Abstract
Microarray technologies have been an increasingly important tool in cancer research in the last decade, and a number of initiatives have sought to stress the importance of the provision and sharing of raw microarray data. Illumina BeadArrays provide a particular problem in this regard, as their random construction simultaneously adds value to analysis of the raw data and obstructs the sharing of those data.
We present a compression scheme for raw Illumina BeadArray data, designed to ease the burdens of sharing and storing such data, that is implemented in the BeadDataPackR BioConductor package (http://bioconductor.org/packages/release/bioc/html/BeadDataPackR.html). It offers two key advantages over off-the-peg compression tools. First it uses knowledge of the data formats to achieve greater compression than other approaches, and second it does not need to be decompressed for analysis, but rather the values held within can be directly accessed.
Keywords: Illumina BeadArray, microarray, compression, open data
Introduction
Background
From identifying deleterious copy number aberrations, 1 to refining classes of tumor,2 to genome-wide associations studies,3 the value of microarrays to cancer research in the last decade is readily apparent. Early on in this trend it became clear that the availability of data,4 and the quality of reporting,5 were crucial factors in the value of microarray experiments. With an understanding of the sometimes subtle influences of normalization, summarization and other pre-processing steps,6,7 came an appreciation of the need for the provision of raw data in order that the validity of conclusions could be assessed.
The Minimum Information about a Microarray Experiment (MIAME) initiative indeed calls for raw microarray data files (including images),8 but anticipates raw microarray data being in a regular format. This poses a problem for randomly constructed arrays such as Illumina’s BeadArray technology,9 where the number and locations of replicates of each probe vary from array to array, and for which it has become standard to report summary data.
Illumina BeadArrays have been used in a number of high-profile cancer studies (eg, for genotyping in SEARCH,10 for methylation and copy number in TCGA,11 and for expression in METABRIC.)12 and it has been shown that the analysis of raw Illumina BeadArray data, rather than the default summarized data, can be advantageous. However there is no data repository oriented towards storing these raw Illumina data, and there has been only limited success in storing such data in standard repositories.13,14 Researchers have thus been largely responsible for hosting their own raw data, but the size of those data inhibits this practice, and the ability of researchers to access raw data.
Here we present a schema and tool for the compression of raw BeadArray data, and illustrate the tool on recently published breast cancer cell-line data. A custom scheme offers two key advantages over standard compression methods such as zip. Firstly, by using our knowledge of the original file structures we can achieve smaller files than can unsupervised routines. Secondly, our file format permits direct analysis of the raw data without the need for a decompression step (although we allow for the decompression of our format into the original files should that be the user’s preference).
It is tempting, as personal storage becomes ever cheaper, to suppose that there is no need for compressed bead-level data. Indeed for a single user analyzing a small experiment there may be limited benefit, although even here the increase in size of experiment that will fit onto a CD or DVD is, if not a necessity, at least a convenience. By contrast, for institutions running tens of thousands of BeadArrays a year and requiring enterprise-level storage and archiving of the data the potential savings are truly substantial. The greatest benefit though comes when sharing data. The value of tools to facilitate data sharing in the context of cancer microarray experiments has previously been noted,15 and in BeadDataPackR we present a tool to overcome obstacles to data sharing for raw Illumina BeadArray data.
The nature of bead-level data
Illumina BeadArrays are scanned upon two occasions: once by the manufacturer to decode and identify the random probe layout, and once by the user to generate data. Raw Illumina BeadArray data consist of a .tif image (typically ~85MB for one HT12 array), a .txt file that gives bead identities, truncated locations and partially processed intensities (typically ~30MB), and a .locs file that gives precise bead locations in grid order (typically ~9MB). Other useful raw data files are also produced, but do not concern us here, as they are small files and generated per chip not per array. Since the compression of .tif files is well-studied,16 we focus on the compression of the .txt and .locs files.
We have previously described these file structures in detail.17 To summarize: a typical .txt file contains four columns of data (seven for two channel arrays) with each bead on the array represented by a row. Beads that were not decoded by Illumina are not commonly included, so the number of lines differs for each array. The first column gives the ID for a bead, whilst the second contains the background corrected intensity of that bead. The third and fourth columns store the X and Y coordinates of the bead center. These are given to seven significant figures, resulting in the fractional parts of the coordinates being given to between two and six decimal places, depending upon the magnitude of the integer part. If the data are from a two channel array, the fifth column contains details of the red channel intensity, with the sixth and seventh holding the coordinate information for that channel.
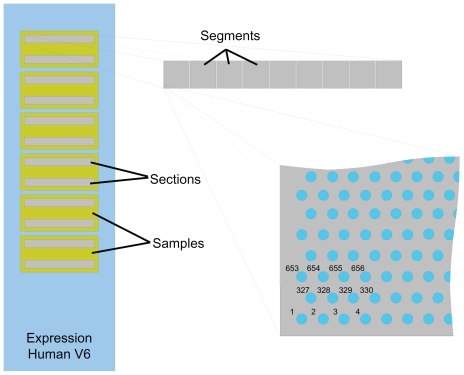
The array is divided into a number of segments, and within each segment the beads are laid out in a hexagonal grid, as illustrated in Figure 1. The .locs file stores the bead center coordinates for every bead on the array, rather than just those that were successfully decoded. The coordinates are stored as pairs of floating point numbers and are grouped by their segments on the array. Within each segment they are stored in grid order.
Figure 1.
Showing the physical layout of a typical Illumina BeadChip (in this case a Whole Genome 6 expression array). Illustrated are the multiple arrays (samples) on the chip, the multiple sections within an array, the multiple segments within a section, and the hexagonal grid structure within the segments (only one corner of a segment is illustrated). The ordering of beads within the .locs file is also indicated on the grid.
The advantages of a bead-level analysis
The activities that one undertakes with bead-level data that show them to be advantageous to summarized data are broadly divisible into five categories: Quality Assessment (QA), Quality Control (QC), alternative preprocessing, including two-channel preprocessing, and true bead-level analyses.
If QA is the only activity for which one uses bead-level data then the ultimate aim is to use Illumina’s summarized data, but only after filtering out arrays that are flagged as being problematic.18 With QC only, the aim is to identify problematic arrays and correct identified flaws before re-summarizing in essentially the same manner as Illumina and continuing with downstream analyses as though the array had been perfect. Such steps might include correcting for mis-registration of the array,17 correcting for gradients across the array, or resolving spatial artifacts.19,20
Illumina’s summarization incorporates steps such as background correction, outlier identification, adjustment for non-specific hybridization and so forth. Thus the analyst may find the flexibility offered by bead-level data to be advantageous. They may for example wish to extract intensities differently,17 use alternative background correction methods,21 transform to a different scale before summarization,22,23 or to normalize the replicate strips of a BeadArray separately.24 Moreover, when using two-channel platforms we may wish to use data from a combination of channels for steps such as outlier removal, or to calculate covariances to feed into summary statistics such as the log-ratio.25
Finally, it should be noted that while the high number of replicate observations offers good estimates of technical variance, and thus uncertainty about the summarized value, this information does not tend to be successfully propagated through the early stages of an analysis (eg, standard forms of normalization) and so a true bead-level analysis (eg, a hierarchical model.) may be required to capture such uncertainty.
The desirable recorded precision of bead coordinates
Due to the floating point representation of bead coordinates in the .locs file, the apparent precision recorded for bead coordinates is to a fraction of a picometer. This is of course not credible, nor is it actually claimed, but we comment upon it to illustrate that one should have no qualms about reducing the recorded precision. Even the reduced precision recorded in the .txt file suggests precision to within a fraction of a nanometer which still stretches credibility.
Further, if one examines how the fractional parts of the coordinate are used to calculate the bead intensity it is clear that such precision is often unnecessary. If the coordinates of a bead center in the scanned image are given as (314.1592 ..., 271.8281 ...) then Illumina’s foreground value for the bead is calculated as a weighted sum of intensities from a 4 × 4 pixel square around those coordinates. The central pixels take uniform weight and the remaining pixels have weights that are calculated as functions of Xf and Yf (the fractional parts of the X and Y coordinates: 0.1592 ... and 0.8281 ... in our example).
In extreme, albeit unlikely, cases, these Xf and Yf values can end up being multiplied by numbers of the order of 104, so changes at the fourth decimal place of the coordinates can noticeably affect the resultant bead intensity. Indeed there exists a scenario where a low-intensity bead, with a proximal high-intensity artifact, will be highly sensitive to the precision of location, but the presence of an artifact makes such beads unreliable.
Beads then fall into two classes: those where the degree of precision is not influential, and those where it is, indicating that the intensity is not reliable. This in itself merits reducing the degree of stored precision, but coupled with the doubts over whether the accuracy of identifying bead centers can warrant the claimed precision, one might confidently reduce the precision of the stored coordinates to minimize the file size. The full precision should only be required to recreate Illumina’s reported intensities (alternative feature extraction schemes should surely not make such demands of precision), and by default we preserve these within the file in any case. Should the full precision be desired, we retain the option of storing it.
We can though anticipate some costs to reducing the precision. The 16 pixels used to calculate the foreground intensity are determined by the bead coordinates, as are 189 pixels used to calculate the background intensity. A subtle consequence of reducing precision is that the ‘correct’ pixels may not be used in foreground and background intensity calculations. In both cases, there is a threshold in location that (if crossed by the bead-center) will result in a different set of pixels being used. It is possible that in rounding bead-center coordinates, we can cross the threshold and the effects of this can be observed when using the default Illumina feature extraction rules.
It would be possible to include an option in BeadDataPackR to always round away from the threshold in such circumstances (except when there are conflicts between the foreground and background thresholds). Such an approach shows some marginal benefits for the default intensity extraction algorithms when near full precision is used (data not shown), but is highly detrimental if the precision is too far reduced. Thus we do not recommend following such an approach, and do not offer this as an option in compression, but instead suggest that more robust intensity calculations (especially for the background) might be used.17
Answering the question of what level of precision should be retained requires knowledge of the intended analysis. If the image files are not going to be provided, then the only value in having the locations is to recreate the network of beads and to look for spatial trends in values. This would require recording locations only to the nearest pixel. Alternatively, the image file may be available, and one may wish to extract bead intensities using a bespoke algorithm. In this case, no matter the sophistication of the algorithm being applied, it is hard to envisage a scenario where claiming knowledge of the bead-center to more than 1/256th of a pixel could be warranted, and much less might be tolerable.
Methods
Compression file structure
For a typical array-section containing approximately a million beads, it is clear that reducing storage by a byte a bead will reduce the overall file size by 1MB. Savings on file sizes are obtained from the trivial (removing carriage returns and tabs ~4MB), the obvious (only recording coordinates once rather than in both the .locs and .txt files ~8MB), the efficient (recording bead IDs only once, along with the original number of occurrences ~4MB) and the fundamental (representing numbers in binary rather than ASCII format ~11MB). Further savings can be achieved by reducing the precision to which locations are stored and, for two-color arrays, recording the red coordinates as differences from the green coordinates.
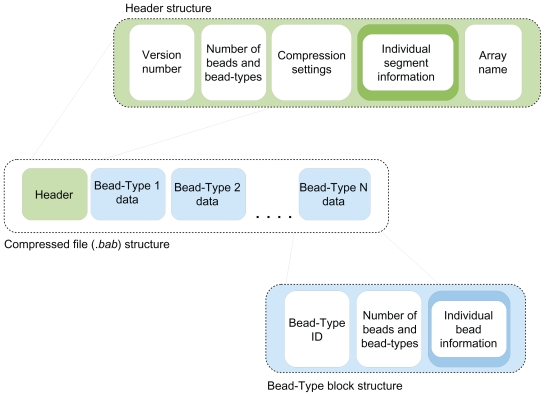
The structure of the compressed file (with extension .bab) consists of a header for the array followed by sequential blocks of data, one for each bead-type on the array. The header contains information about the whole array, such as the total number of beads, as well as the settings used during compression. Each bead-type block begins with the bead-type ID, followed by the number of beads of that type. Following this are the bead intensities, one for each bead of this type, stored as unsigned 16bit integers. Because image processing can sometimes lead to bead intensities that fall outside this range, these are followed by a series of 2bit flags that effectively allow the values to be stored as signed 18bit integers, with a range of −131,071 to 131,071.
The next section of data contains the X and Y coordinates for each bead, although their exact rep-resentation is dependent upon the degree of precision selected. For a single channel array, rather than storing the full precision of the two coordinates within 8 bytes (as they are in the .locs file), the integer parts can be stored within 4 bytes and then 3, 2, 1, or 0 bytes used for the fractional parts. For two color arrays, where we now have four coordinates to store, the fractional parts can occupy between 7 and 0 bytes if we wish to trade off precision for file size.
The final element of each block is an index for each bead, defining its location in the .locs file at a cost of only ~1MB. The file structure is illustrated in Figure 2, and full details are provided in the supplementary material.
Figure 2.
An overview of the structure of the compressed (.bab) files. The structure consists of a header section followed by several blocks of data (one per bead-type). Overviews of the structures of the header and of a ‘bead-type block’ are also given.
Methods
Coordinate precision details
If reducing precision, one has the choice of rounding either to a binary fraction or to a decimal fraction (ie, if using 1 byte for each channel one can either multiply the fractional part of the coordinate by 256 and round to the nearest integer, or multiply by 100 and round to the nearest integer). Naturally the binary representation of a fraction is the most efficient way to approach the task, offering greatest resolution for the cost in storage, however there is one notable merit to using a decimal representation.
If the user is planning to reconstruct the .txt file, and has used the binary representation, then (except in trivial cases) there are two possibilities. Either the .txt file will have to abandon the ‘seven significant figures’ format, or there will be a second loss of resolution when rounding to seven significant figures. The majority of beads will ultimately be rounded to a resolution no greater than if a decimal fraction had been used initially, and through the compounding of errors may end up with the location rounded to a different value than if the rounding had taken place in one procedure.
BeadDataPackR allows the user to choose in which base to store the fractions, and also whether to round the values in the .txt file to have a uniform number of significant figures.
Methods
Grid order reconstruction
The two input files not only contain different numbers of beads, but the order in which the bead information is stored also differs. Thus, to recreate the two input files accurately, it is necessary to store some information relating to their ordering within the .bab file. In grouping the data by bead-type we effectively retain the ordering of the input .txt file, meaning that only the .locs file ordering needs to be stored.
The most straightforward mechanism for achieving this is simply to record an integer index of the .locs file, which is used during reconstruction. For our typical array of approximately a million beads this index can be stored using ~3MB. However, since the .locs file already has a rigid ordering based upon the grid structure of the array, we can exploit this to utilize a smaller index (~1MB) while still being able to recreate the original file accurately.
The .locs file is broken down by array segments, each of which is processed separately. Within each of these segments a linear model is fitted to characterize the relationship between the coordinates and the row and column position of each bead in the grid. Denoting the coordinates of the beads in terms of their grid indices as Gx and Gy, and the coordinates of the bead-centers within the image (in units of pixels) as Px and Py we then fit the model.
where ɛ x and ɛ y are independent normal random variables (so models for Gx and Gy are actually fitted separately). The beta coefficients are then stored within the header of the .bab file.
Additionally, we employ a single byte per bead to store the row and column of the bead modulo 15 to aid reconstruction. During reconstruction of the .locs file each array segment is again processed and the beta coefficients are used to project to putative grid indices Hxand Hy.
Hxand Hy are then corrected to the nearest position that would return the correct row and column modulo 15 as stored in our single byte index. This allows us to detect and correct instances where the predicted location of a bead is out by up to seven rows or columns in either direction. Where there exist beads whose identified locations in the image are so far out that this approach would fail (ie, they would be more than seven rows or columns out), this is identified during compression and the full integer index used instead.
We would note that we choose modulo 15 rather than 16 not only for reasons of symmetry but because we anticipate the potential value in being able to flag a bead as having been located in the image extremely out of position, although we do not currently exploit this. We acknowledge that we do not use the most efficient grid representation, as by representing a hexagonal grid in rectangular coordinates, a half of all locations must be empty. Thus an alternative encoding could allow for the correction of beads that are identified in the image even further from their anticipated grid position than we can currently. In practice, though, we have not seen such beads, and if there did exist such beads then there would be concerns more serious than the matter of data compression.
Methods
R commands
BeadDataPackR is available to download from BioConductor.26,27 We provide two functions in the BeadDataPackR package for R. The function compressBeadData() takes the raw files and produces a .bab file from them, while the function decompress- BeadData() takes a .bab file and reconstructs the .txt and .locs files to the requested degree of precision. While the ability to reconstruct the original files is useful, given that many existing analysis scripts may expect the raw files, this step is not a necessity. Indeed, the structure of the file format means data can be extracted directly, allowing it to form the primary input to analysis tools. This can be advantageous in a number of cases, most notably when one wants to access data relating to a small number of beads on an array. Such a subset of the data can be obtained more quickly and with lower memory requirements when using the .bab format as opposed to reading the original .txt files. If one is performing these actions across a large number of arrays the savings can be substantial.
Methods
Analysis
We applied the BeadDataPackR compression scheme to all of the Illumina array variants to which we have access. In each case, the full range of bead-coordinate precisions was considered in order that the value in reducing that precision might be assessed. For comparison each was also compressed using a standard compression scheme (zip). The only version of Illumina BeadArray for which we have not been successful in applying the software is an early version of the IlluminaWG6- V1 chip. The bead-level data of this type, to which we have access, are in a notably different form.
In addition, to assess the impact of reducing the stored precision of bead coordinates on downstream analyses, we revisit a recently published dataset from a breast cancer study.28 As part of an investigation into the interaction between retinoic acid receptor-α (RARα) and estrogen receptor (ER) in a hormone depleted breast cancer cell-line (MCF-7), the authors ran 12 gene expression Illumina HT12-V3 BeadArrays as three replicates of a 2 × 2 factorial design. The two factors being estrogen, and an siRNA targeting RARα.
For illustration, we consider only the contrast relating to estrogen treatment, and ignore the orthogonal data (eg, ChIP-seq) that the authors used in their analyses. We compare an analysis of the data with the full precision (equivalent to allowing 4 bytes for the fractional parts of the coordinates) to analyses where precision has been lost.
The original files were compressed on five occasions (using 0,1,2,3 or 4 bytes for the storage of the fractional parts of the bead-coordinates) and then decompressed to obtain five sets of .txt files, all using BeadDataPackR. Intensities were extracted from the .tif files and summarized using beadarray29 and a standard analysis of a factorial experiment performed in limma.30
Results
The compression performance of BeadDataPackR for various types of Illumina BeadArray is given in Table 1. It can be seen that with full preservation of location precision, BeadDataPackR reduces file sizes to approximately a third of the originals for all types of BeadArray and always outperforms standard compression. This can come down to a quarter (or less) of the original file-size if we are willing to sacrifice the reported precision in the original files.
Table 1.
Showing the performance of BeadDataPackR for four varieties of single-channel array (HumanWG6-V2, HumanRef8-V2, HumanWG6-V3, Human HT12), and four varieties of dual-channel array (CNV370-Duo, Infinium II, DASL, Human 1M). The sizes of the original files, the zipped files, and the files compressed using BeadDataPackR are given in MB for differing degrees of precision in the storage of the bead-coordinates. These values are representative and will show small variations between arrays of the same type.
| Single-colour |
Two-colour |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WG6-V2 | Ref8-V2 | WG6-V3 | HT12 | CNV370 | Inf. II | DASL | 1M | ||
| Original | 39.0 | 37.5 | 38.4 | 40.2 | 35.6 | 66.9 | 68.6 | 69.1 | |
| Zipped | 18.2 (47%) | 17.4 (46%) | 17.9 (47%) | 18.7 (47%) | 17.6 (49%) | 33.9 (51%) | 34.3 (50%) | 34.2 (49%) | |
| Compressed by BeadDataPackR using X bytes to store fractional parts of coordinates | 8 | 12.1 (34%) | 23.6 (35%) | 23.6 (34%) | 23.1 (33%) | ||||
| 7 | 10.4 (29%) | 20.3 (30%) | 20.3 (30%) | 20.1 (29%) | |||||
| 6 | 9.9 (28%) | 19.1 (29%) | 19.2 (28%) | 19.1 (28%) | |||||
| 5 | 9.3 (26%) | 18.0 (27%) | 18.1 (26%) | 18.0 (26%) | |||||
| 4 | 12.7 (33%) | 12.3 (33%) | 12.5 (33%) | 12.8 (32%) | 8.7 (24%) | 16.9 (25%) | 17.0 (25%) | 17.0 (25%) | |
| 3 | 11.6 (30%) | 11.2 (30%) | 11.5 (30%) | 11.7 (29%) | 8.2 (23%) | 15.8 (24%) | 15.9 (23%) | 16.0 (23%) | |
| 2 | 10.4 (27%) | 10.1 (27%) | 10.4 (27%) | 10.5 (26%) | 7.6 (21%) | 14.7 (22%) | 14.7 (21%) | 15.0 (22%) | |
| 1 | 9.3 (24%) | 9.0 (24%) | 9.3 (24%) | 9.4 (23%) | 7.1 (20%) | 13.6 (20%) | 13.6 (20%) | 14.0 (20%) | |
| 0 | 8.2 (21%) | 7.9 (21%) | 8.2 (21%) | 8.3 (21%) | 6.5 (19%) | 12.5 (19%) | 12.5 (18%) | 13.0 (19%) | |
The results of the investigation into how the stored precision affects downstream analyses are summarized in Table 2. All comparisons are limited to 19443 probes that have been assessed as being perfect and containing no SNPs by a reannotation effort,31 and also as having a GC content of between 40% and 70%. It is clear that some precision can be sacrificed with no noticeable effect on results (eg, when using 2 or 3 bytes), and that even when using 1 byte, performance can probably be considered satisfactory. Since we might distrust results that are not robust to a reduction in claimed precision, 1 byte may suffice.
Table 2.
Impact of reducing the precision of the stored bead-coordinates on the downstream analysis of a 12 array expression experiment. For 19443 well-annotated probes, the mean-squared errors (relative to full precision) for individual beads, for summarized gene intensities, and for log-ratios between arrays are presented. The mean variance from 4 sets of 3 technical replicates is also presented, as is the ‘correct’ proportion (relative to full precision) of the top 1200 returned genes in a differential expression analysis. There are approximately 1200 significantly differentially expressed genes in an analysis using the full precision.
| Number of bytes used for storage | MSE beadlog-intensity | MSE summarized intensities | Mean variance of 3 tech reps | MSE log-ratio of two arrays | Proportion of first 1200 ER-driven genes returned |
|---|---|---|---|---|---|
| 4 | 0 | 0 | 0.0167 | 0 | 1.000 |
| 3 | 4.3 × 10−8 | 9.6 × 10−7 | 0.0169 | 1.9 × 10−6 | 0.999 |
| 2 | 6.4 × 10−6 | 1.6 × 10−5 | 0.0169 | 3.2 × 10−5 | 0.998 |
| 1 | 1.5 × 10−4 | 1.8 × 10−4 | 0.0169 | 3.6 × 10−4 | 0.984 |
| 0 | 2.4 × 10−2 | 2.4 × 10−3 | 0.0179 | 4.8 × 10−3 | 0.931 |
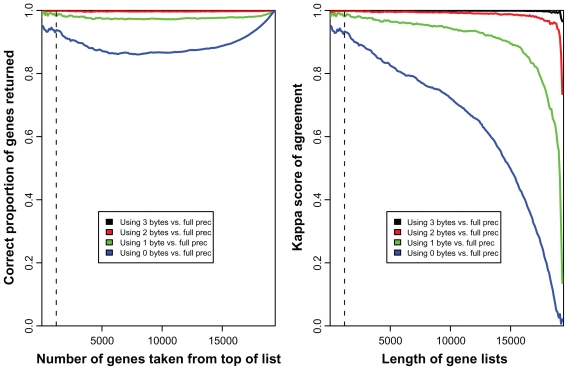
Comparing gene lists of various lengths in Figure 3, the performance of using 2 or 3 bytes to store location coordinates is indistinguishable from full precision in terms of the top x-ranked genes returned. Naturally if we return 19443 probes (the complete set) in our gene-list then the proportion of genes that match will be 1. The decline in performance for longer gene lists can be attributed to the volatility of what is an effectively random ordering of all of the genes for which there is no differential expression.
Figure 3.
Impact of reducing the precision of the stored bead-coordinates on the downstream analysis of a 12 array expression experiment. For 19443 well-annotated probes, top gene-lists are compared across analyses based upon full precision and reduced precision. The left-hand panel gives, for varying lengths of gene list, the proportion of genes from the full-precision analysis that are returned in the reduced-precision analysis. The right-hand panel gives, for varying lengths of gene list, Cohen’s kappa score of agreement for the two partitions (one from the full-precision analysis and one from the reduced-precision analysis). The full-precision analysis suggests that approximately 1200 probes show differential expression and this length of gene list is indicated.
This aspect is highlighted when considering not only the genes that are on the genelist, but looking at agreement beyond chance of the two sets of genes (those that are on the gene-list and those that are not). Here the performances of the different precision levels become more apparent, but within a length of genelist that might be reasonable for this dataset, the conclusion that using 2 or 3 bytes to store location coordinates has no detrimental effect, and that using 1 byte has a minimal effect, holds.
Discussion
We have focused on the problem of compressing the .txt and .locs files and have ignored the image files. Image compression, and in particular that of microarray images is long studied, and we would note that the compression of images using standard tools works well (image files compressed in either bzip2 or gzip format can be read directly using the beadarray package). Also it is entirely reasonable to share the .txt and .locs files but not the images, whereas the images without these files are of no value.
BeadDataPackR is designed primarily to compress bead-level data produced directly from the scanner. It is reasonable that a user may wish to compress data they have generated themselves (for example to include intensities calculated using an alternative method or to distribute values for only a subset of the beads on an array). Although this is possible by generating a .txt file in place of that provided by Illumina, it would be beneficial to be able to generate a .bab file from within standard analysis tools.
The format might also be extended to include multiple arrays (perhaps a whole experiment) in the same file. This would achieve only minimal savings in file size, but due to the file format would allow for streamed access of data from large experiments, rather than having to have all of the data in memory all of the time. Similarly, it may be advantageous to store a linear transformation (of full rank) of the intensities in order that quantities such as the mean for a bead-type could be read directly from the .bab file without increasing file size, yet preserving the ability to extract the individual intensities.
Our aim is to increase access to raw BeadArray data by making it practicable both to share and to download experimental data, and this is achieved by the BeadDataPackR package. It should be noted there is value for the storage of data also. If a typical single color array generates 125MB of data, we can see this reduced to 50MB (with approximately 25MB of savings coming from BeadDataPackR, and 50MB coming from the image compression). Institutes producing and storing terabytes of raw Illumina BeadArray data will naturally see substantial savings on their storage costs, while for the individual investigator, the size of an experiment that will fit onto a CD or DVD is more than doubled.
The savings on size are greater still if we reduce the precision stored, and we have seen that there is scope for such reduction with no impact on the quality of down-stream analyses. If there were a move towards storing/providing only the raw text files and not images, then still further savings could be achieved by abandoning true locations and storing only relative grid positions reducing file sizes for a single array to ~5MB.
Acknowledgements
We thank Simon Tavaré, Jonathan Cairns and Mark Dunning for many helpful discussions. We thank Jason Carroll and Roslin Russell for access to the illustrative data used. We acknowledge the support of The University of Cambridge, Cancer Research UK; Hutchison Whampoa Limited.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Fridlyand J, Snijders AM, Ylstra B, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.S⊘rlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight J. News feature: when the chips are down. Nature. 2001;410:860–1. doi: 10.1038/35073680. [DOI] [PubMed] [Google Scholar]
- 5.Dupoy A, Simon RM. Critical Review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–57. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 6.Tseng GC, Oh M-K, Rohlin L, Liao JC, Wong WH. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Research. 2001;29:2549–57. doi: 10.1093/nar/29.12.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 8.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genetics. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson KL, Kruglyak S, Graige MS, et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–7. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azzato EM, Driver KE, Lesueur F, et al. Effects of common germline genetic variation in cell cycle control genes on breast cancer survival: results from a population-based cohort. Breast Cancer Res. 2008;10:R47. doi: 10.1186/bcr2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papatheodorou I, Chrichton C, Morris L, et al. A metadata approach for clinical data management in translational genomics studies in breast cancer. BMC Medical Genomics. 2009;2:66. doi: 10.1186/1755-8794-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acevado LG, Bleda M, Green R, Farnham PJ. Analysis of the mechanisms mediating tumor-specific changes in gene expression in human liver tumors. Cancer Research. 2008;68:2641–51. doi: 10.1158/0008-5472.CAN-07-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MD, Barbosa-Morais NL, Schmidt D, et al. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–8. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Simon R. BRB-ArrayTools data archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Informatics. 2008;6:9–15. doi: 10.4137/cin.s448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonardi S, Luo Y. Gridding and compression of microarray images. Proc IEEE Comput Syst Bioinform Conf. 2004;2004:122–30. doi: 10.1109/csb.2004.1332424. [DOI] [PubMed] [Google Scholar]
- 17.Smith ML, Dunning MJ, Tavaré S, Lynch AG. Identification and correction of previously unreported spatial phenomena using raw Illumina BeadArray data. BMC Bioinformatics. 2010;11:208. doi: 10.1186/1471-2105-11-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunning MJ, Cairns J, Russell R, Lynch AG. Fortifying the analysis of Illumina data. Proceedings of IASC2008 (4th World conference of the the International Association for Statistical Computing); Yokahama, Japan. 2008 Dec 5–8. [Google Scholar]
- 19.Stokes TH, Han X, Moffitt RA, Wang MD. Extending microarray quality control and analysis algorithms to Illumina chip platform. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:4637–40. doi: 10.1109/IEMBS.2007.4353373. [DOI] [PubMed] [Google Scholar]
- 20.Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG. BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics. 2008;24:2921–2. doi: 10.1093/bioinformatics/btn557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y, Wang X, Story M. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics. 2009;25:751–7. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunning MJ, Barbosa-Morais NL, Lynch AG, Tavaré S, Ritchie ME. Statistical issues in the analysis of Illumina data. BMC Bioinformatics. 2008;9:85. doi: 10.1186/1471-2105-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Research. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi W, Banerjee A, Ritchie ME, Gerondakis S, Smyth G. Illumina WG-6 BeadChip strips should be normalized separately. BMC Bioinformatics. 2009;10:372. doi: 10.1186/1471-2105-10-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch AG, Dunning MJ, Iddawela M, Barbosa-Morais NL, Ritchie ME. Considerations for the processing and analysis of GoldenGate-based two-colour Illumina platforms. Stat Methods Med Res. 2009;18:437–52. doi: 10.1177/0962280208099451. [DOI] [PubMed] [Google Scholar]
- 26.Smith ML, Lynch AG. BeadDataPackR [BioConductor package] [Accessed 2010 Aug 23]. Available at http://bioconductor.org/packages/release/bioc/html/BeadDataPackR.html.
- 27.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross-Innes CS, Stark R, Holmes KA, et al. Cooperative interaction between retinoic acid receptor-a and estrogen receptor in breast cancer. Genes and Development. 2010;24:171–82. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunning MJ, Smith ML, Ritchie ME, Tavaré S. BeadArrayR classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–4. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 30.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 31.Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Research. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]