Abstract
The mammalian reoviruses and rotaviruses have evolved specific mechanisms to evade the Type I interferon (IFN) antiviral response. Rotavirus likely represses the IFN response by at least 4 mechanisms. First, the rotavirus protein NSP1, most likely functioning as an E3 ligase, can induce proteasome-dependent degradation of the transcription factors IRF3, IRF5, and IRF7 to prevent their induction of IFN. Second, NSP1 can induce proteasome-dependent degradation of the ubiquitin ligase complex protein β-TrCP, resulting in stabilization of IκB and concomitant failure of virus to activate NF-κB for induction of IFN. Third, rotavirus may sequester NF-κB in viroplasms. And fourth, rotavirus can prevent STAT1 and STAT2 nuclear translocation. The predominant mechanism for rotavirus inhibition of the IFN response is likely both rotavirus strain-specific and cell type-specific. The mammalian reoviruses also display strain-specific differences in their modulation of the IFN response. Reovirus activates RIG-I and IPS-1 for phosphorylation of IRF3. Reovirus-induced activation of MDA5 also participates in induction if IFN-β, perhaps through activation of NF-κB. Reovirus likely inhibits the IFN response by at least 3 virus strain-specific mechanisms. First, the reovirus μ2 protein can induce an unusual nuclear accumulation of IRF9 and repress IFN-stimulated gene (ISG) expression, most likely by disrupting IRF9 function as part of the heterotrimeric transcription factor complex, ISGF3. Second, the reovirus σ3 protein can bind dsRNA and prevent activation of the latent antiviral effector protein PKR. And third, genetic approaches have identified the reovirus λ2 and σ2 proteins in virus strain-specific modulation of the IFN response, but the significance remains unclear. In sum, members of the family Reoviridae have evolved a variety of mechanisms to subvert the host's innate protective response.
Rotaviruses and Reoviruses
The rotavirus and orthoreovirus (“reovirus”) genera fall in the family Reoviridae and include viruses that infect a wide variety of mammalian species including humans. Rotaviruses are responsible for up to 50% of diarrheal illnesses in infants and young children worldwide and account for a half million or more deaths in this age group annually (Estes and Kapikian 2007). Two vaccines (RotaTeq and Rotarix) are now recommended for routine use in the United States, but they have been available for only the past several years and their use and efficacy in developing countries remain untested (Dennehy 2008; Parashar and Glass 2009). Rotaviruses are also important pathogens in food animals (Saif and others 1994), and reassortment between animal and human rotaviruses as well as direct transmission from animals to humans has contributed to the diversity of rotaviruses associated with human disease (Gentsch and others 2005; Tsugawa and Hoshino 2008; Matthijnssens and others 2009). Reoviruses, in contrast, are not associated with serious human disease, but provide an excellent model for study of viral pathogenesis in neonatal mice (Schiff and others 2007). Studies using reovirus reassortants have identified the viral determinants of tropism and disease in the central nervous system (Weiner and others 1977) and heart (Sherry and Fields 1989; Sherry and Blum 1994), and replication in the gastrointestinal tract for spread to other animals (Keroack and Fields 1986; Bodkin and Fields 1989). The structure of the whole virion has been resolved by cryoelectron microscopy (Dryden and others 1993), and the reovirus core has been solved to 3.6 Å resolution (Reinisch and others 2000). Finally, the recent introduction of a reverse genetics system for reoviruses (Kobayashi and others 2007) currently unavailable for rotaviruses has provided the tool to identify determinants of pathogenesis at the single amino acid level (Danthi and others 2008). Therefore, the identification of mechanisms by which rotaviruses and reoviruses modulate the host innate response will provide complementary insights toward our goal of controlling disease.
Rotaviruses are comprised of 11 double-stranded RNA (dsRNA) gene segments encoding 13 mature proteins, and are assembled into a nonenveloped triple-layered infectious particle (Estes and Kapikian 2007). Reoviruses, in contrast, are comprised of 10 dsRNA gene segments encoding 12 proteins and are assembled into a nonenveloped particle composed of a core and outer capsid (Schiff and others 2007). Moreover, the 2 genera have remarkably different replication strategies. Synthesis and assembly for both viruses is exclusively cytoplasmic, although some viral proteins have been detected in the nucleus. Both viruses also induce the formation of distinctive cytoplasmic structures associated with their replication and assembly, known as viroplasms for rotavirus and viral inclusion bodies (VIBs) for reovirus. However, during replication the immature rotavirus subviral particle buds through the membrane of the endoplasmic reticulum and acquires a transient envelope, which is subsequently lost before maturation and exit from the cell. There is no analogous step for reovirus. Not surprisingly then, given the different structures and replication strategies, rotavirus and reovirus do not have clearly identifiable homologous genes or proteins apart from assignments based on enzymatic function. And as discussed later, there is currently no evidence that rotavirus and reovirus share mechanisms in their modulation of the host interferon (IFN) response.
General Viral Modulation of the IFN Response
Viral infection of most cells can induce expression and secretion of type I IFN (IFN-α or IFN-β) as described by Gale and Sen in this special issue of the Journal of Interferon and Cytokine Research. Viruses are recognized by cell sensors, which stimulate a cascade of events that activate transcription factors IFN regulatory factor-3 (IRF3) and NF-κB for induction of IFN-β and -α4. These cytokines are secreted and bind to the IFN-α/β receptor to activate Jak1 and Tyk2 (kinases), which phosphorylate and activate transcription factors STAT1 and STAT2 to form a heterotrimeric transcription factor complex (ISGF3) with a third transcription factor, IRF9. ISGF3 translocates to the nucleus and binds to IFN-stimulated response elements (ISREs) to induce transcription of hundreds of IFN-stimulated genes (ISGs), some of which are antiviral. ISG IRF7 is itself a latent transcription factor, which when activated by virus-induced phosphorylation forms a homodimer or heterodimer with IRF3, to further induce IFN-β and IFN-α4 in a positive amplification loop. The IFN pathway can involve other cell factors and is cell type-specific (van Boxel-Dezaire and others 2006). This rapid innate response provides critical protection, as evidenced by the increased virulence of many viruses in mice lacking the IFN-α/β receptor (Muller and others 1994; Shresta and others 2004; Ida-Hosonuma and others 2005; Koerner and others 2007; Ryman and others 2007).
In turn, viruses have evolved many mechanisms to sabotage this potent protective response ((Randall and Goodbourn 2008), and other reviews in this special issue). For example, they can inactivate factors upstream of IRF3 and NF-κB (Foy and others 2005; Barral and others 2007; Childs and others 2007; Mibayashi and others 2007; Alff and others 2008; Lu and others 2008), degrade STATs (Ulane and others 2005), prevent STAT phosphorylation (Guo and others 2005; Devaux and others 2007; Johnson and others 2008) or nuclear translocation (Reid and others 2006; Frieman and others 2007), decrease (Leonard and Sen 1996) or prevent IRF9 nuclear translocation (Barnard and McMillan 1999), and inhibit general nucleocytoplasmic transport (Porter and others 2006). Many antiviral ISGs are latent until activated by viral dsRNA. Viruses can express dsRNA-binding proteins (Langland and Jacobs 2004; Hakki and Geballe 2005), like reovirus protein σ3 (Imani and Jacobs 1988; Yue and Shatkin 1997), or dsRNA decoys to inhibit viral activation of these host antiviral proteins (Sharp and others 1993; Lei and others 1998), which can also participate in further induction of IFN-β (Malathi and others 2007). Accordingly, virus strain-specific differences in subversion of the IFN response can determine virulence (Seo and others 2002; Donelan and others 2003). Deletion of viral genes encoding IFN repressors provides a promising vaccine strategy and repressor proteins present attractive targets for therapeutic intervention (Borden and others 2007). This review will focus on our current understanding of rotavirus and reovirus modulation of the IFN response, with particular attention to virus strains and cell types given the precedents that both are relevant to this interaction.
Rotavirus Modulation of the IFN Response
Rotavirus infection induces IFN in humans (De Boissieu and others 1993) and animals (Chaplin and others 1996), and pretreatment with IFN can reduce diarrhea in calves (Schwers and others 1985); therefore the IFN response likely plays an important role in protection against natural disease. Investigations in mice suggest IFN can play a protective, exacerbating, or irrelevant role in rotavirus infection and disease (Petersen and others 1997; Angel and others 1999; Vancott and others 2003; Shivakumar and others 2004; Feng and others 2008), and that the role for IFN is virus strain-specific (Feng and others 2008). As discussed later, rotaviruses likely inhibit the IFN response by at least 4 mechanisms: (1) NSP1-mediated degradation of IRF proteins to subvert induction of IFN; (2) NSP1-mediated repression of NF-κB activation to subvert induction of IFN; (3) sequestration of NF-κB in viroplasms; and (4) inhibition of IFN-stimulated STAT nuclear accumulation (Fig. 1). Moreover, repression of the IFN response is dependent on both the rotavirus strain and the cell type.
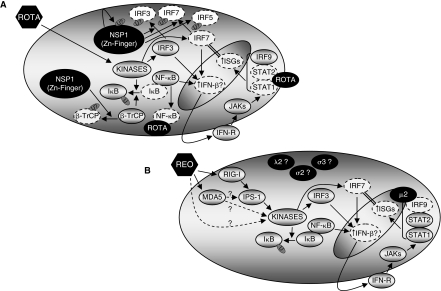
FIG. 1.
Rotavirus and reovirus modulation of the Type I interferon (IFN) response. (A) Rotavirus likely inhibits the IFN response by at least 4 virus strain-specific mechanisms. Rotavirus induces phosphorylation (activation) and nuclear translocation of IFN regulatory factor-3 (IRF3), and induces phosphorylation of IκB to release NF-κB from an inactive complex. (1) The rotavirus NSP1 protein can induce proteasome-mediated degradation of IRF3, IRF5, and IRF7 to subvert their induction of IFN-β. (2) NSP1 can induce proteasome-mediated degradation of β-TrCP, which normally participates in ubiquitination of IκB to induce its proteasome-mediated degradation. NSP1 can therefore stabilize IκB and repress NF-κB. (3) Rotavirus infection can result in sequestration of NF-κB in viroplasms, although it is unclear whether this has functional consequences. (4) Rotavirus can prevent nuclear translocation of STAT1 and STAT2, resulting in inhibition of IFN-stimulated induction of IFN-stimulated gene (ISG) expression. Since IRF7 is an IFN-stimulated gene (ISG), and IRF7 is critical for the positive amplification loop for IFN-β expression, this may also reduce IFN-β expression. (B) Reovirus likely inhibits the IFN response by at least 3 virus strain-specific mechanisms. Reovirus activates RIG-I and IPS-1 for phosphorylation of IRF3. Reovirus-induced activation of MDA5 also participates in induction of IFN-β, perhaps through activation of NF-κB. (1) The reovirus μ2 protein can induce an unusual nuclear accumulation of IRF9 and repress IFN-stimulated induction of ISGs, most likely by disrupting IRF9 function as part of the heterotrimeric transcription factor complex, ISGF3. As discussed for rotavirus, repression of IRF7 expression may also reduce IFN-β expression. (2) The reovirus σ3 protein can bind double-stranded RNA (dsRNA), and prevent activation of the latent antiviral effector protein PKR. To date, there is no evidence that σ3 modulates activation of sensors involved in induction of IFN. (3) Genetic approaches have identified the reovirus λ2 and σ2 proteins in virus strain-specific modulation of the IFN response, but the significance remains unclear.
Rotavirus Modulation of Induction of IFN
The first evidence that rotavirus proteins might interact with cell proteins to modulate the IFN response resulted from a yeast 2-hybrid screen to identify proteins binding to the rotavirus NSP1 protein (Graff and others 2002). NSP1 is a nonstructural protein with a highly conserved zinc finger domain, which is found throughout the cytoplasm as well as associated with the cytoskeleton (Estes and Kapikian 2007). Graff and others found that the NSP1 from both bovine (B641) and murine (EW) rotavirus strains interacted with IRF3 in MA104 cells and they suggested that this interaction might repress rotavirus induction of IFN. Indeed, Barro and Patton found that NSP1 induces proteasome-mediated degradation of IRF3 to repress IFN signaling (Barro and Patton 2005). Specifically, IRF3 bound NSP1 in Caco-2 cells infected with a simian rotavirus strain (SA11-4F). While SA11-4F reduced IRF3 in infected Caco-2 cells, a derivative virus expressing an NSP1 with the C-terminal 17 amino acids deleted (SA11-5S) did not. This strain and another expressing a truncated NSP1 (SA11-30-1A, deleted for 71 amino acids) activated an IFN reporter construct better than the parental wild-type viruses did in FRhL2 cells. The strains expressing truncated NSP1 proteins also induced IRF3 dimerization in Caco-2 cells and nuclear localization of a GFP-tagged IRF3 in FRhL2 cells, while the parental wild-type viruses did not. Cotransfection of constructs expressing wild-type NSP1 but not C-terminally deleted NSP1 degraded GFP-IRF3 in 293T cells, but GFP-IRF3 was spared if the cells were incubated with the proteasome inhibitor MG132. Finally, the viruses with a truncated NSP1 generated smaller plaques than parental wild-type strains did in FRhL2 cells, and siRNA-mediated reduction of NSP1 in SA11-4F-infected MA10 cells reduced plaque size, suggesting that the C-terminal domain of NSP1 modulates viral replication. Together, the data demonstrated that the C-terminal domain of NSP1 induces proteasome-mediated degradation of IRF3 to repress viral induction of IFN, and suggested that this enhances rotavirus replication.
Given the strong conservation of protein domains across IRFs, Barro and Patton used the same panel of simian rotaviruses to ask whether rotavirus NSP1 also modulates IRF7, critical for amplification of IFN, and IRF5, inducing IFN and other cytokines in B lymphocytes and some dendritic cells (Barro and Patton 2007). Supernatants from FRhL2 cells infected with parental wild-type viruses but not viruses with a truncated NSP1 inhibited VSV-GFP expression in FRhL2 cells, and this inhibition was reversed by incubation with anti-IFN-β antibodies. Similar to previous results for IRF3, transfected constructs expressing full-length but not C-terminally truncated NSP1 degraded IRF7, and IRF7 was spared in the presence of MG132. Moreover, co-immunoprecipitations suggested a physical interaction between NSP1 and IRF7. Comparable experiments suggested NSP1-induced degradation of IRF5. Finally, siRNA-mediated inhibition of either IRF3 or IRF7 increased growth of parental wild-type virus but not virus expressing a C-terminally truncated NSP1 in Caco-2 cells. Collectively, the results demonstrated that rotavirus NSP1 induces proteasome-mediated degradation of IRF3, IRF5, and IRF7 (Fig. 1). Given the role of IRF5 in induction of other antiviral responses, this NSP1 function could have broad effects on viral replication and spread.
How might rotavirus NSP1 induce degradation of IRF proteins? Graff and others found that IRF3 was degraded in MA104 cells infected with a bovine (B641) but not a porcine (OSU) rotavirus strain, and that the OSU strain induced IRF3 phosphorylation instead (Graff and others 2007). Transfected B641 NSP1 was much more effective than OSU NSP1 at binding to and inducing degradation of IRF3, and site-directed mutagenesis of the zinc finger in the B641 NSP1 reduced both activities. Together, the data suggest that NSP1 must bind to IRF3 to induce its degradation and that the zinc finger is critical for this function. Interestingly, mutations in the zinc finger also increased both B641 and OSU NSP1 stability, suggesting NSP1 autoregulation of stability. Stability of both NSP1 proteins was increased in the presence of MG132, consistent with results from others for NSP1 from a simian rotavirus strain (RRV) (Pina-Vazquez and others 2007). Graff and others speculated that the NSP1 zinc finger might be part of an unconventional RING finger domain involved in E3 ubiquitin ligase activity (Graff and others 2007). E3 ligases often regulate their own stability through self-ubiquitination and the zinc finger is required for E3 ligase activity. Consistent with their speculation, OSU NSP1 was sensitive to proteasome-mediated degradation, but failed to induce proteasome-mediated IRF3 degradation presumably because of poor binding to IRF3. In sum, the data suggest that NSP1 regulates its own stability as well as that of IRF3 through proteasome-mediated degradation, that this function requires a zinc finger consistent with a proposed E3 ligase activity, and that this function is virus strain-specific.
Rotavirus NSP1 modulation of IRF3 is also cell type-specific. Douagi and others found that a simian rotavirus strain (RRV) induced degradation of IRF3 in mouse embryo fibroblasts (MEFs) but not in mouse myeloid dendritic cells (mDCs) (Douagi and others 2007). RRV infected mDCs efficiently as evidenced by immunofluorescent microscopy, but while 1 structural protein (VP6) was expressed at high levels, a nonstructural protein (NSP4) was expressed poorly and there was no production of progeny virus. RRV infection of these mDCs induced IFN through both IRF3-dependent and IRF3-independent pathways, indicating at least some activation of IRF3. It is possible that NSP1 is expressed poorly in RRV-infected mDCs allowing activation of IRF3 and induction of IFN; NSP1 levels were not measured. Alternatively, mDCs may protect IRF3 from NSP1-induced degradation.
Rotavirus NSP1 can also modulate NF-κB, a second important transcription factor involved in viral induction of IFN-β. Graff and others found that a swine rotavirus strain (OSU) induced IRF3 phosphorylation and nuclear translocation in MA104 cells but surprisingly, OSU failed to induce IFN-β expression (Graff and others 2009). Transfected OSU NSP1 repressed poly I:C induction of reporters for IFN-β and NF-κB in 293-TLR3 cells, suggesting that OSU NSP1 represses induction of IFN-β by subverting NF-κB. Indeed, infection with OSU or a bovine rotavirus strain (NCDV) failed to activate the p50 or p65 subunits of NF-κB in MA104 cells, in contrast to infection with a bovine rotavirus strain (A5-16) expressing a C-terminally truncated NSP1. Neither OSU nor NCDV degraded these NF-κB subunits. Instead, while A5-16 induced efficient phosphorylation and degradation of IκBα to release active NF-κB, OSU and NCDV induced phosphorylation but only minimal degradation of IκBα and inhibited TNF-α-induced degradation of IκBα. In addition, OSU or NCDV but not A5-16 infection induced degradation of β-TrCP, a protein in the SCFβ-TrCP E3 ligase complex that targets IκBα for proteasome-mediated degradation. Incubation in MG132 inhibited this degradation. Finally, coimmunoprecipitations demonstrated that OSU and NCDV NSP1 proteins bound β-TrCP. Together the data suggest that NSP1 can bind β-TrCP to stimulate its proteasome-mediated degradation, thereby stabilizing IκBα to repress NF-κB and subvert viral induction of IFN-β (Fig. 1). This NSP1 effect is rotavirus strain-specific. The authors suggest that substrate specificity of NSP1 may determine whether degradation of IRF3 or β-TrCP is the dominant mechanism for rotavirus subversion of the IFN-β response, with NCDV NSP1 preferentially targeting IRF3 and OSU NSP1 preferentially targeting β-TrCP. Interestingly, there is also apparently an NSP1-independent mechanism for rotavirus modulation of NF-κB: regardless of whether the infecting rotavirus activated the p65 subunit of NF-κB (A5-16) or not (NCDV and OSU strains), p65 remaining in the cytoplasm was found localized to viroplasms, suggesting a specific sequestration of this transcription factor (Fig. 1). Holloway and others found that simian (RRV) and human (Wa) rotavirus strains allowed TNF-α-stimulated phosphorylation (activation) but not nuclear translocation of p65 (Holloway and others 2009). These data suggest that modulation of NF-κB is likely to be common among rotavirus strains across animal species, but virus strain-specific.
Rotavirus Modulation of IFN Signaling
Rotaviruses can also modulate IFN signaling, by inhibiting STAT1 and STAT2 nuclear translocation. Holloway and others found that a simian (RRV) rotavirus strain inhibited IFN-α- and IFN-γ-stimulated gene expression in MA104 cells (IFN-α and IFN-γ) and Caco-2 cells (IFN-α; IFN-γ was not tested) (Holloway and others 2009). Human (Wa) rotavirus also inhibited IFN-α- and IFN-γ-stimulated gene expression in MA104 cells, but Wa was less effective than RRV. Neither virus induced degradation of STAT1 or STAT2, nor did they inhibit their IFN-stimulated phosphorylation. Instead, RRV inhibited IFN-stimulated nuclear translocation of STAT1 and STAT2 in both MA104 and Caco-2 cells (Fig. 1). Simian (SA11), human (Wa), and bovine (UK) strains also inhibited at least IFN-α-stimulated STAT2 translocation in MA104 cells (other conditions were not tested), suggesting that this repression mechanism is common among rotavirus strains across animal species. Transfection of MA104 cells with constructs expressing RRV NSP1, NSP3, or NSP4 failed to repress IFN-α signaling, suggesting that none of these NSPs individually are sufficient to mediate repression.
Reovirus Modulation of the IFN Response
The IFN response is a critical determinant of protection against reovirus-induced disease in 2 neonatal mouse models. First, the IFN response determines reovirus strain-specific differences in induction of myocarditis, paralleling virus strain-specific differences in viral cytopathic effect (Baty and Sherry 1993) and modulation of the IFN response in primary cultures of cardiac myocytes (Sherry and others 1998). Second, the IFN response is critical for protection against viral replication in the central nervous system, paralleling reovirus activation of STAT1 in primary cortical neuron cultures (Goody and others 2007). Finally, the capacity to repress Type I IFN signaling correlates with the capacity to induce myocarditis (Zurney and others 2009), suggesting that subversion of the IFN response can determine reovirus disease. As discussed later, reoviruses likely inhibit the IFN response by at least 3 mechanisms: (1) μ2-mediated inhibition of IFN signaling; (2) σ3-mediated sequestration of dsRNA; and (3) unknown mechanisms associated with λ2 and α2 (Fig. 1).
Reovirus Modulation of Induction of IFN
Reovirus induction of IFN was first described over 40 years ago (Oie and Leh 1968). Reovirus dsRNA remains a popular reagent to investigate signals for activation of sensors triggering IFN synthesis (Kato and others 2008), although the reovirus replication strategy would suggest that reovirus dsRNA is either rarely or never exposed during natural infection (Schiff and others 2007). An early comprehensive study of reovirus induction of IFN suggested that infectious virus and ultraviolet light (UV)-inactivated virus may stimulate different cell responses for induction of IFN (Lai and Joklik 1973). One possibility is that UV-inactivated virus is unstable, and that exposed dsRNA is recognized by cell sensors that have been described using naked dsRNA (Kato and others 2008). Lai and Joklik also found that empty viral particles lacking dsRNA and noninfectious viral cores failed to induce IFN, suggesting that reovirus entry processes themselves are unlikely to induce IFN (Lai and Joklik 1973). Finally, their use of temperature-sensitive mutants of reovirus indicated that all mutants, even those involved in a late stage of virion morphogenesis, were defective in induction of IFN. Although they concluded that induction of IFN required the formation of progeny virions, studies since then suggest that most reovirus proteins play roles in multiple stages of the replication cycle and therefore the mutants could have been defective in earlier events critical for induction of IFN.
Later studies took advantage of the segmented reovirus genome to identify genes associated with virus strain-specific differences in induction of IFN. Using a panel of reassortant viruses, Sherry and others found that the S2, M1, and L2 genes from strain T3D were associated with efficient induction of IFN-α/β in primary cultures of murine cardiac myocytes and fibroblasts (Sherry and others 1998). The S2-encoded σ2 protein is an abundant structural protein in the virion core, the L2-encoded λ2 protein spans the virion from the core to the virion surface and expresses multiple enzymatic functions associated with mRNA capping, and the M1-encoded μ2 protein is a minor core protein associated with the λ3 polymerase and expressing RNA-binding and NTPase activities (Schiff and others 2007). After entry and partial uncoating, the reovirus core synthesizes single-stranded RNA (ssRNA) copies from the genomic dsRNA template; later in infection a subviral particle composed of the core and other reovirus proteins synthesizes both dsRNA and ssRNA (Schiff and others 2007). This would suggest that the capacity to induce IFN is a consequence of viral RNA synthesis, and indeed, the same reovirus genes are correlated with the rate of RNA synthesis in infected cardiac myocytes (Sherry and others 1996). However, it is also possible that the capacity to induce IFN is determined by interactions of these reovirus proteins with cell proteins, either as part of the virion or as protein in the cell. For example, while μ2 is only a minor component of the virion, it is expressed abundantly in infected cells where it induces hyperacetylation of microtubules and determines the morphology of reovirus VIBs (Parker and others 2002). The specific roles played by these reovirus proteins in induction of IFN remains to be determined.
More recent studies have dissected reovirus modulation of cell RNA sensors (RIG-I and MDA5) and transcription factors (IRF3 and NF-κB) involved in induction of IFN (Fig. 1). Holm and others (2007) found that reovirus T3D activation of an IRF3/IRF7 reporter construct in 293T cells required genomic dsRNA, similar to earlier studies on induction of IFN (Lai and Joklik 1973). T3D activated IRF3/IRF7 in the presence of ribavirin suggesting that viral RNA synthesis is not required; however, ribavirin actually enhanced IRF3/IRF7 activation slightly suggesting that interpretation of these results is complex. Reovirus T3D activation of the IRF3/IRF7 reporter construct in 293T cells required RIG-I and IPS-1 but not MDA5, as shown by cotransfection experiments where a dominant negative inhibitor of RIG-I or siRNA specific for RIG-I or IPS-1 but not MDA5 repressed this activation. T3D activation of IRF3/IRF7 in HeLa cells was also repressed by siRNA for RIG-I but not MDA5. In contrast, T3D activation of an NF-κB reporter construct in 293T cells required neither RIG-I nor IPS-1, suggesting that reovirus induction of IFN-β involves both RIG-I-dependent and RIG-I-independent pathways. The use of 2 different pathways is supported in studies by Loo and others, who found that T3D induction of ISGs was reduced in MEFs lacking either RIG-I or MDA5, and that T3D induction of ISGs was almost completely ablated in RIG-I-null MEFs treated with siRNA specific for MDA5 (Loo and others 2008). Together the data suggest that T3D is recognized by RIG-I for activation of IRF3, and MDA5 perhaps for activation of NF-κB. The demonstration that the length of the dsRNA target can determine sensor preference provides one possible mechanism for reovirus stimulation of both RIG-I and MDA5. Indeed, stimulation of cells with long reovirus dsRNA gene segments activates MDA5 while short reovirus dsRNA gene segments activate RIG-I (Kato and others 2008); however, it is unclear how these gene segments might be exposed during reovirus replication (Schiff and others 2007). Identification of the reovirus components that may activate RIG-I and MDA5 during natural infection awaits future studies.
Reovirus Modulation of IFN Signaling and Antiviral Effects
Reovirus strains differ in their sensitivity to the antiviral effects of IFN (Jacobs and Ferguson 1991), and again, a genetic approach was used to identify the underlying viral genetic determinants. Sherry and others found that the S2, M1, and L2 genes from strain T3D were associated with sensitivity to the antiviral effects of IFN-α/β in primary cultures of murine cardiac myocytes and fibroblasts (Sherry and others 1998). Interestingly, these are the same 3 genes that were associated with reovirus strain-specific differences in induction of IFN-α/β (discussed earlier). One plausible explanation is that reovirus strain-specific differences in exposure of viral dsRNA determine both events. Viral dsRNA exposure could readily impact activation of RIG-I and MDA5 for induction of IFN-α/β, but could also impact activation of latent proteins involved in IFN-stimulated antiviral responses, such as PKR and 2′,5′-oligoadenylate synthetase. The S2, M1, and L2 genes encode viral core proteins which, as discussed earlier, may modulate viral RNA synthesis. Alternatively, these viral core proteins may determine core stability and leakage of the genomic dsRNA resulting in cytoplasmic exposure. It seems likely that the reovirus dsRNA-binding protein σ3, encoded by the S4 gene, would prevent exposure of dsRNA, since it can substitute for a similar role of E3L in vaccinia virus (Beattie and others 1995). Indeed, σ3 can inhibit dsRNA-mediated activation of PKR (Imani and Jacobs 1988; Yue and Shatkin 1997), and reovirus strain-specific differences in inhibition of host cell protein synthesis is associated with the S4 gene (Sharpe and Fields 1982). The failure to identify the S4 gene as associated with induction of or sensitivity to IFN can be interpreted only to mean that σ3 does not determine reovirus strain-specific differences in these events, not that σ3 is irrelevant to these events. A second possible explanation for the identification of the same 3 genes as determinants of reovirus induction of and sensitivity to IFN is that the 2 events are interdependent and therefore, modulation of one will impact the other. For example, if reovirus resistance to IFN reflected the repression of IFN signaling, then resistant viruses would prevent IFN induction of IRF7 expression and interrupt the positive amplification loop for induction of IFN. Accordingly, reoviruses resistant to IFN would also induce less IFN. The identification of one of these 3 genes as encoding a repressor of IFN signaling (the M1 gene; (Zurney and others 2009)) suggests that this may be the case, as discussed later.
Reoviruses can repress IFN-α/β signaling, this repression is virus strain-specific, and it involves a novel mechanism not previously described. Zurney and others found that reovirus T1L repressed IFN-α/β induction of the ISGs IRF7 and STAT1 in mouse L929 cells, but T3D did not (Zurney and others 2009). Experiments using a panel of reovirus reassortants identified the T1L M1 and S2 genes as associated with this repression. However, experiments using recombinant viruses with single gene substitutions generated by reverse genetics demonstrated that the T1L M1 gene was a determinant of repression while the S2 gene was uninvolved and had merely cosegregated with the M1 gene in the reassortant panel. The T1L, but not the T3D, M1 gene repressed IFN-α/β induction of an ISG reporter construct when cotransfected into HEK293 cells, indicating that other reovirus genes or cell responses stimulated by infection were not required for this repression. T1L infection did not induce STAT1, STAT2, or IRF9 degradation and did not inhibit STAT1 or STAT2 translocation to the nucleus. However, T1L but not T3D infection resulted in an unusual nuclear accumulation of IRF9. A recombinant virus comprised of a T1L backbone with an M1 gene substituted from T3D failed to induce IRF9 nuclear accumulation, demonstrating that the T1L M1 gene is required for this effect. In addition, a recombinant virus comprised of a T3D backbone and an M1 gene substituted from T1L-induced IRF9 nuclear accumulation, demonstrating that the T1L M1 gene is sufficient for this effect. Together, the results suggest that the T1L M1 gene-encoded μ2 protein induces an unusual nuclear accumulation of IRF9 associated with reovirus subversion of the IFN response (Fig. 1). IRF9 contains a nuclear localization signal and can shuttle to the nucleus in the absence of stimulation, but lacks a nuclear export signal and instead associates with STAT2 for exit from the nucleus (Lau and others 2000; Banninger and Reich 2004; Reich 2007). Therefore, the T1L μ2 protein might disrupt IRF9–STAT2 interactions, resulting in IRF9 nuclear accumulation and concomitant disruption of the IRF9–STAT2–STAT1 interactions required for IFN signaling. As discussed earlier, μ2 is a minor viral core protein involved in viral RNA synthesis, but it is also abundantly expressed in infected cells and is an important determinant of VIB formation (Parker and others 2002). Interestingly, μ2 is also found in the nucleus of infected cells (Parker and others 2002; Kobayashi and others 2009), despite the exclusively cytoplasmic reovirus replication strategy. Future studies will address whether μ2 represses IFN signaling by disrupting IRF9–STAT2 interactions either in the cytoplasm or nucleus, or whether this repression involves another mechanism.
Conclusions
While the evidence to date suggests that rotavirus and reovirus use different mechanisms to subvert the IFN response (Fig. 1), many avenues remain unexplored. What are the cell sensors that recognize rotavirus? Does reovirus modulate IRFs other than IRF9? Rotavirus and reovirus modulation of cell proteins is virus strain-specific, and likely cell type-specific as well (Zurney and others 2007), and therefore results should be interpreted with caution. Nonetheless, future studies may reveal new commonalities between these 2 genera and offer insights into controlling diseases caused by dsRNA viruses.
Acknowledgments
B.S. thanks Susan Irvin for excellent discussions and critical review of the manuscript. This work was supported by NIH grant AI062657.
References
- Alff PJ. Sen N. Gorbunova E. Gavrilovskaya IN. Mackow ER. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1–TRAF3 complex formation. J Virol. 2008;82(18):9115–9122. doi: 10.1128/JVI.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel J. Franco MA. Greenberg HB. Bass D. Lack of a role for type I and type II interferons in the resolution of rotavirus-induced diarrhea and infection in mice. J Interferon Cytokine Res. 1999;19(6):655–659. doi: 10.1089/107999099313802. [DOI] [PubMed] [Google Scholar]
- Banninger G. Reich NC. STAT2 nuclear trafficking. J Biol Chem. 2004;279(38):39199–39206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- Barnard P. McMillan NA. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology. 1999;259(2):305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- Barral PM. Morrison JM. Drahos J. Gupta P. Sarkar D. Fisher PB. Racaniello VR. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81(8):3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro M. Patton JT. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci USA. 2005;102(11):4114–4119. doi: 10.1073/pnas.0408376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro M. Patton JT. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J Virol. 2007;81(9):4473–4481. doi: 10.1128/JVI.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baty CJ. Sherry B. Cytopathogenic effect in cardiac myocytes but not in cardiac fibroblasts is correlated with reovirus-induced acute myocarditis. J Virol. 1993;67(10):6295–6298. doi: 10.1128/jvi.67.10.6295-6298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E. Denzler KL. Tartaglia J. Perkus ME. Paolettie E. Jacobs BL. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69(1):499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodkin DK. Fields BN. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J Virol. 1989;63(3):1188–1193. doi: 10.1128/jvi.63.3.1188-1193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC. Sen GC. Uze G. Silverman RH. Ransohoff RM. Foster GR. Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin PJ. Entrican G. Gelder KI. Collins RA. Cloning and biologic activities of a bovine interferon-alpha isolated from the epithelium of a rotavirus-infected calf. J Interferon Cytokine Res. 1996;16(1):25–30. doi: 10.1089/jir.1996.16.25. [DOI] [PubMed] [Google Scholar]
- Childs K. Stock N. Ross C. Andrejeva J. Hilton L. Skinner M. Randall R. Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359(1):190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Danthi P. Kobayashi T. Holm GH. Hansberger MW. Abel TW. Dermody TS. Reovirus apoptosis and virulence are regulated by host cell membrane penetration efficiency. J Virol. 2008;82(1):161–172. doi: 10.1128/JVI.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boissieu D. Lebon P. Badoual J. Bompard Y. Dupont C. Rotavirus induces alpha-interferon release in children with gastroenteritis. J Pediatr Gastroenterol Nutr. 1993;16(1):29–32. doi: 10.1097/00005176-199301000-00006. [DOI] [PubMed] [Google Scholar]
- Dennehy PH. Rotavirus vaccines: an overview. Clin Microbiol Rev. 2008;21(1):198–208. doi: 10.1128/CMR.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P. von Messling V. Songsungthong W. Springfeld C. Cattaneo R. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology. 2007;360(1):72–83. doi: 10.1016/j.virol.2006.09.049. [DOI] [PubMed] [Google Scholar]
- Donelan NR. Basler CF. Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77(24):13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douagi I. McInerney GM. Hidmark AS. Miriallis V. Johansen K. Svensson L. Karlsson Hedestam GB. Role of interferon regulatory factor 3 in type I interferon responses in rotavirus-infected dendritic cells and fibroblasts. J Virol. 2007;81(6):2758–2768. doi: 10.1128/JVI.01555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden KA. Wang G. Yeager M. Nibert ML. Coombs KM. Furlong DB. Fields BN. Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122(5):1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes MK. Kapikian AZ. Rotaviruses. In: Knipe DM, editor; Howley PM, editor. Fields virology. 5th. Philadelphia, PA: Lippincott-Raven; 2007. pp. 1917–1974. [Google Scholar]
- Feng N. Kim B. Fenaux M. Nguyen H. Vo P. Omary MB. Greenberg HB. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J Virol. 2008;82(15):7578–7590. doi: 10.1128/JVI.00391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy E. Li K. Sumpter R., Jr Loo YM. Johnson CL. Wang C. Fish PM. Yoneyama M. Fujita T. Lemon SM. Gale M., Jr Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102(8):2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M. Yount B. Heise M. Kopecky-Bromberg SA. Palese P. Baric RS. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007;81(18):9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR. Laird AR. Bielfelt B. Griffin DD. Banyai K. Ramachandran M. Jain V. Cunliffe NA. Nakagomi O. Kirkwood CD. Fischer TK. Parashar UD. Bresee JS. Jiang B. Glass RI. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis. 2005;192(Suppl 1):S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Goody RJ. Beckham JD. Rubtsova K. Tyler KL. JAK-STAT signaling pathways are activated in the brain following reovirus infection. J Neurovirol. 2007;13(4):373–383. doi: 10.1080/13550280701344983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JW. Ettayebi K. Hardy ME. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 2009;5(1):e1000280. doi: 10.1371/journal.ppat.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JW. Ewen J. Ettayebi K. Hardy ME. Zinc-binding domain of rotavirus NSP1 is required for proteasome-dependent degradation of IRF3 and autoregulatory NSP1 stability. J Gen Virol. 2007;88(Pt 2):613–620. doi: 10.1099/vir.0.82255-0. [DOI] [PubMed] [Google Scholar]
- Graff JW. Mitzel DN. Weisend CM. Flenniken ML. Hardy ME. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J Virol. 2002;76(18):9545–9550. doi: 10.1128/JVI.76.18.9545-9550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT. Hayashi J. Seeger C. West Nile virus inhibits the signal transduction pathway of alpha interferon. J Virol. 2005;79(3):1343–1350. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M. Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol. 2005;79(12):7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway G. Truong TT. Coulson BS. Rotavirus antagonizes cellular antiviral responses by inhibiting the nuclear accumulation of STAT1, STAT2 and NF-{kappa}B. J Virol. 2009;83(10):4942–4951. doi: 10.1128/JVI.01450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm GH. Zurney J. Tumilasci V. Leveille S. Danthi P. Hiscott J. Sherry B. Dermody TS. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J Biol Chem. 2007;282(30):21953–21961. doi: 10.1074/jbc.M702112200. [DOI] [PubMed] [Google Scholar]
- Ida-Hosonuma M. Iwasaki T. Yoshikawa T. Nagata N. Sato Y. Sata T. Yoneyama M. Fujita T. Taya C. Yonekawa H. Koike S. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J Virol. 2005;79(7):4460–4469. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imani F. Jacobs B. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc Natl Acad Sci USA. 1988;85(21):7887–7891. doi: 10.1073/pnas.85.21.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BL. Ferguson RE. The Lang strain of reovirus serotype 1 and the Dearing strain of reovirus serotype 3 differ in their sensitivities to beta-interferon. J Virol. 1991;65(9):5102–5104. doi: 10.1128/jvi.65.9.5102-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE. Song B. Knipe DM. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology. 2008;374(2):487–494. doi: 10.1016/j.virol.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Mikamo-Satoh E. Hirai R. Kawai T. Matsushita K. Hiiragi A. Dermody TS. Fujita T. Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keroack M. Fields BN. Viral shedding and transmission between hosts determined by reovirus L2 gene. Science. 1986;232(4758):1635–1638. doi: 10.1126/science.3012780. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. Antar AA. Boehme KW. Danthi P. Eby EA. Guglielmi KM. Holm GH. Johnson EM. Maginnis MS. Naik S. Skelton WB. Wetzel JD. Wilson GJ. Chappell JD. Dermody TS. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe. 2007;1(2):147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. Ooms LS. Chappell JD. Dermody TS. Identification of functional domains in reovirus replication proteins muNS and mu2. J Virol. 2009;83(7):2892–2906. doi: 10.1128/JVI.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner I. Kochs G. Kalinke U. Weiss S. Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J Virol. 2007;81(4):2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M-HT. Joklik WK. The induction of interferon by temperature-sensitive mutants of reovirus. UV-irradiated reovirus, and subviral reovirus particles. Virology. 1973;51(1):191–204. doi: 10.1016/0042-6822(73)90379-6. [DOI] [PubMed] [Google Scholar]
- Langland JO. Jacobs BL. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology. 2004;324(2):419–429. doi: 10.1016/j.virol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Lau JF. Parisien JP. Horvath CM. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc Natl Acad Sci USA. 2000;97(13):7278–7283. doi: 10.1073/pnas.97.13.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M. Liu Y. Samuel CE. Adenovirus VAI RNA antagonizes the RNA-editing activity of the ADAR adenosine deaminase. Virology. 1998;245(2):188–196. doi: 10.1006/viro.1998.9162. [DOI] [PubMed] [Google Scholar]
- Leonard GT. Sen GC. Effects of adenovirus E1A protein on interferon-signaling. Virology. 1996;224(1):25–33. doi: 10.1006/viro.1996.0503. [DOI] [PubMed] [Google Scholar]
- Loo YM. Fornek J. Crochet N. Bajwa G. Perwitasari O. Martinez-Sobrido L. Akira S. Gill MA. Garcia-Sastre A. Katze MG. Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LL. Puri M. Horvath CM. Sen GC. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J Biol Chem. 2008;283(21):14269–14276. doi: 10.1074/jbc.M710089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K. Dong B. Gale M., Jr Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448(7155):816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J. Potgieter CA. Ciarlet M. Parreno V. Martella V. Banyai K. Garaicoechea L. Palombo EA. Novo L. Zeller M. Arista S. Gerna G. Rahman M. Van Ranst M. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J Virol. 2009;83(7):2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M. Martinez-Sobrido L. Loo YM. Cardenas WB. Gale M., Jr Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81(2):514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U. Steinhoff U. Reis LFL. Hemmi S. Pavlovic J. Zinkernagel RM. Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Oie HK. Leh PC. Reovirus type 2: production of and sensitivity to interferon in human amnion cells. Proc Soc Exp Biol Med. 1968;127(4):1210–1213. doi: 10.3181/00379727-127-32912. [DOI] [PubMed] [Google Scholar]
- Parashar UD. Glass RI. Rotavirus vaccines—early success, remaining questions. N Engl J Med. 2009;360(11):1063–1065. doi: 10.1056/NEJMp0810154. [DOI] [PubMed] [Google Scholar]
- Parker JSL. Broering TJ. Kim J. Higgins DE. Nibert ML. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol. 2002;76(9):4483–4496. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. Bruns E. Kuske M. von Wussow P. Treatment of extrahepatic biliary atresia with interferon-alpha in a murine infectious model. Pediatr Res. 1997;42(5):623–628. doi: 10.1203/00006450-199711000-00013. [DOI] [PubMed] [Google Scholar]
- Pina-Vazquez C. De Nova-Ocampo M. Guzman-Leon S. Padilla-Noriega L. Post-translational regulation of rotavirus protein NSP1 expression in mammalian cells. Arch Virol. 2007;152(2):345–368. doi: 10.1007/s00705-006-0850-8. [DOI] [PubMed] [Google Scholar]
- Porter FW. Bochkov YA. Albee AJ. Wiese C. Palmenberg AC. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc Natl Acad Sci USA. 2006;103(33):12417–12422. doi: 10.1073/pnas.0605375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE. Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Reich NC. STAT dynamics. Cytokine Growth Factor Rev. 2007;18(5–6):511–518. doi: 10.1016/j.cytogfr.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP. Leung LW. Hartman AL. Martinez O. Shaw ML. Carbonnelle C. Volchkov VE. Nichol ST. Basler CF. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80(11):5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM. Nibert ML. Harrison SC. Structure of the reovirus core at 3.6A resolution. Nature. 2000;404(6781):960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Ryman KD. Meier KC. Gardner CL. Adegboyega PA. Klimstra WB. Non-pathogenic Sindbis virus causes hemorrhagic fever in the absence of alpha/beta and gamma interferons. Virology. 2007;368(2):273–285. doi: 10.1016/j.virol.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Saif LJ. Rosen B. Parwani A. Animal rotaviruses. In: Kapikian AZ, editor. Virus infection of the gastrointestinal tract. New York: Marcel Dekker; 1994. pp. 279–367. [Google Scholar]
- Schiff LA. Nibert ML. Tyler KL. Orthoreoviruses and their replication. In: Knipe DM, editor; Howley PM, editor. Fields virology. 5th. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 1853–1915. [Google Scholar]
- Schwers A. Vanden Broecke C. Maenhoudt M. Beduin JM. Werenne J. Pastoret PP. Experimental rotavirus diarrhoea in colostrum-deprived newborn calves: assay of treatment by administration of bacterially produced human interferon (Hu-IFN alpha 2) Ann Rech Vet. 1985;16(3):213–218. [PubMed] [Google Scholar]
- Seo SH. Hoffmann E. Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8(9):950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- Sharp TV. Schwemmle M. Jeffrey I. Laing K. Mellor H. Proud CG. Hilse K. Clemens MJ. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21(19):4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AH. Fields BN. Reovirus inhibition of cellular RNA and protein synthesis: role of the S4 gene. Virology. 1982;122(2):381–391. doi: 10.1016/0042-6822(82)90237-9. [DOI] [PubMed] [Google Scholar]
- Sherry B. Baty CJ. Blum MA. Reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than generation of infectious virus in cardiac myocytes. J Virol. 1996;70(10):6709–6715. doi: 10.1128/jvi.70.10.6709-6715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry B. Blum MA. Multiple viral core proteins are determinants of reovirus-induced acute myocarditis. J Virol. 1994;68(12):8461–8465. doi: 10.1128/jvi.68.12.8461-8465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry B. Fields BN. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J Virol. 1989;63(11):4850–4856. doi: 10.1128/jvi.63.11.4850-4856.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry B. Torres J. Blum MA. Reovirus induction of and sensitivity to beta-interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J Virol. 1998;72(2):1314–1323. doi: 10.1128/jvi.72.2.1314-1323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar P. Campbell KM. Sabla GE. Miethke A. Tiao G. McNeal MM. Ward RL. Bezerra JA. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest. 2004;114(3):322–329. doi: 10.1172/JCI21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S. Kyle JL. Snider HM. Basavapatna M. Beatty PR. Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78(6):2701–2710. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa T. Hoshino Y. Whole genome sequence and phylogenetic analyses reveal human rotavirus G3P[3] strains Ro1845 and HCR3A are examples of direct virion transmission of canine/feline rotaviruses to humans. Virology. 2008;380(2):344–353. doi: 10.1016/j.virol.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulane CM. Kentsis A. Cruz CD. Parisien JP. Schneider KL. Horvath CM. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J Virol. 2005;79(16):10180–10189. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH. Rani MR. Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25(3):361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Vancott JL. McNeal MM. Choi AH. Ward RL. The role of interferons in rotavirus infections and protection. J Interferon Cytokine Res. 2003;23(3):163–170. doi: 10.1089/107999003321532501. [DOI] [PubMed] [Google Scholar]
- Weiner HL. Drayna D. Averill DR., Jr Fields BN. Molecular basis of reovirus virulence: role of the S1 gene. Proc Natl Acad Sci USA. 1977;74(12):5744–5748. doi: 10.1073/pnas.74.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z. Shatkin AJ. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology. 1997;234(2):364–371. doi: 10.1006/viro.1997.8664. [DOI] [PubMed] [Google Scholar]
- Zurney J. Howard KE. Sherry B. Basal expression levels of IFNAR and Jak-STAT components are determinants of cell-type-specific differences in cardiac antiviral responses. J Virol. 2007;81(24):13668–13680. doi: 10.1128/JVI.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurney J. Kobayashi T. Holm GH. Dermody TS. Sherry B. Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J Virol. 2009;83(5):2178–2187. doi: 10.1128/JVI.01787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]