Abstract
In this study we evaluate the suitability of two methods of RNA conservation in blood samples, PAXgene and RNAlater, in combination with variable shipping conditions for their application in multicenter studies and biobanking. RNA yield, integrity, and purity as well as levels of selected mRNA and microRNA species were analyzed in peripheral human blood samples stabilized by PAXgene or RNAlater and shipped on dry ice or at ambient temperatures from the study centers to the central analysis laboratory. Both examined systems were clearly appropriate for RNA stabilization in human blood independently of the shipping conditions. The isolated RNA is characterized by good quantity and quality and well suited for downstream applications like quantitative RT-PCR analysis of mRNA and microRNA. Superior yield and integrity values were received using RNAlater. It would be reasonable to consider the production and approval of blood collection tubes prefilled with RNAlater to facilitate the use of this excellent RNA stabilization system in large studies.
Keywords: RNA, PAXgene, RNAlater, biobank, marker, blood, pre-analytical variation
Introduction
A major aim in cancer research is the identification and validation of appropriate tumor markers detectable in easily accessible body fluids. Human blood is a preferred source of markers due to the minimal invasive character of sample collection and the vascularization of most tissues (including tumors). For marker discovery the analysis of mRNA expression signatures in peripheral human blood has been widely used showing to be a promising technique.1 Once markers with potential diagnostic relevance are identified they need to be validated in large prospective studies, commonly conducted in multicenter studies. The different participating study centers are usually separated by varying distances from a central analysis laboratory or biobank. Study centers face the challenge of having limited access to research laboratory support and might be unable to perform RNA isolation immediately after blood collection.2 Additionally, isolation and analysis of RNA in a central laboratory avoid inter-laboratory variations.3 Such a study design results in unique challenges due to pre-analytical variations occurring before marker analyses and includes factors like method of blood collection, shipping conditions, and time delay between sample collection and analysis.4 RNA stability plays a crucial role due to pre-analytical variations, especially degradation by endogenous RNases and unintentional expression of individual genes after blood drawing.5 Altered expression signatures based on pre-analytical variations lead to false assessments of potential markers. Therefore, evaluation of influences on marker integrity and standardization of sample collection, shipping, and storage conditions in biobanking is necessary to ensure reliable and reproducible analyses of markers.2
Discovery of microRNAs (miRNAs) has opened new opportunities for markers in the diagnosis of cancer.6 MiRNAs are small (18–26 nucleotides) noncoding RNAs playing a central role in the regulation of gene expression.7 Altered miRNA expression has been reported in several human malignancies and differences between tumor tissues and their normal counterparts could be exploited for diagnostics and prognosis.8
The introduction of blood collection systems containing stabilizing additives has significantly improved the RNA quantity and quality of blood samples collected in multicenter studies.9,10 RNA stabilization systems have the advantage that there is no need to isolate the RNA immediately. Instead, the collected blood samples can be stored at more accessible temperatures in the study centers before shipment to the central analysis laboratory or biobank resulting in reduced pre-analytical variability. A well-described method for RNA stabilization in human blood is the PAXgene system.9,11–13 In contrast to mRNAs less is known about the ability of PAXgene to stabilize miRNAs in peripheral blood samples. Recently, it has been reported that miRNAs could be isolated from PAXgene-stabilized blood and that quantity and quality might be sufficient for downstream applications.14 However, failure of PAXgene to stabilize specific transcripts was also reported.15 Therefore, alternative methods for RNA stabilization in blood need to be evaluated. RNAlater is a common stabilization reagent for RNA in cells and tissues increasingly being used in genomic studies and biobanking.16 To our knowledge, no study analyzing the usability of RNAlater for RNA stabilization in human peripheral blood was conducted so far.
The aim of our study was to evaluate the performance of PAXgene and RNAlater for RNA stabilization in peripheral blood and the influences of shipping conditions and subsequent storage on RNA yield, integrity, purity, and amount of specific mRNA and miRNA species.
Material and Methods
Sample collection and shipping conditions
In March and April 2009 peripheral blood samples from 43 male former uranium miners (mean age 78 years, range 76–89 years) exposed to radon and other carcinogens were collected in three participating study centers. As study centers served local medical offices assigned to exammine former uranium miners in Schneeberg, Plauen, and Niederdorf in Thuringia, Germany. All volunteers provided written informed consent and permission was obtained from the ethics committee of the Bavarian Chamber of Physicians (reference number 08082).
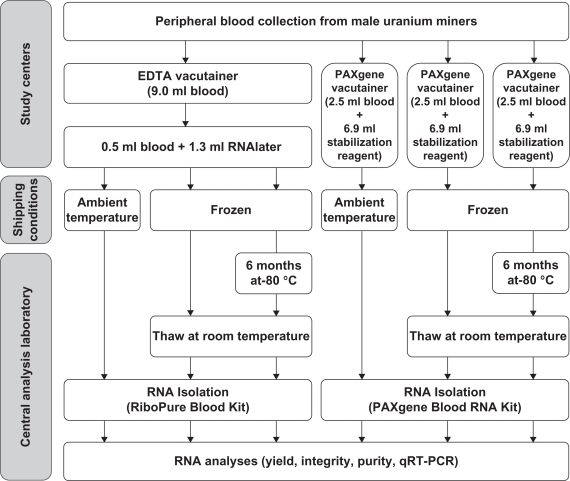
The workflow of the study design is presented in Figure 1. In brief, from each donor peripheral blood was collected in a 9.0 ml EDTA vacutainer (Becton Dickinson, Heidelberg, Germany) and three 2.5 ml PAXgene Blood RNA vacutainers (PreAnalytiX, Hombechtikon, Switzerland). Aliquots (0.5 ml each) of EDTA blood were mixed with 1.3 ml RNAlater (Ambion, Austin, TX, USA) immediately after blood drawing. Until shipment, two blood+RNAlater aliquots (RNAlater/frozen, RNAlater/frozen+stored) and two PAXgene vacutainers (PAXgene/frozen, PAXgene/frozen+stored) were stored on dry ice17 and the remaining PAXgene vacutainer (PAXgene/ambient) and blood+RNAlater aliquot (RNAlater/ambient) were stored at ambient temperature. Shipment of blood samples from the study centers to the central analysis laboratory was performed frozen on dry ice by courier service over night (PAXgene/frozen, PAXgene/frozen+stored, RNAlater/frozen, RNAlater/frozen+stored) and at ambient temperatures by regular mail with possible delay in delivery (PAXgene/ambient, RNAlater/ambient). After arrival at the central analysis laboratory PAXgene/frozen+stored and RNAlater/frozen+stored samples were stored at −80 °C for six months until processing. All other blood samples were processed immediately.
Figure 1.
Workflow of the study design.
RNA Isolation
Frozen samples were thawed at room temperature and RNA was isolated immediately after thawing using the PAXgene Blood RNA Kit (Qiagen, Hilden, Germany) for the PAXgene samples and the RiboPure Blood Kit (Ambion) for the RNAlater samples. RNA was isolated according to the manufacturers’ instructions and stored in elution buffers supplied with the isolation kits at −80 °C until analysis.
RNA Yield, Purity, and Integrity
Concentration and purity of RNA were quantified by measuring the absorbance at 260 nm (A260) and 280 nm (A280) using a NanoDrop ND-100 spectrophotometer (Thermo Scientific, Waltham, MA, USA). RNA yields were calculated from RNA concentrations relative to one milliliter of blood. Qualitative analyses of RNA were performed with the Agilent 2100 Bioanalyzer using RNA 6000 Nano and RNA 6000 Pico assays (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s instructions. The Bioanalyzer system is based on capillary electrophoresis, which has become an established and common tool18 allowing separation with high resolution as well as quantification with high accuracy19 and is therefore suitable for reliable quality control of RNA.20 According to Schroeder et al RNA integrity numbers (RIN) from 0–10 (low to high RNA quality) were calculated using the Agilent software.21
Quantitative Real Time-PCR (qRT-PCR)
The ATM gene (ataxia-telangiectasia mutated) was selected as target because altered expression was observed after exposure to ionizing radiation.22 RNA was analyzed using the TaqMan reverse transcription reagents and the TaqMan gene expression assays for ATM (order number Hs00175892) and GAPDH (order number Hs99999905) according to the manufacturer’s instructions. All primers and reagents were obtained from Applied Biosystems (Foster City, CA, USA). For cDNA synthesis samples were incubated at 42 °C for 80 min followed by an incubation at 95 °C for 5 min. Using the miRGen database23 miRNA-26a and miRNA-26b were determined as possible regulators due to their potential binding capacity to the ATM mRNA. MiRNAs were analyzed using the commercial Taq-Man microRNA assays for miRNA-26a (order number 4373070) and miRNA-26b (order number 4373069) obtained from Applied Biosystems and performed as described previously.24 For cDNA synthesis samples were incubated at 16 °C for 30 min, 42 °C for 30 min, and at 85 °C for 5 min. Quantitative real time-PCR (qRT-PCR) was performed using a MJ Research PTC-200 thermal cycler (Global Medical Instrumentation, Inc., Ramsey, MN, USA) and an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems). Samples were initially denatured at 95 °C for 10 min, followed by 40 sequential cycles at 95 °C for 15 sec and at 60 °C for 1 min. Non-template controls were included in all assays. Raw Ct values of all samples are listed in Supplementary Table 1.
Table 1.
Median values and inter-quartile ranges (IQR) of RNA yield, integrity, and purity subjected to stabilization systems, shipping conditions, and subsequent storage.
| N |
RNA yield (μg/ml blood) |
RNA integrity (RIN) |
RNA purity (A260/A280) |
||||
|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | ||
| PAXgene/frozen | 41 | 1.6 | 0.8–2.1 | 7.4 | 4.2–8.3 | 2.08 | 2.04–2.13 |
| RNAlater/frozen | 43 | 7.5 | 5.0–9.3 | 8.5 | 8.1–9.1 | 1.95 | 1.89–2.02 |
| PAXgene/ambient | 43 | 1.4 | 0.8–2.1 | 6.9* | 5.4–7.8* | 2.06 | 1.96–2.10 |
| RNAlater/ambient | 43 | 4.4 | 2.7–6.2 | 8.2 | 7.7–8.7 | 1.99 | 1.87–2.16 |
| PAXgene/frozen+stored | 41 | 2.2 | 1.5–3.0 | 7.8 | 7.4–8.3 | 2.08 | 2.04–2.12 |
| RNAlater/frozen+stored | 43 | 6.2 | 5.0–8.4 | 8.6 | 8.1–9.0 | 1.92 | 1.84–2.04 |
Note:
N = 42 (Determination of RIN failed in one sample).
The cycle threshold (Ct) is defined as the number of cycles required for the FAM signal to cross the threshold in qRT-PCR. For Ct estimation a fixed threshold of 0.2 was used.25 Targets with Ct > 35 are considered to be under the detection limit26 and marked as 35 for calculation.27 Samples were analyzed in duplicate and ratios of ATM mRNA, miRNA-26a, and miRNA-26b to GAPDH mRNA were calculated using mean Ct values.
Statistical Analysis
Median values and inter-quartile ranges (IQR) were calculated for RNA yield, purity, and integrity stratified by stabilization systems and shipping conditions. Differences between the paired observations were determined by Wilcoxon signed-rank test. A repeated measurement analysis of variance (ANOVA) of stabilization system and shipping conditions as main effects and their interaction was performed with ranks for RNA yield, integrity, and Ct values. P-values were adjusted for multiple testing by Bonferroni-Holm correction.
Results
Sample collection and shipment
From 258 anticipated blood samples of 43 participants (six per donor) two samples for PAXgene/frozen+stored could not be drawn. All samples shipped by courier service from the participating study centers to the central analysis laboratory were delivered on the next day without any delay. Five PAXgene/ambient and five RNAlater/ambient samples shipped by regular mail arrived with a delay of four days whereas all other samples shipped by mail (N = 38) were delivered not later than two days after sending.
RNA Recovery
RNA could be isolated from 254 of 256 delivered blood samples, including all RNAlater/frozen, RNAlater/ambient, RNAlater/frozen+stored, PAXgene/ambient, and PAXgene/frozen+stored samples. Isolation of two PAXgene/frozen samples failed due to imperfect filling of the vacutainers.
Median values and IQR of RNA yield, integrity, and purity subjected to stabilization system, shipping conditions, and subsequent storage are presented in Table 1. Highest yield (7.5 μg/ml blood, IQR 5.0–9.3 μg/ml blood) was observed in RNAlater/frozen samples and lowest yield (1.4 μg/ml blood, IQR 0.8–2.1 μg/ml blood) in PAX-gene/ambient samples. Highest RIN value was 8.6 (IQR 8.1–9.0) for RNAlater/frozen+stored samples and lowest RIN value was 6.9 (IQR 5.4–7.8) for PAXgene/ambient samples. Median purity of the isolated RNA ranged from 1.92 for RNAlater/frozen+stored (IQR 1.84–2.04) to 2.08 for PAXgene/frozen and PAXgene/frozen + stored (IQR 2.04–2.13 and 2.04–2.12, respectively).
Significant influence of the stabilization systems on RNA yield and integrity was demonstrated, whereas shipping conditions had a significant impact on yield only. No clear interaction of stabilization systems and shipping conditions could be found. Overall, higher yields and RIN values were observed in RNAlater-stabilized samples compared to PAXgene-stabilized samples independent of shipping conditions or storage (P < 0.0001). Frozen shipping of RNAlater samples resulted in higher yield compared to shipping at ambient temperature (7.5 vs. 4.4 μg/ml; P < 0.0001). Using PAXgene a higher average yield was achieved for frozen+stored samples in comparison to frozen samples (2.2 vs. 1.6 μg/L; P = 0.0002).
Median values and IQR of RNA yield, integrity, and purity of samples shipped at ambient temperatures due to different delivery times are presented in Table 2. Differences could not be evaluated because of the small number of samples.
Table 2.
Median values and inter-quartile ranges (IQR) of RNA yield, integrity, and purity of PAXgene- and RNA later-stabilized blood samples shipped at ambient temperature by regular mail and with different delivery times.
| Sample | Delivery time | N |
RNA yield (μg/ml blood) |
RNA integrity (RIN) |
RNA purity (A260/A280) |
|||
|---|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | |||
| PAXgene | 1–2 days | 38 | 1.6 | 0.8–2.1 | 7.3* | 5.6–7.8* | 2.01 | 1.96–2.09 |
| 4 days | 5 | 1.2 | 0.9–1.2 | 6.1 | 5.4–6.5 | 2.11 | 2.07–2.11 | |
| RNAlater | 1–2 days | 38 | 4.4 | 2.6–6.2 | 8.2 | 7.7–8.7 | 2.01 | 1.90–2.17 |
| 4 days | 5 | 4.6 | 4.2–5.9 | 8.5 | 8.4–8.6 | 1.84 | 1.80–1.87 | |
Note:
N = 37 (Determination of RIN failed in one sample).
RNA Analyses
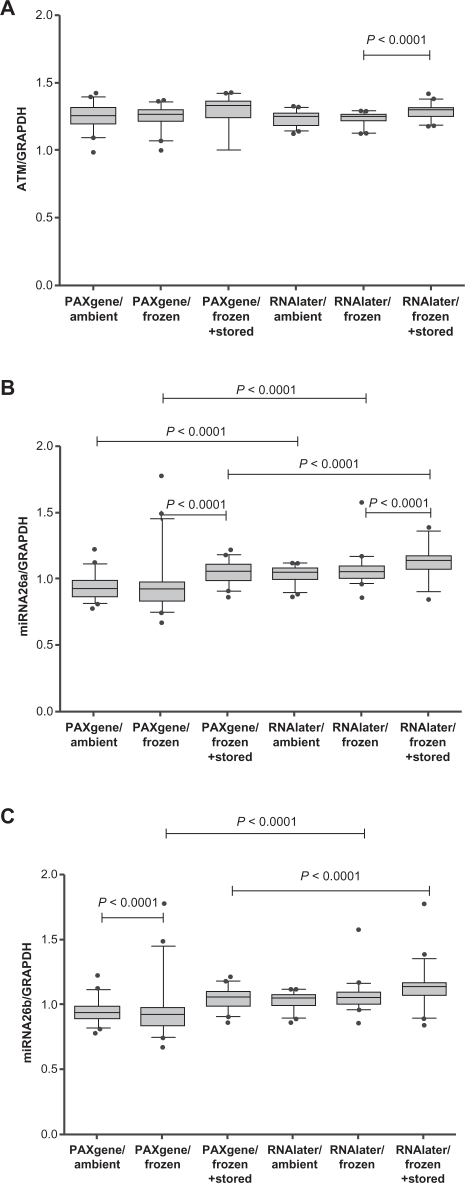
Amounts of ATM mRNA were not influenced by stabilization system or shipping conditions, whereas for miRNA-26a and miRNA-26b a significant impact of the stabilization systems was observed. The interaction of stabilization systems and shipping conditions had a significant impact only for miRNA-26b. ATM mRNA amounts were altered between RNAlater/frozen and RNAlater/frozen+stored samples (P <0.0001) (Fig.2A). Different amounts of miRNA-26a were revealed for PAXgene/frozen vs. RNAlater/frozen (P < 0.0001), PAXgene/ambient vs. RNAlater/ambient (P < 0.0001), PAXgene/frozen vs. PAXgene/frozen+stored (P < 0.0001), PAXgene/frozen+stored vs. RNAlater/frozen+stored (P < 0.0001), and RNAlater/frozen vs. RNAlater/frozen+stored (P < 0.0001), (Fig. 2B). For miRNA-26b altered amounts were observed for PAXgene/frozen vs. PAXgene/ambient (P < 0.0001), PAXgene/frozen vs. RNAlater/frozen (P < 0.0001), and PAXgene/frozen+stored vs. RNAlater/frozen+ stored (P < 0.0001), (Fig. 2C).
Figure 2.
Normalized levels of ATM (A), miRNA-26a (B), and miRNA-26b (C) depending on stabilization systems, shipping conditions, and subsequent storage.
Differences of ATM mRNA, miR-26a, and miR-26b amounts due to different delivery times could not be evaluated because of the small number of samples (data not shown).
Discussion
A crucial challenge of gene expression analyses for marker discovery is sample handling after blood collection when immediate RNA isolation is not possible.2 For multicenter studies with blood-based samples the choice of stabilization systems and shipping conditions is important because RNA degrades time-dependently during sample shipment to the central analysis laboratory.28 Additionally, sufficient sample integrity during short-term and long-term storage in biobanks should be warranted.
We analyzed two systems regarding their performance to stabilize mRNA and miRNA in human peripheral blood and the influence of sample shipment under frozen conditions per courier service and at ambient temperatures by regular mail, with and without additional delay in delivery.
The obtained RNA yields in RNAlater-stabilized samples are consistently higher than in PAXgene-stabilized samples. Discrepancies might be due to susceptibility of PAXgene to an imperfect ratio of stabilization reagent to blood caused by insufficient filling of the vacutainers. Disparities result in poor RNA isolation or even in failed RNA isolation as happened in two cases during this study. A simple strategy to check the filling degree of the PAXgene vacutainers can be weighing. PAXgene vacutainers filled completely with 2.5 ml blood have a weight of 19.35 g and for successful RNA isolation a tolerance range of ±0.5 g might be acceptable (personal communication T. Voss, Qiagen, Germany). However, when strictly adhering to the manufacturers’ protocols both stabilization systems provide sufficient RNA yields for downstream applications.
Analysis of RNA integrity and purity served as tool to determine the overall quality of stabilization, preparation, and purification that can be influenced by degradation.3 Regarding purity, ratios >1.8 are considered suitable for gene expression analyses.20 Good RNA purities were obtained throughout for all stabilization systems, shipping conditions, and subsequent storage, reflecting efficient isolation and purification procedures. For RNA quality it was suggested that RIN values should be at least >5 and values >8 are perfect for downstream applications.18,29 Samples stabilized with RNAlater provided anaverage integrity >8 while PAXgene resulted in slightly lower RIN values. Based on the obtained results isolated RNA stabilized by either PAXgene or RNAlater is highly appropriate for qRT-PCR.
While the quality of RNA differed by stabilization system, no differences of ATM mRNA levels could be observed in RNAlater and PAXgene stabilized blood samples shipped frozen or at ambient temperatures, indicating that both systems and shipping conditions are highly feasible for stabilization of mRNA. However, it was reported that PAXgene failed to stabilize certain transcripts.15 Therefore, before starting multicenter studies it will be meaningful to analyze the intended stabilization system under the selected shipping condition with the transcripts of interest. Because only one mRNA species was analyzed in our study, a failed stabilization of other mRNAs by RNAlater or PAXgene cannot be ruled out. Surprisingly, despite a general excellent stability of miRNAs in human blood30 miRNA-26a and miRNA-26b appeared to be more susceptible to stabilization systems and shipping conditions, similar to the failed stabilization of certain mRNAs observed by Kagedal et al.15 Indeed, it was demonstrated recently that individual miRNAs possess different stabilities31 while little is known about the specific stabilities of miRNA-26a and miRNA-26b. In this context it should be noted that the isolation procedures used in this study were not specifically adapted for miRNAs. Therefore, some variations of miRNA levels might be due to shortcomings of the isolation procedures. Also, because different RNA isolation procedures were used, the differences between the two stabilization systems might be due, in part, to the different RNA kits. Meanwhile, optimized isolation protocols for PAXgene and RNAlater became available that have the potential to enhance the recovery rates of miRNAs in the stabilized samples. Recently, Gaartz et al successfully used a specific miRNA isolation kit for PAXgene-stabilized blood samples.32
Our results clearly indicate that both stabilization systems are well suited for the application in multicenter studies. Particularly, shipping of collected samples by regular mail at ambient temperatures is simple, saves cost, and greatly reduces the workload for participating study centers that are usually not accustomed to perform scientific studies. Based on our results the use of PAXgene or RNAlater facilitates the stabilization of RNA for several days at ambient temperatures without consistent alterations in yield, integrity, and purity of total RNA and levels of individual mRNA and miRNA species.
Regarding the storage of samples in biobanks, both stabilization systems seem to be suitable at least for short-term storage (up to six months) as indicated by the results of this study. Regarding long-term storage PAXgene samples could be stored for up to 50 months at −80 °C without any alterations in RNA levels according to the manufacturers’ protocol. For long-term storage of RNAlater-stabilized blood samples nothing is known yet.
With regard to the handling in fieldwork, the use of PAXgene is favorable because the staff in study centers does not need experience in sample processing. As blood-collection systems already prefilled with RNAlater are not available yet, the blood collecting tubes need to be opened to mix the blood with RNAlater. Besides requiring technical expertise this additional step possesses a potential risk of infection. Based on the good performance of RNAlater to stabilize RNA in human peripheral blood, the production and approval of blood collection tubes prefilled with RNAlater would support the use of this excellent isolation system in large studies.
In conclusion, our study shows that PAXgene and RNAlater were both appropriate for RNA stabilization in blood independently of the shipping conditions. Both systems facilitate the isolation of stabilized RNA from blood, which is characterized by good quantity and quality and is well suited for qRT-PCR of mRNA and miRNA. To our knowledge, this is the first study investigating the use of RNAlater for stabilization of RNA in human peripheral blood. It could be shown that RNAlater performs well and constitutes an alternative to existing systems, provided that a user-friendly handling, by i.e. prefilled collection tubes, will be possible in the future. However, evaluation of a large panel of mRNAs and miRNAs is necessary to validate RNAlater for the use of RNA stabilization in blood.
Acknowledgments
This work was supported by a grant (3608S04532) from The Federal Office for Radiation Protection. We thank J. Schreiber for skilled technical support. We are grateful to K.-D. Bergert, D. Hahn, and G. Meichsner for collaboration in obtaining human blood samples.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100(4):1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matheson LA, Duong TT, Rosenberg AM, Yeung RS. Assessment of sample collection and storage methods for multicenter immunologic research in children. J Immunol Methods. 2008;339(1):82–9. doi: 10.1016/j.jim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Langebrake C, Gunther K, Lauber J, Reinhardt D. Preanalytical mRNA stabilization of whole bone marrow samples. Clin Chem. 2007;53(4):587–93. doi: 10.1373/clinchem.2006.078592. [DOI] [PubMed] [Google Scholar]
- 4.Fraser CG. Inherent biological variation and reference values. Clin Chem Lab Med. 2004;42(7):758–64. doi: 10.1515/CCLM.2004.128. [DOI] [PubMed] [Google Scholar]
- 5.Benoy IH, Elst H, van Dam P, et al. Detection of circulating tumour cells in blood by quantitative real-time RT-PCR: effect of pre-analytical time. Clin Chem Lab Med. 2006;44(9):1082–7. doi: 10.1515/CCLM.2006.210. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila Pa) 2009;2(9):807–13. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 9.Rainen L, Oelmueller U, Jurgensen S, et al. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48(11):1883–90. [PubMed] [Google Scholar]
- 10.Thach DC, Lin B, Walter E, et al. Assessment of two methods for handling blood in collection tubes with RNA stabilizing agent for surveillance of gene expression profiles with high density microarrays. J Immunol Methods. 2003;283(1–2):269–79. doi: 10.1016/j.jim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Chai V, Vassilakos A, Lee Y, Wright JA, Young AH. Optimization of the PAXgene blood RNA extraction system for gene expression analysis of clinical samples. J Clin Lab Anal. 2005;19(5):182–8. doi: 10.1002/jcla.20075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pahl A, Brune K. Gene expression changes in blood after phlebotomy: implications for gene expression profiling. Blood. 2002;100(3):1094–5. doi: 10.1182/blood-2002-03-0813. [DOI] [PubMed] [Google Scholar]
- 13.Ovstebo R, Lande K, Kierulf P, Haug KB. Quantification of relative changes in specific mRNAs from frozen whole blood—methodological considerations and clinical implications. Clin Chem Lab Med. 2007;45(2):171–6. doi: 10.1515/CCLM.2007.035. [DOI] [PubMed] [Google Scholar]
- 14.Kruhoffer M, Dyrskjot L, Voss T, et al. Isolation of microarray-grade total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. J Mol Diagn. 2007;9(4):452–8. doi: 10.2353/jmoldx.2007.060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kagedal B, Lindqvist M, Farneback M, Lenner L, Peterson C. Failure of the PAXgene Blood RNA System to maintain mRNA stability in whole blood. Clin Chem Lab Med. 2005;43(11):1190–2. doi: 10.1515/CCLM.2005.206. [DOI] [PubMed] [Google Scholar]
- 16.Florell SR, Coffin CM, Holden JA, et al. Preservation of RNA for functional genomic studies: a multidisciplinary tumor bank protocol. Mod Pathol. 2001;14(2):116–28. doi: 10.1038/modpathol.3880267. [DOI] [PubMed] [Google Scholar]
- 17.Beekman JM, Reischl J, Henderson D, et al. Recovery of microarray-quality RNA from frozen EDTA blood samples. J Pharmacol Toxicol Methods. 2009;59(1):44–9. doi: 10.1016/j.vascn.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28(19):1601–13. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- 19.Cerkovnik P, Perhavec A, Zgajnar J, Novakovic S. Optimization of an RNA isolation procedure from plasma samples. Int J Mol Med. 2007;20(3):293–300. [PubMed] [Google Scholar]
- 20.Becker C, Hammerle-Fickinger A, Riedmaier I, Pfaffl MW. mRNA and microRNA quality control for RT-qPCR analysis. Methods. 2010;50(4):237–43. doi: 10.1016/j.ymeth.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innes CL, Heinloth AN, Flores KG, et al. ATM requirement in gene expression responses to ionizing radiation in human lymphoblasts and fibroblasts. Mol Cancer Res. 2006;4(3):197–207. doi: 10.1158/1541-7786.MCR-05-0154. [DOI] [PubMed] [Google Scholar]
- 23.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35(Database issue):D149–55. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guthrie JL, Seah C, Brown S, Tang P, Jamieson F, Drews SJ. Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J Clin Microbiol. 2008;46(11):3798–9. doi: 10.1128/JCM.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ning B, Dial S, Sun Y, Wang J, Yang J, Guo L. Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen. 2008;13(3):194–201. doi: 10.1177/1087057108315513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst T, Hoffmann J, Erben P, Hehlmann R, Hochhaus A, Muller MC. Suitability of the PAXgene system to stabilize bone marrow RNA in imatinib-resistant patients with chronic myeloid leukemia. Clin Chem Lab Med. 2008;46(3):318–22. doi: 10.1515/CCLM.2008.086. [DOI] [PubMed] [Google Scholar]
- 29.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27(2–3):126–39. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3(11):e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bail S, Swerdel M, Liu H, et al. Differential regulation of microRNA stability. RNA. 2010;16(5):1032–9. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaarz A, Debey-Pascher S, Classen S, et al. Bead array-based microrna expression profiling of peripheral blood and the impact of different RNA isolation approaches. J Mol Diagn. 2010;12(3):335–44. doi: 10.2353/jmoldx.2010.090116. [DOI] [PMC free article] [PubMed] [Google Scholar]