Abstract
Epigenetic modifications must underlie lineage-specific differentiation as terminally differentiated cells express tissue-specific genes, but their DNA sequence is unchanged. Hematopoiesis provides a well-defined model to study epigenetic modifications during cell-fate decisions, as multipotent progenitors (MPPs) differentiate into progressively restricted myeloid or lymphoid progenitors. While DNA methylation is critical for myeloid versus lymphoid differentiation, as demonstrated by the myeloerythroid bias in Dnmt1 hypomorphs1, a comprehensive DNA methylation map of hematopoietic progenitors, or of any multipotent/oligopotent lineage, does not exist. Here we examined 4.6 million CpG sites throughout the genome for MPPs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), and thymocyte progenitors (DN1, DN2, DN3). Dramatic epigenetic plasticity accompanied both lymphoid and myeloid restriction. Myeloid commitment involved less global DNA methylation than lymphoid commitment, supported functionally by myeloid skewing of progenitors following treatment with a DNA methyltransferase inhibitor. Differential DNA methylation correlated with gene expression more strongly at CpG island shores than CpG islands. Many examples of genes and pathways not previously known to be involved in choice between lymphoid/myeloid differentiation have been identified, such as Arl4c and Jdp2. Several transcription factors, including Meis1, were methylated and silenced during differentiation, suggesting a role in maintaining an undifferentiated state. Additionally, epigenetic modification of modifiers of the epigenome appears to be important in hematopoietic differentiation. Our results directly demonstrate that modulation of DNA methylation occurs during lineage-specific differentiation and defines a comprehensive map of the methylation and transcriptional changes that accompany myeloid versus lymphoid fate decisions.
Hematopoietic stem cells (HSC) can self renew for life and differentiate into all myeloid and lymphoid blood lineages2 (Fig. 1a). Recent evidence suggests that DNA methylation plays a direct role in regulating both HSC self-renewal and commitment to lymphoid versus myeloid fates1,3. Although the frequencies of myeloid progenitors and differentiated cells were normal in Dnmt1-hypomorphic mice, lymphoid-restricted CLPs and their downstream thymic T cell progenitors (DN1, DN2 and DN3) were diminished. In the bone marrow of Dnmt1-hypomorphs, lymphoid, but not myeloid, transcripts were reduced, and promoters of two myeloerythroid genes were hypomethylated in HSCs. These observations support a critical role for DNA methylation in lymphocyte development, possibly through regulation of gene expression.
Figure 1. Examples of known lineage-related genes showing differential DNA methylation between lymphoid and myeloid progenitors.
a, Hematopoietic progenitors included in this study. Dashed-arrow indicates existence of intermediate progenitors. DMR in b, Lck and c, Mpo. Upper panels: top half: CpG methylation (p); lower half: CpG dinucleotides (black tick marks), CpG density (curve), CpG islands (orange lines) and the gene annotation (see online Methods). Middle panels: methylation of individual CpGs (in the red boxes), mean values connected by lines. Bottom panels: mRNA expression levels, normalized to the highest expression among the populations (mean ± s.d., n=3; 5 for MPPFL− for microarrays).
Here we have examined genome-wide methylation profiles of the mouse hematopoietic system, because it provides the first opportunity to examine differential methylation of a hierarchical progression of purified cell populations with well-characterized differentiation potentials (Fig. 1a). Eight populations, ranging from uncommitted MPP through oligopotent progenitors specified during myeloid versus lymphoid fate decisions, were FACS-purified and subjected to Comprehensive High-throughput Array-based Relative Methylation (CHARM) analysis (Fig. 1a and Supplementary Fig. 1). This approach investigated the methylation status of CpGs throughout the mouse genome using an algorithm favoring regions of higher CpG density (including all CpG islands4), but without bias for CpG location relative to genes5. Using CHARM, we recently found that differential methylation occurs more frequently in CpG island “shores” (regions within 2kb of an islands) than in CpG islands during multiple cellular differentiation processes6,7. Additionally, mRNA of each population was subjected to microarray and RT-PCR analyses to generate gene expression data. Thus, we were able to directly compare differentially methylated regions (DMRs) throughout the genome with expression levels of nearby genes for all eight populations.
This analysis revealed DMRs in numerous genes known to play a role in lymphoid or myeloid fate specification. For example, Lck, the src family kinase member responsible for initiating signaling downstream of the T cell receptor (TCR)8, was transcriptionally upregulated from DN1 to DN3, consistent with its role in pre-TCR signal transduction (Fig. 1b). Interestingly, as Lck transcription was upregulated, CpGs in exon 1 through intron 2 were progressively demethylated (Fig. 1b). Similarly, myeloid specification from MPP through GMP was accompanied by transcriptional upregulation and progressive hypomethylation of Mpo, which encodes an enzyme central to the microbicidal activity of neutrophils9 (Fig. 1c). Additionally, Cxcr2, which encodes a chemokine receptor responsible for neutrophil chemotaxis10, was upregulated during myeloid commitment from CMP through GMP, while the gene was demethylated (Supplementary Fig. 2a). Furthermore, Gadd45α, which is implicated in myeloid development11, was found to be concomitantly upregulated and demethylated in the CMP to GMP transition (Supplementary Fig. 2b). Gadd45α can actively demethylate DNA in different model systems12,13; thus, hypomethylation of Gadd45α during myelopoiesis may promote further hypomethylation of genes regulating myeloid commitment; however, the role of Gadd45α in promoting demethylation is still controversial14. Taken together, these data indicate that CHARM analysis correctly identifies DMRs in known lymphoid and myeloid specifying genes, each confirmed by pyrosequencing and gene expression analysis, making it a valuable tool for identifying candidate genes important for lymphoid or myeloid fate specification.
Viewed globally, CHARM analysis revealed striking epigenetic plasticity, resulting in increased overall methylation upon lymphoid relative to myeloid commitment (Table 1). Most DMRs distinguishing MPPFL+ cells from CLP lost methylation during this step of early lymphoid commitment, but upon the subsequent transition to DN1, 15-fold more DMRs showed gain, as opposed to loss, of methylation. Similarly in the earliest step of myeloid commitment from MPPFL+ to CMP there were substantially more hypermethylated than hypomethylated DMRs, but nearly all DMRs showed loss of methylation on transition from CMP to GMP. Comparing DN1 to GMP, two populations similarly differentiated towards lymphoid and myeloid fates, respectively, there were 8-fold more DMRs with higher-level methylation in DN1 cells, suggesting a skewing toward greater methylation in lymphoid compared to myeloid hematopoiesis. These observations might explain why Dnmt1-hypomorphic mice, which are unable to properly maintain CpG methylation, have normal myeloid, but diminished lymphoid development1,3.
Table 1.
Numbers of differentially methylated regions (DMRs) identified in myeloid versus lymphoid commitment
| Comparisons (Group 1 vs. Group 2) | Numbers of DMRsa |
DMRs within 2 kb of a transcriptional start site | Locations of DMRs relative to CpG islands (%) |
|||
|---|---|---|---|---|---|---|
| Group 1>Group 2 | Group 1<Group 2 | Islands | Shoresc | >2 kbc | ||
| MPPFL− vs. MPPFL+ | 18 | 112 | 69 | 56.1 | 35.4 | 8.5 |
| MPPFL+ vs. CLP | 302 | 46 | 182 | 52.4 | 39.0 | 8.6 |
| CLP vs. DN1 | 203 | 3017 | 2433 | 17.8 | 80.7 | 1.5 |
| DN1 vs. DN2 | 3923 | 174 | 3125 | 18.9 | 80.3 | 0.7 |
| DN2 vs. DN3 | 47 | 12 | 13 | 5.0 | 78.3 | 16.7 |
| MPPFL+ vs. CMP | 149 | 1412 | 1129 | 17.3 | 81.2 | 1.5 |
| CMP vs. GMP | 768 | 11 | 554 | 16.8 | 81.1 | 2.1 |
| DN1 vs. GMP | 1011 | 122 | 773 | 16.8 | 78.2 | 5.0 |
Area cutoff of 2.0 was used to calculate the number of DMRs (see online Methods).
Shores were defined for this table as overlapping islands by <50% and/or extending up to 2 kb from an island.
Regions 2 kb from an island.
To test the hypothesis that reduced methylation preferentially promotes myeloid as opposed to lymphoid differentiation, we turned to an in vitro assay system that promotes both myeloid and lymphoid development15,16. In the presence of 5-aza-2′-deoxycytidine, the percentage of myeloid progeny increased at the expense of lymphoid progeny for MPPFL+, CLP, DN1 and DN2, but not DN3, which remained lymphoid committed (Supplementary Fig. 3a–b). This myeloid skewing was most pronounced in DN1 cells, perhaps indicating that the large number of methylated DMRs in DN1 compared to CLP is critical for lymphoid specification (Table 1). We conclude that inhibiting DNA methylation promotes myeloid versus lymphoid specification, providing a mechanism for the myeloid skewing observed in Dnmt1 hypomorphs1.
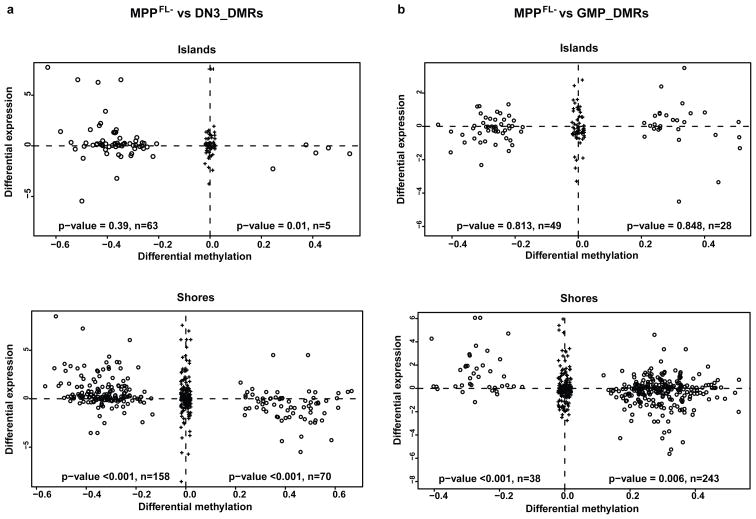
Consistent with our previous studies6,7, most DMRs were in CpG island shores (Table 1). The exceptions were for MPPFL− vs. MPPFL+, and MPPFL+ vs. CLP, in which most DMRs were in CpG islands: interestingly, both of these transitions are involved in early differentiation. Differential DNA methylation and gene expression showed a statistically significant inverse relationship particularly at CpG island shores (Fig. 2 and Supplementary Fig. 4). As the CHARM array design was targeted toward CpG density but not gene architecture per se, we also created a new array that included all promoters, and hybridized DNA from three of the groups studied earlier. Analysis showed a similar statistically significant inverse relationship between differential DNA methylation and gene expression, again particularly at CpG island shores (Supplementary Fig. 4g–h). Thus, CpG island shores are the regions with the most variability in DNA methylation between hematopoietic populations, and this variability correlates best with changes in gene expression. [However, not all DNA methylation changes correlated with changes in gene expression: for example, Tha1 is demethylated during lymphoid specification (see CHARM plots on http://charm.jhmi.edu/hsc), but is expressed at high levels from MPP through DN3. In converse, and as expected since there are multiple mechanisms for epigenetic regulation, we also identified lineage-specifying genes with changes in expression levels, but not in DNA methylation, such as Gata3 and Hes1 (see microarrays deposited in GEO).
Figure 2. Gene expression correlates strongly with DMRs at shores.
DMRs within 2kb of gene TSSs (black circles) were divided into two groups: Island (inside, cover, or overlap more than 50% of a CpG island), and Shores (up to 2000bp away from a CpG island). After RMA preprocessing, the log2 ratios of the gene expression differences (from leftto right) were plotted against Δp (left group minus right group). Black pluses represent random DMR-gene pairs more than 2kb apart. Wilcoxon rank-sum tests were performed to test the null hypothesis. a, MPPFL− vs. DN3_DMRs. b, MPPFL− vs. GMP_DMRs.
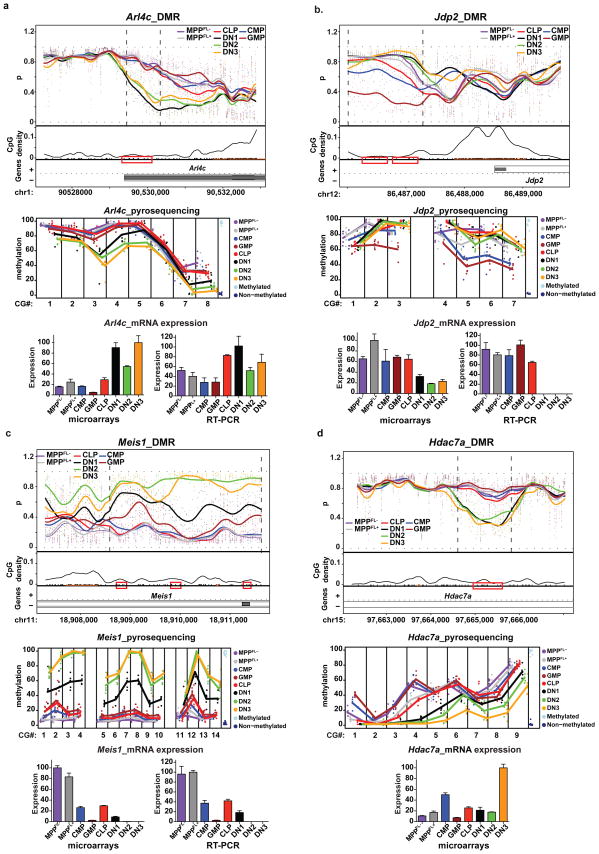
Many novel genes with the potential to contribute to myeloid/lymphoid fate specification were revealed by comparing CHARM-identified DMRs with gene expression data. For example, Arl4c, a member of the ADP-ribosylation factor family of GTP-binding proteins, was upregulated and hypomethylated in DN1-3 thymocytes (Fig. 3a). Arl4c may play a role in vesicular transport17, but its role in lymphoid specification is unknown. Multiple other genes with DMRs suggestive of a role in lymphoid development, such as Smad7, Gcnt2 and Cited2, were also identified (Supplementary Fig. 5). Smad7, which negatively regulates TGF-beta signaling, is selectively upregulated and hypomethylated at the earliest stages of thymocyte development, suggesting a role in promoting lymphopoiesis (Supplementary Fig. 5a). However, it causes myeloid lineage skewing when overexpressed in human cord blood progenitors18. Gcnt2 transcripts were downregulated in thymocyte progenitors, and the locus became hypermethylated progressively in DN1-3 progenitors (Supplementary Fig. 5b), consistent with a role for Gcnt2 in enabling the myeloid potential that is lost during final lymphoid lineage commitment at the DN3 stage19,20.
Figure 3. CHARM identified genes with previously unknown functions in lymphoid/myeloid lineage commitment and pluripotency maintenance.
a and b, Examples of DMRs with methylation changes in lymphoid/myeloid progenitors. a, the DMR in Arl4c. b, the DMR in Jdp2. C, the DMR in Meis1. D, the DMR in Hdac7a. The CHARM plots, pyrosequencing, Affymetrix GeneChip, and RT-PCR data are organized and displayed as in Fig. 1b.
Novel potential regulators of myelopoiesis were also identified. The Jdp2 locus was hypomethylated and its transcript was upregulated in CMP and GMP relative to thymocyte progenitors (Fig. 3b). Jdp2 is thought to repress transcription by recruiting histone deacetylases and regulating nucleosome assembly21. Dach1 was also hypomethylated and expressed from MPPFL− through GMP, but was silenced in CLP and DN1-3 thymocyte progenitors (Supplementary Fig. 5d), suggesting it may contribute to myelopoiesis. Dach1 has been implicated in transcriptional repression through association with histone deacetylases and its drosophila homolog is known to play a role in gonadal, limb, and ocular development22. Thus, Jdp2 and Dach1 may feedback on the epigenome to control expression of tissue specific genes, but their role in hematopoiesis remains uncharacterized.
Our analyses also revealed a set of genes that were progressively hypermethylated and transcriptionally silenced as differentiation progressed towards both myeloid and lymphoid fates, suggesting a role in maintenance of a multipotent state. Meis1, 2900052L18Rik, Hlf, Hoxa9 and Prdm16 are all such candidates (Fig. 3c and Supplementary Fig. 6). Meis1 is known to be required for hematopoiesis and megakaryocyte lineage development23 and may function cooperatively with Hoxa9 to regulate hematopoiesis24. Furthermore, both Hlf and Prdm16 have been implicated in hematopoiesis25,26.
Lastly, epigenetic chromatin modifiers, including Hdac7a and Dnmt3b, were also differentially methylated during hematopoietic differentiation, suggesting feed-forward mechanisms that could expand and lock in epigenetic programming necessary for cell fate commitment (Fig. 3d and Supplementary Figure 7). Hdac7, which encodes a histone deacetylase and represses transcription, was demethylated and upregulated in DN1-DN3 thymocytes (Fig. 3d). Since Hdac7 is highly expressed in DN3 cells, which can no longer be reprogrammed toward a myeloid fate by ectopic IL-2R signaling19, it may actively repress genes responsible for maintaining myeloid lineage potential19. In contrast, Dnmt3b, a methyltransferase responsible for de novo CpG methylation, is hypermethylated and downregulated progressively in CMPs and GMPs (Supplementary Figure 7). Dnmt3a and Dnmt3b were shown to be essential for HSC self-renewal, but their roles in lineage commitment remain inconclusive27. Downregulation of Dnmt3b in myeloid committed cells could prevent new DNA methylation, helping to maintain the observed hypomethylated state associated with myelopoiesis. In addition, the upregulation of Dnmt3b in DN1 independent of DNA methylation changes might explain the dramatic acquisition of DNA methylation from CLP to DN1 (Table 1).
In summary, these data provide a comprehensive map of the methylome during myeloid and lymphoid commitment from hematopoietic progenitors. To facilitate the general accessibility of the methylome for these hematopoietic progenitors, we also provide here a novel web platform with which the methylation status of any genomic locus of interest can be easily queried to generate output methylation plots. In addition to identifying candidate genes for further investigation, the data suggest several important themes for the epigenetics of lineage-specific differentiation. First, myelopoiesis and lymphopoiesis achieve markedly different methylation endpoints in differentiation, with lymphopoiesis depending much more heavily on the acquisition of DNA methylation marks, and myelopoiesis depending much more on their loss. Besides providing a mechanism for the proposed DNMT1-dependence of lymphopoiesis, these results may also explain the therapeutic specificity of DNA demethylating drug treatment of myelodysplasia, in which malignant cells arrested in early development may be induced to differentiate by DNA demethylation28. In addition, the results show a remarkable dynamic plasticity in methylation during lineage development. The changes are evocative of Waddington’s illustrations of hills and valleys in the epigenetic landscape of development. We have recently proposed that development depends on dynamic stochastic variation in the epigenetic landscape in a given genetic environment29, and the maturation of undifferentiated progenitors to progressively more differentiated states could restrict that variation. Support for this idea is provided in an accompanying manuscript in this journal examining the epigenetic memory in iPS cells derived from fibroblasts and blood30. In that paper, lymphocyte-derived iPS cells cluster with CLP but not the myeloid lineage using DNA methylation differences we identified, suggesting the existence of lymphocyte memory in these iPS cells consistent with the DNA methylation profiles described in this paper.
Methods Summary
Flow cytometry
Bone marrow cells and thymocytes were stained with monoclonal antibodies, then analyzed and sorted using a FACSAria. Antibody details are provided in Online Methods.
CHARM DNA methylation analysis
Genomic DNA was isolated from samples, fractionated, digested, purified, labeled and subject to CHARM array analysis as previously described7. Details are provided in Online Methods.
Bisulfite pyrosequencing
Genomic DNA was isolated from cells, treated with bisulfite and amplified by PCR. DNA methylation was measured by quantitative pyrosequencing. Details are provided in Online Methods.
Affymetrix microarray expression analysis
Genome-wide gene expression analysis was performed using the Affymetrix GeneChip Mouse Genome 430 2.0 Array. Details are provided in Online Methods.
OP9:OP9DL1 stromal co-cultures
50 double sorted progenitors were cultured in wells containing confluent 1:1 OP9:OP9DL1 stromal cells in the presence of cytokines. At day 6, the cells in each well were stained and analyzed by flow cytometry. Details are provided in Online Methods.
Quantitative PCR
Cells were sorted into TRIzol, RNA was isolated and cDNA was synthesized. Real-time PCR was performed using SYBR Green reagents. Details are provided in Online Methods.
DNA methylation query website
DNA methylation in any region from the CHARM array can be plotted at http://charm.jhmi.edu/hsc. Details are provided in Online Methods.
Supplementary Material
Acknowledgments
We thank L. Jerabek for laboratory management, C. Richter and N. Teja for antibody production, A. Mosley, J. Dollaga, and D. Escoto for animal care, E. Zuo and the Stanford PAN facility for microarray processing, and E. Briem and A. N. Allen for CHARM array processing. This investigation was supported by National Institutes of Health grants R37CA053458 and P50HG003233 (to A.P.F), R01AI047457 and R01AI047458 (to I.L.W.), and a grant from the Thomas and Stacey Siebel Foundation (to I.L.W). L.I.R.E. was supported by Special Fellow Career Development award from the Leukemia and Lymphoma Society; J.S. was supported by a fellowship from the California Institute for Regenerative Medicine (T1-00001); D.J. Rossi was supported by National Institutes of Health grant R00AGO29760; M.A.I. was supported by National Institutes of Health grant CA09151 and a fellowship from the California Institute for Regenerative Medicine (T1-00001); T.S. was supported by a fellowship from the National Institutes of Health (F32AI058521).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions: H.J. performed genome-scale and gene-specific DNA methylation analysis; L.E. and J.S. performed cell-sorting, generated microarray datasets, and performed gene expression analysis; P.L. assisted L.E., and H.L. assisted H.J.; A.D. and H.J. performed statistical analysis with P.M., M.A and R.A.I; D.J.R., M.A.I., T.S., H.K., and L.H. generated microarray datasets; A.P.F. and I.L.W. designed the experiment, and A.P.F., H.J., L.E., and J.S. wrote the paper with the assistance of ].K.K., and G.Q.D.
Author Information
Reprints and permissions information is available at http://npg.nature.com/reprintsandpermissions. L.W. has stock in Amgen and is co-founder of Cellerant Inc. and Stem Cells Inc. He is also a consultant for Stem Cells Inc. The other authors declare no competing financial interests.
References
- 1.Broske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 2.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 3.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 5.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molina TJ, et al. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 9.Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 10.Cacalano G, et al. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science. 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 11.Gupta SK, Gupta M, Hoffman B, Liebermann DA. Hematopoietic cells from gadd45a-deficient and gadd45b-deficient mice exhibit impaired stress responses to acute stimulation with cytokines, myeloablation and inflammation. Oncogene. 2006;25:5537–5546. doi: 10.1038/sj.onc.1209555. [DOI] [PubMed] [Google Scholar]
- 12.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 13.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel N, et al. Conserved DNA methylation in Gadd45a(−/−) mice. Epigenetics. 2009;4:98–99. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 16.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 17.Wei SM, Xie CG, Abe Y, Cai JT. ADP-ribosylation factor like 7 (ARL7) interacts with alpha-tubulin and modulates intracellular vesicular transport. Biochem Biophys Res Commun. 2009;384:352–356. doi: 10.1016/j.bbrc.2009.04.125. [DOI] [PubMed] [Google Scholar]
- 18.Chadwick K, Shojaei F, Gallacher L, Bhatia M. Smad7 alters cell fate decisions of human hematopoietic repopulating cells. Blood. 2005;105:1905–1915. doi: 10.1182/blood-2004-03-0881. [DOI] [PubMed] [Google Scholar]
- 19.King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci U S A. 2002;99:4508–4513. doi: 10.1073/pnas.072087899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo M, et al. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 21.Jin C, et al. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat Struct Mol Biol. 2006;13:331–338. doi: 10.1038/nsmb1063. [DOI] [PubMed] [Google Scholar]
- 22.Popov VM, et al. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol Metab. 21:41–49. doi: 10.1016/j.tem.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillay LM, Forrester AM, Erickson T, Berman JN, Waskiewicz AJ. The Hox cofactors Meis1 and Pbx act upstream of gata1 to regulate primitive hematopoiesis. Dev Biol. doi: 10.1016/j.ydbio.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 24.Hu YL, Fong S, Ferrell C, Largman C, Shen WF. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol. 2009;29:5181–5192. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crable SC, Anderson KP. A PAR domain transcription factor is involved in the expression from a hematopoietic-specific promoter for the human LMO2 gene. Blood. 2003;101:4757–4764. doi: 10.1182/blood-2002-09-2702. [DOI] [PubMed] [Google Scholar]
- 26.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claus R, Almstedt M, Lubbert M. Epigenetic treatment of hematopoietic malignancies: in vivo targets of demethylating agents. Semin Oncol. 2005;32:511–520. doi: 10.1053/j.seminoncol.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Feinberg AP, Irizarry RA. Evolution in Health and Medicine Sackler Colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. doi: 10.1038/nature09342. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.