed by R.A. Fisher42. Such an approach is being applied, for example, to the analysis of quantitative traits associated with VMRs39.
The apparent additional complexity epigenomics brings to genetics may seem daunting. But I don't think Vesalius would have been intimidated, and I know Victor would have been delighted.
Acknowledgments
I thank E. Pujadas, K. Reddy, and R. Ohlsson for comments on the manuscript. This work was supported by NIH Grant 5R37CA054358.
References
- 1.Vesalius A. De humani corporis fabrica libri septem. J. Oporini; Basel: 1543. [Google Scholar]
- 2.McKusick VA. The anatomy of the human genome: a neo-Vesalian basis for medicine in the 21st century. JAMA. 2001;286:2289–2295. doi: 10.1001/jama.286.18.2289. [DOI] [PubMed] [Google Scholar]
- 3.Proudfoot NJ, Shander MH, Manley JL, Gefter ML, Maniatis T. Structure and in vitro transcription of human globin genes. Science. 1980;209:1329–1336. doi: 10.1126/science.6158093. [DOI] [PubMed] [Google Scholar]
- 4.Crossley M, Orkin SH. Regulation of the beta-globin locus. Curr Opin Genet Dev. 1993;3:232–237. doi: 10.1016/0959-437x(93)90028-n. [DOI] [PubMed] [Google Scholar]
- 5.Viville S, Surani MA. Towards unravelling the Igf2/H19 imprinted domain. Bioessays. 1995;17:835–838. doi: 10.1002/bies.950171004. [DOI] [PubMed] [Google Scholar]
- 6.Boggs BA, et al. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 8.van Steensel B, Dekker J. Genomics tools for the unraveling of chromosome architecture. Nat Biotechnol. 2010;28 doi: 10.1038/nbt.1680. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expressionof cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 10.Sandhu KS, et al. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–2603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 12.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacFarlane LA, Gu Y, Casson AG, Murphy PR. Regulation of fibroblast growth factor-2 by an endogenous antisense RNA and by argonaute-2. Mol Endocrinol. 24:800–812. doi: 10.1210/me.2009-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatinstate to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith ST, et al. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz YB, et al. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 22.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, et al. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 25.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi A, et al. Differential methylation of tissue-and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saterlee J, Schubeler D, Ng H. Tackling the epigenome: challenges and opportunities for collaborative efforts. Nat Biotechnol. 2010;28 doi: 10.1038/nbt1010-1039. in press. [DOI] [PubMed] [Google Scholar]
- 28.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28 doi: 10.1038/nbt.1685. in press. [DOI] [PubMed] [Google Scholar]
- 29.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 31.Wade N. New York Times, Edn. The NewYork Times Company; New York: Jun 12, 2010. [Google Scholar]
- 32.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Petronis A, Paterson AD, Kennedy JL. Schizophrenia: an epigenetic puzzle? Schizophr Bull. 1999;25:639–655. doi: 10.1093/oxfordjournals.schbul.a033408. [DOI] [PubMed] [Google Scholar]
- 34.Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- 35.Gondor A, Ohlsson R. Chromosome crosstalk in three dimensions. Nature. 2009;461:212–217. doi: 10.1038/nature08453. [DOI] [PubMed] [Google Scholar]
- 36.Kerkel K, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs JR, et al. Abundant quantitative trait Loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107 (Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feinberg AP, et al. Personalized epigenomic signatures stable over time and covarying with body mass index. Science Transl Med. 2010 doi: 10.1126/scitranslmed.3001262. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernstein BE, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28 doi: 10.1038/nbt1010-1045. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barton NH, Briggs DEG, Eisen JA, Goldstein DB, Patel NH. Evolution. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2007. [Google Scholar]