Abstract
Besides the established role of interleukin-12 (IL-12) and IL-18 on interferon-γ (IFN-γ) production by natural killer (NK), T, and B cells, the effects of these cytokines on macrophages are largely unknown. Here, we investigated the role of IL-12/IL-18 on nitric oxide (NO) and tumor necrosis factor-α (TNF-α) production by CD11b+ adherent peritoneal cells, focusing on the involvement of endogenously produced IFN-γ. C57BL/6 cells released substantial amounts of NO when stimulated with IFN-γ or lipopolysaccharide (LPS), but failed to respond to IL-12 or IL-18 or both. However, IL-12/IL-18 pretreatment was able to program these cells to release 6–8-fold more NO and TNF-α in response to LPS or Trypanosoma cruzi stimulation, with NO levels directly correlating with macrophage resistance to intracellular parasite growth. Analysis of IL-12/IL-18-primed cells from mice deficient in IFN-γ, IFNGR, and IFN regulatory factor-1 (IRF-1) revealed that these molecules were essential for LPS-induced NO release, but TNF-α production was IFN-γ independent. Conversely, the myeloid differentiation factor 88 (MyD88)-dependent pathway was indispensable for IL-12/IL-18-programmed LPS-induced TNF-α production, but not for NO release. Contaminant T and NK cells largely modulated the IL-12/IL-18 programming of LPS-induced NO response through IFN-γ secretion. Nevertheless, a small population of IFN-γ+ cells with a macrophage phenotype was also identified, particularly in the peritoneum of chronically T. cruzi-infected mice, reinforcing the notion that macrophages can be an alternative source of IFN-γ. Taken together, our data contribute to elucidate the molecular basis of the IL-12/IL-18 autocrine pathway of macrophage activation, showing that endogenous IFN-γ plays an important role in programming the NO response, whereas the TNF-α response occurs through an IFN-γ-independent pathway.
Introduction
Macrophages, monocytes, and dendritic cells (DCs) are the major sources of interleukin-12 (IL-12),1–3 a heterodimeric cytokine composed of p35 and p40 subunits. The central role of this cytokine in the development of immune responses was evidenced by data showing that treatment of mice with rIL-12 or IL-12 cDNA induces and sustains in vivo generated effector/memory Th1 cells,4 upregulates the synthesis of antigen-specific complement-fixing antibodies,5 and protects against tumors and infectious diseases.6,7 Conversely, IL-12p40 gene knockout (IL-12p40KO) mice have inadequate Th1 responses8 and increased susceptibility to infections in which protection is primarily mediated by interferon-γ (IFN-γ), such as leishmaniasis,9 Chagas' disease,10 and tuberculosis.11 The ability of IL-12 to direct the differentiation pattern of T cells indicates that this cytokine bridges innate and adaptive immunity, influencing the development of immune responses and, therefore, the degree of susceptibility to infection.12
It is generally accepted that the central role of IL-12 in host defense against many intracellular pathogens arises from its capacity to stimulate IFN-γ secretion by natural killer (NK) and T cells, which in turn activates phagocytes to control parasite growth.13 Nonetheless, in recent years, macrophages have been recognized as competent cells regarding the ability to respond to IL-12, which has led to the notion that this cytokine can induce macrophage activation through an autocrine pathway. Indeed, it has been shown that macrophages not only express β1 and β2 chains from IL-12 receptor (IL-12R), but also respond to IL-12 by producing IFN-γ, tumor necrosis factor-α (TNF-α), and nitric oxide (NO).14–24 IL-12 has also been implicated in programming the macrophage response to lipopolysaccharide (LPS) by upregulating the production of TNF-α.25 IL-18, a cytokine secreted by several cell types, including macrophages, originally designated as IFN-γ-inducing factor (IGIF),26 has been shown to act in synergism with IL-12 to stimulate IFN-γ production by T cells,27 NK cells,28 B cells,27 macrophages,16,18,21 and DCs.29,30 Although IL-18 per se does not seem to induce IFN-γ secretion by these cells, it can improve the response to IL-12 in different ways. In macrophages, the synergic effect of IL-18 depends on nuclear translocation of Stat4 that is attained only in the presence of both cytokines,18 whereas in DCs, IL-18 upregulates the activity of p38, a member of the MAP kinase (MAPK) superfamily, culminating with IFN-γ secretion.29
Another feature attributed to IL-12 is the ability to down-regulate the expression of transforming growth factor-β1 (TGF-β1) mRNA in monocytes and bone marrow cells.31 Overall, IL-12 directly influences the macrophage activation profile, driving them to react against foreign stimuli with a response dominated by proinflammatory cytokines. In this context, we have shown previously that macrophages from IL-12p40KO mice have an activation bias, spontaneously secreting large amounts of TGF-β, and responding with weak NO production to rIFN-γ.32,33 Moreover, IL-12p40KO macrophages are more permissive to the growth of the intracellular protozoan Trypanosoma cruzi than are wild-type cells and have an impaired ability to ingest opsonized Plasmodium chabaudi-infected erythrocytes. Thus, IL-12 might affect host defense against pathogens by at least two distinct pathways: (1) inducing IFN-γ production by several cell types and (2) programming macrophages prior to the onset of immune responses, rendering these cells prone to react with an effector (NO-dominant) rather than a tolerant (TGF-β-dominant) profile.
In the attempt to further characterize the effects of IL-12 and IL-18 on macrophages, we have focused our attention on the role of endogenously produced IFN-γ in the ability of IL-12/IL-18 to program the responses of CD11b+ adherent peritoneal cells (CD11b+ aPECs). Taken together, our data highlight the IL-12/IL-18 autocrine pathway of macrophage activation and reinforce the notion that at least in some circumstances, macrophages can be an alternative source of IFN-γ.
Materials and Methods
Mice and parasites
Six to eight-week-old C57BL/6, 129/SV, IFN-γKO, IFN-GRKO, IL-12p40KO, MyD88KO, IRF-1KO, CD14KO, and iNOSKO male mice were bred in our animal facilities at the University of São Paulo, under standard pathogen-free conditions. IFN-γKO, IFNGRKO, IL-12p40KO, iNOSKO, and CD14KO mice are originally from Jackson Laboratories (Bar Harbor, ME), MyD88KO34 mice were kindly provided by Dr Bernard Ryffel (Centre National de la Recherche Scientifique, Orléans, France), and IRF-1KO35 mice were kindly provided by Dr Luiz Fernando Reis (Ludwig Institute for Cancer Research, São Paulo, Brazil). T. cruzi trypomastigotes of the Sylvio-X10/4 strain were purified from a monkey epithelial cell line (LLC-MK2). C57BL/6 mice were infected i.p. with 103 trypomastigotes 3 months before the experiments.
PEC suspensions
Four to six mice were injected i.p. with 5 mL of 3% starch (Sigma, St. Louis, MO). Five days later, cells were obtained by peritoneal lavage with chilled RPMI 1640 (Sigma).
Purification of cell populations
CD11b+ PECs were purified by magnetic beads coupled with anti-CD11b (Mac-1, M1/70) monoclonal antibodies (mAb), according to the manufacturer's instructions (Miltenyi Biotec GMBH, Bergisch Gladbach, Germany). Cells with size (FSC) and granularity (SSC) characteristics of macrophages were sorted by flow cytometry using a FacsVantage (Becton Dickinson, Mountain View, CA) with CELLQUEST software (Becton Dickinson). CD3+NK1.1+ and CD3−NK1.1− cells were purified with phycoerythrin (PE)-labeled anti-NK1.1 (PK136) and anti-CD3 (145-2C11) mAbs (PharMingen, San Diego, CA), followed by magnetic beads coupled with anti-PE mAb (Miltenyi Biotec).
Phenotypic analysis of purified macrophages
Cells (106) stained with FITC, PE, or Cy-Chrome-labeled mAb to CD11b (Mac-1, M1/70), CD3 (145-2C11), and NK1.1 (PK136) (PharMingen) and F4/80 mAb (Caltag Laboratories, Burlingame, CA) were analyzed by flow cytometry using a Facscalibur (Becton Dickinson) with CELLQUEST software.
CD11b+ aPEC culture conditions
Purified CD11b+ PECs (2 × 105) were incubated for 4 h in RPMI 1640 supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL), 2-mercaptoethanol (50 μM), l-glutamine (2 mM), sodium pyruvate (1 mM), and 3% heat-inactivated fetal bovine serum (FBS). All supplements were purchased from Life Technologies (Rockville, MD). Nonadherent cells were removed by three vigorous washes with warm and FBS-free medium. To evaluate the direct effect of LPS, IFN-γ IL-12, or IL-18, cells were cultured with 0.1–50 μg/mL LPS (Sigma), 2.5–20 ng/mL rIFN-γ (PharMingen), or 0.5–10 ng/mL rIL-12 (PharMingen) with or without 0.5–10 ng/mL rIL-18 (MBL International Corporation, Woburn, MA), and the supernatants was harvested 48 h later. To evaluate the priming effect of IL-12, IL-18, and IFN-γ, cells were cultured for 18 h with rIL-12 (2.5 ng/mL), rIL-18 (2.5 ng/mL), or rIFN-γ (2.5 ng/mL), washed with medium, and then stimulated for additional 48 h with LPS (1 μg/mL) or T. cruzi (5 parasites/macrophage).
Detection of NO and TNF-α in culture supernatants
Culture supernatants were assayed for NO by the Griess reaction. Briefly, 50 μL supernatant was incubated with 50 μL Griess reagent for 5 min at ambient temperature, and the NO−2 concentration was determined by measuring the optical density (OD) at 550 nm in reference to a standard NaNO2 solution. A modified bioassay was used to quantify TNF-α.36 TNF-α-sensitive L929 cells (ATCC, Rockville, MD) were incubated in supplemented RPMI 1640 overnight at 37°C in a 5% CO2 atmosphere at a density of 5.5 × 104 cells in flat-bottomed 96-well plates. After medium removal, 100 μL of serial dilutions of cell culture supernatants (diluted from 1:2 to 1:1024) were added to each well. Four hours later, 10 μL medium containing actinomycin D was added to each well at a final concentration of 5 μg/mL. After 20 h in culture, viable L929 cells were stained with 20 μL/well of 0.75% crystal violet in 30% acetic acid for 15 min, rinsed, and dried. Methanol was used to solubilize the crystal violet, and the absorbance was read at 630 nm with a Vmax-Kinetic Microplate Reader (Molecular Devices, Sunnyvale, CA). Percent cytotoxicity was calculated by reference to control monolayers incubated in medium only. One cytotoxic unit corresponded to the sample dilution in which 50% of L929 cells were killed. To obtain TNF-α-positive serum, mice were infected with BCG, and 2 weeks later 20 μg LPS was given intravenously (iv). TNF-α-rich serum was collected 90 min after LPS inoculation.
Killing of intracellular T. cruzi parasites
The killing assay for intracellular T. cruzi parasites has been described previously.32 CD11b+ PECs (2 × 105) were added to tissue culture chambers (Lab-Tek Chamber Slide, Nunc, Rochester, NY) and incubated for 4 h in supplemented RPMI medium. Adherent cells were then cultured for 24 h with rIL-12 (2.5 ng/mL), rIL-18 (2.5 ng/mL), rIFN-γ (2.5 ng/mL), or medium, infected with T. cruzi at 5:1 ratio for 120 min, and washed six times to remove extracellular parasites. Cultures were kept for an additional 48 h with cytokines or medium. After this period, cells were stained with Giemsa to count intracellular amastigotes.
Detection of intracellular IFN-γ
CD11b+ aPECs (106) from normal and chronically T. cruzi-infected C57BL/6 mice were cultured for 48 h at 37°C in a 5% CO2 atmosphere, according to the manufacturer's instructions, in the presence or absence of rIL-12 and rIL-18 (10 ng/mL of each). Monensin-containing Golgistop (PharMingen) was added in the last 6 h of culture. After washing, cells were surface stained with FITC-conjugated or Cy-Chrome-conjugated mAbs to CD3 and NK1.1 (PharMingen) and F4/80 mAbs (Caltag). Cells were then fixed with the Cytofix/Cytoperm buffer (PharMingen) and incubated with PE-labeled mAb to IFN-γ (XMG-1.2) (PharMingen) diluted in Perm/Wash buffer (PharMingen). Cells were analyzed by flow cytometry using a Facscalibur with CELLQUEST software.
Confocal microscopy
For analysis of intracellular IFN-γ and TNF-α, CD11b+ PECs (2 × 105) were added to tissue culture chambers (Lab-Tek Chamber Slide) and incubated for 4 h in supplemented RPMI medium. Adherent cells were then cultured for 18 h with rIL-12 (2.5 ng/mL), rIL-18 (2.5 ng/mL), or medium, washed, and then stimulated for an additional 6 h with LPS (1 μg/mL) in the presence of monensin-containing Golgistop at 37°C in a 5% CO2 atmosphere, according to the manufacturer's instructions. After washing, cells were surface stained with FITC-conjugated mAbs to CD11b and MHC II. Cells were then fixed with the Cytofix/Cytoperm buffer and incubated with PE-labeled mAb to IFN-γ (XMG-1.2) and TNF-α (MP6-XT22) diluted in Perm/Wash buffer. All reagents were purchased from PharMingen. A CLSM 410 confocal microscopy (Carl Zeiss Inc., Oberlochen, Germany) was used to reveal the surface and intracellular staining with Carl Zeiss CLSM Image Browser software (version 3.1).
Statistical analysis
Statistical analysis was performed by unpaired ANOVA and Tukey's multiple comparison tests using GraphPad Prism 3 software. Differences between two groups were considered significant when the p value was <0.05 (5%).
Results
Role of endogenous IFN-γ in NO response of CD11b+ aPECs
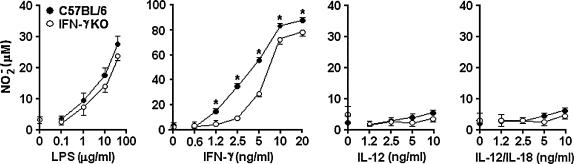
Initially, we compared the NO production of CD11b+ aPECs obtained from C57BL/6 and IFN-γKO mice. The rationale of these experiments was to determine the influence of endogenously produced IFN-γ in the ability of these cells to release NO in response to different stimuli. Therefore, CD11b+ aPECs of both mouse strains were cultured for 48 h with increasing concentrations of LPS, rIFN-γ, rIL-12, or rIL-12 plus rIL-18. As shown in Figure 1, C57BL/6 and IFN-γKO cells released significant amounts of NO in response to IFN-γ. At low concentrations of IFN-γ, however, higher NO production occurred with C57BL/6 cells compared with IFN-γKO cells. When LPS was used as a stimulus, C57BL/6 and IFN-γKO cells responded at similar levels. In this case, however, an optimal response was achieved only with high LPS doses. In contrast, IL-12 was unable to induce NO release, even at high concentrations and in the presence of IL-18. We concluded that IL-12 alone or combined with IL-18 is not capable of stimulating NO production by peritoneal macrophages, a population that under our experimental conditions comprises 98%–99% of CD11b+ aPECs (data not shown). In addition, our data have shown that endogenous IFN-γ is not required for NO release by these cells when the stimulus is LPS or exogenous IFN-γ.
FIG. 1.
NO production by CD11b+ aPECs from C57BL/6 and IFN-γKO mice. Five days after starch inoculation, C57BL/6 and IFN-γKO PECs were harvested and positively selected for CD11b expression using magnetic beads. CD11b+ PECs (2 × 105) were allowed to adhere for 4 h and then washed and cultured with medium (time 0) or stimulated with LPS, rIFN-γ, rIL-12, or rIL-12 plus rIL-18. NO was measured in 48-h supernatants as described in Materials and Methods. Experiments were repeated three times, with the same pattern of results. Data represent the mean ± SD of the different experiments. *p < 0.05, compared with values from IFN-γKO cells.
Role of endogenous IFN-γ in IL-12/IL-18 programming of LPS-induced NO response
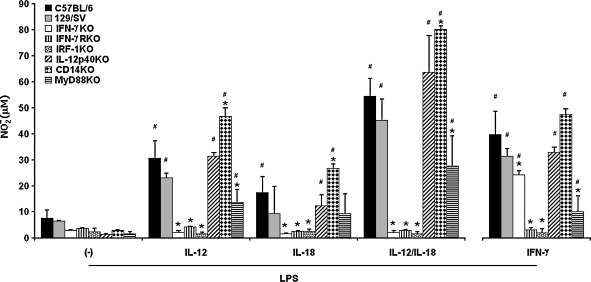
Next, we evaluated the role of IL-12 and IL-18 in macrophage programming for LPS-induced NO response, focusing again on endogenous IFN-γ. Therefore, NO levels were quantified in culture supernatants of CD11b+ aPECs from C57BL/6, 129/SV, IFN-γKO and IFNGRKO mice treated with rIL-12 or rIL-18 or both for 18 h and then stimulated for 48 h with 1 μg/mL LPS, a dose that induces CD11b+ aPECs to release low amounts of NO (Fig. 1). Additionally, the NO response was evaluated in IL-12p40KO and IRF-1KO cells to estimate the role of endogenous IL-12 and IL-23 and the contribution of IFN regulatory factor-1 (IRF-1), a transcriptional activator of genes involved in the IFN responses.37 Data in Figure 2 showed that IL-12 programming of C57BL/6 and 129/SV cells increased the LPS-induced NO production by 3–4-fold. Conversely, this response was completely abolished in IFN-γKO, IFNGRKO, and IRF-1KO cells but not in IL-12p40KO cells. IL-18 pretreatment also primed C57BL/6 cells, although the effect was less pronounced compared with that of IL-12. The minor role of endogenous IL-12 or IL-23 in IL-18 programming of LPS-induced NO response was suggested by data showing that this response was marginally reduced in IL-12p40KO cells. Remarkably, a strong priming effect was observed when IL-12 and IL-18 were added together, resulting in the release of 7–8 times more NO by C57BL/6 and 129/SV cells compared with unprimed controls. Again, IFN-γKO, IFNGRKO, and IRF-1KO cells were unable to release NO, whereas IL-12p40KO cells behaved similarly to C57BL/6 and 129/SV cells. As expected, IFN-γ pretreatment partially rescued the LPS-induced NO response in IFN-γKO cells, but IFNGRKO and IRF-1KO cells remained unresponsive. Complementary results also showed that the priming effect occurred at levels similar to those described when C57BL/6 cells were maintained with IL-12 or IL-18 or both for 6 h, 12 h, 48 h, or 96 h (data not shown). In conclusion, IL-12/IL-18 pretreatment is able to program CD11b+ aPECs to produce NO in response to LPS, and a strong synergism occurs between both cytokines. This priming effect is dependent on endogenous IFN-γ, IFN-γR, and IRF-1.
FIG. 2.
IL-12/IL-18 programming of LPS-induced NO production in CD11b+ aPECs from C57BL/6, 129/SV, IFN-γKO, IFNGRKO, IRF-1KO, IL-12p40KO, MyD88KO, and CD14KO mice. C57BL/6, 129/SV, IFN-γKO, IFNGRKO, IRF-1KO, IL-12p40KO, MyD88KO, and CD14KO CD11b+ aPECs (2 × 105) were cultured for 18 h in medium without ((−)) or with rIL-12, rIL-18, rIL-12 plus rIL-18, or rIFN-γ (2.5 ng/mL of each). After washing, cells were stimulated for an additional 48 h with LPS (1 μg/mL). NO was measured in supernatants as described in Materials and Methods. Experiments were repeated three times, with the same pattern of results. Data represent the mean ± SD of the different experiments. *p < 0.05, compared with values from C57BL/6 cells; #p < 0.05, compared with values from cells kept only in culture medium.
Role of myeloid differentiation factor 88 (MyD88) and CD14 in IL-12/IL-18 programming of LPS-induced NO response
To characterize the toll-like receptor (TLR) signaling pathway involved in the LPS-induced NO response programmed by IL-12 or IL-18 or both, we also analyzed CD11b+ aPECs from MyD88KO and CD14KO mice. As shown in Figure 2, IL-12/IL-18-primed MyD88KO cells were able to respond to LPS, but the NO levels were significantly lower than those obtained in C57BL/6 cells. In contrast, the amounts of NO released by CD14KO cells were higher than those produced by C57BL/6 cells. These findings suggest that macrophage programming is partially dependent on MyD88 and that IL-12/IL-18 pretreatment bypasses the requirement for CD14.
Effects of IL-12/IL-18 programming on T. cruzi-induced NO response
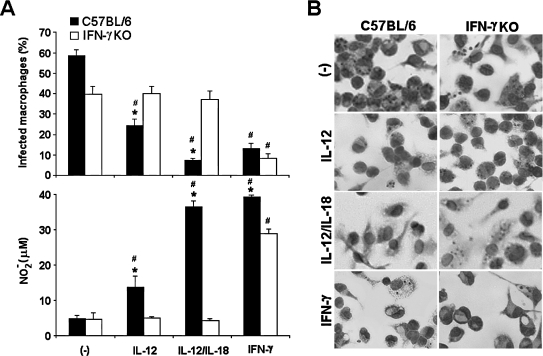
T. cruzi, the etiologic agent of Chagas' disease, is an intracellular parasite whose killing by macrophages is dependent on TLR2 and TLR9 activation38 and NO release.39 In a previous study, we demonstrated the involvement of IL-12 in the resistance of macrophage against this infection by showing that macrophages from IL-12p40KO mice are more permissive to T. cruzi replication than are wild-type cells.32 Our next step was to determine if IL-12/IL-18 programming of CD11b+ aPECs would upregulate the NO response to T. cruzi and, in consequence, restrain the intracellular infection. Parasite growth was inhibited in IL-12/IL-18-primed C57BL/6 cells, an effect that clearly correlated with the NO levels in culture supernatants (Fig. 3). Remarkably, IL-12/IL-18 pretreatment was even more efficient than IFN-γ for protecting C57BL/6 cells against T. cruzi infection. When IL-12 was used alone, even if the NO response was less pronounced, a significant decrease in the percentage of infected macrophages could still be observed. We also found that IFN-γKO cells failed to develop a NO response to T. cruzi, even after IL-12/IL-18 pretreatment. Our main conclusion from these results is that IL-12/IL-18 programming of CD11b+ aPECs protects macrophages from T. cruzi infection. Endogenous IFN-γ mediates this protective effect probably through an NO-dependent mechanism.
FIG. 3.
IL-12/IL-18 programming of T. cruzi-induced NO response and of macrophage microbicidal activity. C57BL/6 and IFN-γKO CD11b+ aPECs (2 × 105) were cultured for 24 h in medium without ((−)) or with rIL-12, rIL-12 plus rIL-18, or rIFN-γ (2.5 ng/mL of each). Cells were then infected with T. cruzi at a 5:1 ratio for 120 min, washed to remove extracellular parasites, and kept for an additional 48 h with cytokines or medium. After this period, cells were stained with Giemsa to count intracellular amastigotes (A and B). NO was measured in supernatants as described in Materials and Methods (A). Experiments were repeated three times, with the same pattern of results. Data represent the mean ± SD of the different experiments. *p < 0.05, compared with values from IFN-γKO cells; #p < 0.05, compared with values from cells kept only in culture medium.
Role of endogenous IFN-γ in IL-12/IL-18 programming of LPS-induced TNF-α response
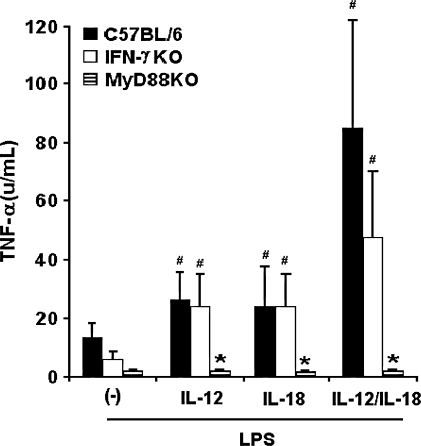
The ability of IL-12 to program the TNF-α response has been described previously.25 In an attempt to verify the role of endogenous IFN-γ in IL-12/IL-18 programming of LPS-induced TNF-α production, we have analyzed CD11b+ aPECs from C57BL/6 and IFN-γKO mice. In C57BL/6 cells, priming with IL-12 or IL-18 led to an increased TNF-α secretion in response to LPS, but combination of these cytokines greatly enhanced this effect (Fig. 4). Interestingly, different from the NO response, pretreatment of IFN-γKO cells with IL-12 or IL-18 or both caused considerable TNF-α production after LPS stimulation. Taken together, these results show that IL-12 and IL-18 are able to program the LPS-induced TNF-α response of CD11b+ aPECs through an IFN-γ-independent pathway.
FIG. 4.
IL-12/IL-18 programming of LPS-induced TNF-α response in CD11b+ aPECs from C57BL/6, IFN-γKO, and MyD88KO mice. C57BL/6, IFN-γKO, and MyD88KO CD11b+ aPECs (2 × 105) were cultured for 18 h in medium without ((−)) or with rIL-12, rIL-18, rIL-12 plus rIL-18, or rIFN-γ (2.5 ng/mL of each). After washing, cells were stimulated for additional 6 h with LPS (1 μg/mL). TNF-α was measured in supernatants as described in Materials and Methods. Experiments were repeated three times, with the same pattern of results. Data represent the mean ± SD of the different experiments. *p < 0.05, compared with values from C57BL/6 cells; #p < 0.05, compared with values from cells kept only in culture medium
Role of MyD88 in LPS-induced TNF-α response programmed by IL-12 or IL-18 or both
To evaluate the involvement of the MyD88-dependent pathway in IL-12/IL-18-primed LPS-induced TNF-α response, CD11b+ aPECs from MyD88KO mice were also analyzed. Data in Figure 4 showed that MyD88KO cells failed to secrete TNF-α in response to LPS, even after IL-12/IL-18 priming. Therefore, we concluded that different from the NO response, TNF-α production programmed by IL-12/IL-18 exclusively occurs through the MyD88-signaling pathway.
Contribution of lymphocytes in IL-12/IL-18 programming of LPS-induced NO and TNF-α responses
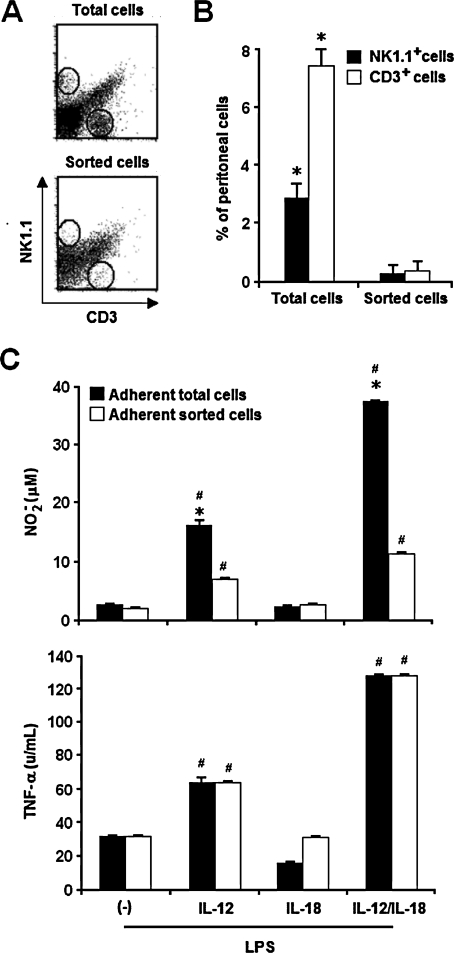
Although several reports have described macrophages as IFN-γ-producing cells, this matter deserves further investigation, as minute contamination with T and NK cells has been implicated in IFN-γ production in IL-12/IL-18-stimulated macrophage cell cultures.40 Therefore, experiments were performed to determine the contribution of lymphocytes to the LPS-induced NO and TNF-α responses of IL-12/IL-18-primed aPECs. In these studies, total C57BL/6 cells showing granularity and size characteristics of lymphocytes were removed from the PEC suspensions using a cell sorter. This procedure eliminated 91.7% of NK1.1+ cells and 95.1% of CD3+ cells (Figs. 5A,B). Total and sorted cells were then allowed to adhere, washed, and cultured with rIL-12 or rIL-18 or both for 18 h and stimulated with LPS for an additional 6 h for TNF-α analysis and 48 h for NO analysis. It is worth noting that in these experiments, cells were not purified according to CD11b expression. As shown in Figure 5C, adherent sorted cells were able to produce NO in response to LPS when primed with IL-12 and IL-12/IL-18. However, in these cases, NO levels reached 43.1% and 30.2% in relation to those of adherent total cells, respectively. In contrast, we observed that LPS-stimulated total and adherent sorted cells produced an equivalent amount of TNF-α, attaining maximum levels after IL-12/IL-18 priming. Thus, according to our data, lymphocytes are not necessary for the IL-12/IL-18 programming of LPS-induced TNF-α response, but they clearly modulate the NO response.
FIG. 5.
IL-12/IL-18 programming of LPS-induced NO and TNF-α responses in highly purified macrophages. (A) Cells with a macrophage size were sorted by flow cytometry from C57BL/6 PECs. (B) Percentage of NK1.1+ and CD3+ cells were determined before and after sorting. (C) CD11b+ aPECs (2 × 105) (sorted and nonsorted) of C57BL/6 mice were cultured for 12 h in medium without ((−)) or with rIL-12 (2.5 ng/mL), rIL-18 (2.5 ng/mL) or both. After washing, cells were stimulated for an additional 6 h (for TNF-α detection) or 48 h (for NO detection) with LPS (1 μg/mL). NO and TNF-α were measured in supernatants as described in Materials and Methods. Experiments were repeated three times, with the same pattern of results. Data represent the mean ± SD of the different experiments. *p < 0.05, compared with values from sorted cells; #p < 0.05, compared with values from cells kept only in culture medium.
Contribution of CD3+NK1.1+ and CD3−NK1.1− cells to IL-12/IL-18 programming of LPS-induced NO response
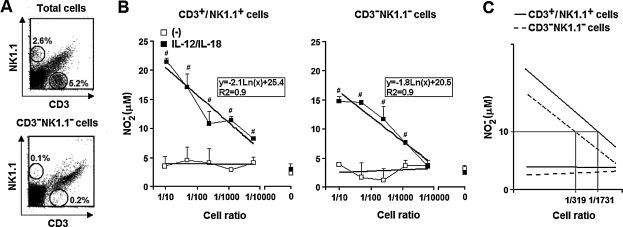
The aim of the next experiments was to determine if peritoneal cells other than T and NK cells could supply the IFN-γ required for IL-12/IL-18 programming of the LPS-induced NO response of CD11b+ aPECs. For these experiments, PECs of iNOSKO mice were used as sources of IFN-γ-producing cells (unable to release NO) and PECs of IFN-γKO mice supplied IFN-γ-responding cells (able to release NO but not IFN-γ). Initially, iNOSKO PECs were separated according to NK1.1 and CD3 expression in CD3−NK1.1− and CD3+/NK1.1+ cells. CD3−NK1.1− (iNOSKO) cells presented around 0.3% of contamination with NK1.1+ and CD3+ cells (Fig. 6A), whereas these cell populations comprised 86.4% of CD3+NK1.1+ (iNOSKO) cells (data not shown). CD3−NK1.1− and CD3+NK1.1+ (iNOSKO) cells were titrated in 96-well culture plates containing IFN-γKO aPECs. Cells were then cultured with rIL-12 plus rIL-18 for 18 h, washed, and stimulated with LPS for an additional 48 h. As shown in Figure 6B, considerable NO levels were detected in the supernatants of LPS-stimulated IFN-γKO cells that had been cultured with CD3−NK1.1− and CD3+NK1.1+ (iNOSKO) cells in the presence of IL-12/IL-18. Moreover, there was a positive correlation between the NO levels and the cell ratios, that is, the number of CD3−NK1.1− or CD3+NK1.1+ (iNOSKO) cells divided by the numbers of IFN-γKO cells. From the tendency titration curves, we compared the capacity of IL-12/IL-18-stimulated CD3−NK1.1− and CD3+NK1.1+ (iNOSKO) cells to provide the IFN-γ needed for programming IFN-γKO cells to release 10 μM of NO−2 in response to LPS. We found that the frequencies of CD3+NK1.1+ and CD3−NK1.1− cells required for obtaining this priming effect were 1/1731 and 1/319, respectively (Fig. 6C). Thus, assuming that half of IFN-γKO cells were adherent cells (105 cells/well), 58 CD3+NK1.1+ cells/well and 313 CD3−NK1.1− cells/well would be necessary to reach 10 μM of NO−2. Furthermore, considering that T or NK cells comprise around 0.3% of CD3−NK1.1− cells, <1 contaminant CD3+ or NK1.1+ cell would be expected among 313 CD3−NK1.1− cells, a number 50 times lower than the one predictable if the priming effect of CD3−NK1.1− cells were due to CD3+NK1.1+ cell contamination. Besides this, considering the tendency titration curves, a very high CD3+NK1.1+ cell contamination (of around 8%) would be necessary to attain 20 μM of NO−2. These data corroborate the notion that cell populations other than T and NK cells may contribute to the IFN-γ production required for programming IFN-γKO cells to release NO in response to LPS.
FIG. 6.
Contribution of CD3+NK1.1+ and CD3−NK1.1− (iNOSKO) cells in the IL-12/IL-18 programming of LPS-induced NO response of adherent peritoneal IFN-γKO cells. (A) iNOSKO PECs were separated according to the expression of NK1.1 and CD3 in CD3−NK1.1− and CD3+NK1.1+ cells as described in Materials and Methods. Percentages of NK1.1+ and CD3+ cells were determined before and after cell separation. (B) aPECs (2 × 105) from IFN-γKO mice were cultured for 12 h in medium without ((−)) or with rIL-12 (2.5 ng/mL) plus rIL-18 (2.5 ng/mL) in the presence of titrated numbers of CD3−NK1.1− and CD3+NK1.1+ (iNOSKO) PECs. After washing, cells were stimulated for an additional 48 h with LPS (1 μg/mL). NO was measured in supernatants as described in Materials and Methods. Cell ratios were obtained by dividing the number of CD3−NK1.1− or CD3+NK1.1+ (iNOSKO) cells by the numbers of IFN-γKO cells. Tendency titration curves calculated by the method of least squares were used to obtain the correspondent mathematic equations. (C) The cell ratios expected to attain a NO−2 level of 10 μM in the culture supernatants were determined by using the mathematic equations obtained from tendency titration curves. Experiments were repeated three times, with the same pattern of results. Data represent the mean ± SD of the different experiments. #p < 0.05, compared with cells kept only in culture medium.
Analysis of IFN-γ production by IL-12/IL-18-stimulated macrophages
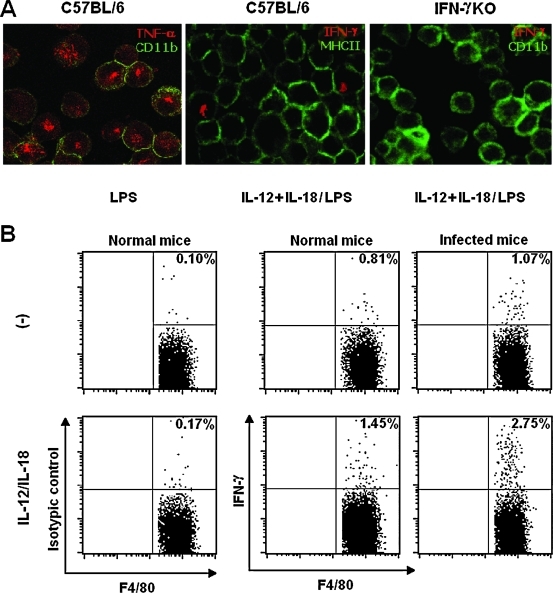
As there are conflicting results in the literature regarding the capacity of peritoneal macrophages to produce IFN-γ,41 experiments were carried out to clarify this issue. When the phenotype of IFN-γ-producing cells was assessed by confocal microscopy, we found a few CD11b+ aPECs expressing intracellular IFN-γ and surface MHC II. Positive staining for intracellular IFN-γ could be detected among C57BL/6 CD11b+ aPECs primed with rIL-12 plus rIL-18 for 18 h and then stimulated with LPS for 6 h in the presence of Golgistop containing monensin (Fig. 7A). IFN-γKO CD11b+ aPECs treated in the same manner were used as a negative control. Intracellular TNF-α staining of LPS-stimulated C57BL/6 CD11b+ aPECs was the positive control. Moreover, low levels of IFN-γ protein (1.5 ± 0.3 ng/mL) could be detected in culture supernatants from IL-12/IL-18-primed LPS-stimulated C57BL/6 CD11b+ aPECs, whereas those from unprimed macrophages were negative. To address IFN-γ production using a different approach, C57BL/6 CD11b+ aPECs cultured with or without rIL-12 plus rIL-18 were surface-labeled for F4/80, CD3, and NK1.1 and analyzed for intracellular IFN-γ by flow cytometry. Adherent PECs from chronically T. cruzi-infected C57BL/6 mice were also evaluated, as we previously observed high NO production by these cells after LPS stimulation. Data in Figure 7B showed an increase of intracellular IFN-γ in F4/80+ CD3−NK1.1− cells after IL-12/IL-18 stimulation. Interestingly, when cells were obtained from chronically T. cruzi-infected mice, the percentage of IFN-γ-producing cells attained 2.75% of F4/80+ CD3−NK1.1− cells. Intracellular IFN-γ was also detected in IL-12/IL-18-stimulated CD3+ and NK1.1+ cells, but CD45R(B220)+ cells showed no staining for this cytokine (data not shown). Among IL-12/IL-18-stimulated CD11b+ aPECs from noninfected mice, 1.2% of the cells were positive for CD3, with 12% of them coexpressing IFN-γ, whereas NK1.1+ cells corresponded to 0.3% of the cells, with 6% of them containing intracellular IFN-γ. This represents a total of 0.16% contamination with IFN-γ+ lymphocytes (data not shown). Thus, among CD11b+ aPECs, the numbers of IFN-γ+ F4/80+ CD3−NK1.1− cells were 9-fold higher than those of IFN-γ+ CD3+/NK1.1+ contaminant cells. In conclusion, our studies have identified MHC II+ and F4/80+ cells, as well as CD3+ and NK1.1+ cells, as sources of IFN-γ among IL-12/IL-18-stimulated CD11b+ aPECs.
FIG. 7.
IFN-γ production by CD11b+ aPECs. (A) CD11b+ aPECs (2 × 105) from C57BL/6 and IFN-γKO mice were cultured in medium without ((−)) or with rIL-12 and rIL-18 (2.5 ng/mL of each) for 18 h. After washing, cells were cultured for 6 h with LPS (1 μg/mL) in the presence of Golgistop containing monensin. Fixed cells were stained with fluorochrome-labeled antibodies to CD11b, MHC II, TNF-α, and IFN-γ and analyzed by confocal microscopy. (B) CD11b+ aPECs (2 × 105) from noninfected and chronically T. cruzi-infected C57BL/6 mice were kept in culture medium without ((−)) or with rIL-12 plus rIL-18 (2.5 ng/mL of each) for 48 h. Golgistop containing monensin was added in the last 6 h. Fixed cells were stained with fluorochrome-labeled antibodies to F4/80, CD3, NK1.1, and IFN-γ and analyzed by flow citometry. Gated F4/80+ CD3−NK1.1− cells are shown. Experiments were repeated three times, with the same pattern of results.
Discussion
Although IL-12 and IL-18 have been shown to induce secretion of IFN-γ, TNF-α, and NO by macrophages,14–24 it is still a matter of debate whether these responses are entirely carried out by macrophages or require the cooperation of other cell types.41 Here, we have investigated the role of endogenous IFN-γ in macrophage programming by IL-12 and IL-18. Initially, we found that IL-12 and IL-18 cannot stimulate NO release by CD11b+ aPECs, but these cytokines are very effective and act synergistically in programming macrophages for a robust production of NO and TNF-α in response to LPS. Accordingly, IL-12/IL-18 pretreatment also primed macrophages for NO production in response to T. cruzi infection, showing a direct correlation with resistance to intracellular parasite replication. We have also found that endogenous IFN-γ, IFNGR, and IRF-1 are essential for IL-12/IL-18 programming of LPS-induced NO production, whereas TNF-α production is independent of endogenous IFN-γ. Regarding the IL-12/IL-18 priming effect for LPS-induced responses, we have observed that the MyD88-dependent pathway of TLR4 signaling is indispensable for TNF-α production, whereas for NO release, the MyD88 requirement is only partial. Concerning the involvement of lymphocytes in IL-12/IL-18 priming, we have found that for TNF-α secretion, contaminant lymphocytes play no role, whereas for NO release, they do play a significant role, as they contribute to providing IFN-γ. Most importantly, we have identified macrophages as an alternative source of IFN-γ among CD11b+ aPECs, particularly in the peritoneum of chronically T. cruzi-infected mice.
Previous studies have shown that peritoneal and bone marrow-derived macrophages stimulated with IL-12 and IL-18 are able to produce NO and that this response is dependent on IFN-γ.42,43 We could not reproduce these results, as we did not detect NO production in response to IL-12/IL-18 stimulation. One likely explanation for these differences is that our macrophage cultures contained lower numbers of IFN-γ-producing cells. This could be due to the fact that we used unprimed macrophages purified according to CD11b expression to minimize lymphocyte contamination. Thus, it seems that IL-12 does not induce NO release by unprimed peritoneal macrophages, even in the presence of considerable amounts of IL-18. In contrast, a few IFN-γ-producing cells among CD11b+ aPECs seem to be sufficient to allow IL-12/IL-18 programming for the NO response to LPS or T. cruzi. This priming effect is undoubtedly an interesting phenomenon, as it renders macrophages highly efficient at the moment of their encounter with pathogens. In addition, considering that iNOS gene promoter contains an NF-κB-binding sequence44 and an IRF-1-binding sequence,45,46 our data indicate that the IRF-1 transcription factor is essential for IL-12/IL-18 programming of LPS-induced NO response, despite the fact that high doses of LPS can directly induce NO production by macrophages through activating NF-κB. In other words, a likely description for the priming phenomenon is that IL-12/IL-18 pretreatment induces the secretion of IFN-γ, which acts through IRF-1 and programs macrophages for enhanced NO production.
Different from the NO response, IL-12/IL-18 pretreatment seems to directly program the LPS-induced TNF-α production in peritoneal macrophages. This conclusion was suggested by our results showing a similar TNF-α response in the presence or in the absence of IFN-γ or contaminant lymphocytes. The involvement of IFN-γ in TNF-α secretion has been investigated previously by treating macrophages with anti-IFN-γ antibodies. These studies showed that the TNF-α response to LPS occurs independently of IFN-γ,15 whereas this cytokine is required for TNF-α production in response to IL-12/IL-18.20 Thus, it looks as if both TLR4-dependent and IFN-γ-dependent pathways participate in TNF-α secretion. In some circumstances, the IFN-γ pathway seems to predominate, as suggested by data showing abrogation of the synergistic effect of IL-12 on TNF-α secretion in BCG-infected IFN-γKO macrophages.22 Conversely, in the priming effect described here, IL-12/IL-18 pretreatment could enhance the amount of TNF-α induced by LPS in an IFN-γ-independent manner by upregulating TNF transcripts, which could result from transcriptional and posttranscriptional modifications, as previously described for phorbolactivated human monocytes.47
Macrophage activation through TLR4 causes NF-κB activation by the MyD88-dependent pathway and in IRF-3 activation by the TRIF (TOLL/IL-IR[TIR] domain containing adapter inducing IFN-β)-dependent pathway.48 TLR4, upon recognition of LPS, can induce the production of proinflammatory cytokines, such as TNF-α, IL-1β, and IL-6, in an MyD88-dependent manner, and in turn, these cytokines might modulate NO production. TLR4 signaling also leads to rapid IRF-3 activation, resulting in IFN-α/β release and to delayed NF-κB activation with production of NO and several chemokines.49 Our results confirm the major role of an MyD88-dependent pathway for the LPS-induced TNF-α response programmed by IL-12/IL-18, whereas for NO release, both arms of the TLR4 signaling pathway seem to cooperate. NF-κB and IRF-1 transcription factors may act together on their specific binding sequences in the iNOS gene promoter for optimal NO production.45,46 Our data showing MyD88-independent LPS-induced NO production in IL-12/IL-18-primed macrophages could be explained either by the delayed NF-κB activation or by the secretion of IFN-α/β. However, our results clearly showed that signaling via IFNGR is essential for NO production, indicating the major role of IFN-γ in this model. Thus, the involvement of IFN-α/β in NO production might be indirect in this process probably by enhancing IFN-γ production via Stat1 activation.50 In addition, although CD14 is known to improve the interaction of LPS with TLR4,48 in our experiments, this coreceptor was not required for the LPS-induced NO response of IL-12/IL-18-primed macrophages. A possible explanation for this finding is that IL-12/IL-18 priming increases the number or avidity of TLR4 molecules in macrophages, bypassing the requirement for CD14.
The low numbers of IFN-γ-producing cells within IL-12/IL-18-stimulated CD11b+ aPECs make their identification a hard task.41 Here, we have addressed this issue by three different approaches. The first approach, in which lymphocytes were eliminated from cell preparations, clearly showed a significant role for these cells in NO production. The second approach, which compared the priming capacity of CD3+NK1.1+ and CD3−NK1.1− cells, indicated that the IFN-γ supplied by cells other than T and NK cells contributes to enhancing the NO response programmed by IL-12/IL-18. Moreover, by evaluating intracellular IFN-γ in IL-12/IL-18-stimulated CD11b+ aPECs, we have identified a small fraction of IFN-γ-producing cells with a macrophage phenotype, which was significantly increased in chronically T. cruzi-infected mice. Thus, it seems that in some circumstances, as the one shown here, macrophages could also secrete IFN-γ. Classic sources of IFN-γ are NK cells, CD4+ cells, and CD8+ T cells, but NK T cells, γδ T cells, and B cells have also been shown to secrete this cytokine.41 More recently, myeloid cells, such as macrophages and DCs, were also reported to produce IFN-γ.40 Thus, in principle, macrophages, DCs, and B cells from the peritoneal cavity could be alternative sources of IFN-γ after IL-12/IL-18 stimulation. We have found that the small fraction of CD3−NK1.1− cells expressing intracellular IFN-γ was positive for F4/80, a macrophage marker not expressed by DCs and B cells. Although MHC II+ IFN-γ+ cells visualized by confocal microscopy could still be B cells, the few contaminant CD45R (B220)+ cells did not show intracellular IFN-γ, as revealed by flow cytometry. Based on these considerations, we concluded that a small proportion of macrophages among CD11b+ aPECs is able to secrete IFN-γ when stimulated with IL-12/IL-18.
Our finding that IFN-γ+F4/80+ CD3−NK1.1− cells were increased in chronically T. cruzi-infected mice not only corroborates the idea that in some situations macrophages can produce IFN-γ but also indicates that this capacity can be acquired under conditions of infection. This is an interesting possibility, as IFN-γ mRNA and protein have been detected in macrophages obtained from mice and humans infected with mycobacteria, Legionella, Salmonella, and Chlamydia.15,19,51,52 Unknown additional signals generated in vivo could be required for making macrophages capable of secreting IFN-γ in response to IL-12/IL-18. As a consequence of this, the proportion of IFN-γ-secreting cells within IL-12/IL-18-stimulated peritoneal macrophages would vary according to the immunologic status of the animals. This is an interesting point because it could be the reason why in our experiments only a minor fraction of IL-12/IL-18-stimulated F4/80+ cells showed intracellular IFN-γ. This hypothesis could explain the fact that this population is increased in chronically T. cruzi-infected mice. Regarding the additional signals that could be necessary to differentiate macrophages in IFN-γ-secreting cells, IL-2 and IL-15 emerge as reliable possibilities. IL-2 has been shown to induce expression and secretion of IFN-γ by peritoneal macrophages, an effect synergized by simultaneous activation with IL-12.24 IL-15, in association with IL-12, is a potent stimulus for IFN-γ production by a recently described population of DCs showing cytotoxic activity.53 The necessity of IL-2 or IL-15 or both for IFN-γ secretion by macrophages could be an alternative explanation for the results of Schleicher et al.,40 showing the absence of IFN-γ protein, with detectable levels of IFN-γ mRNA, in IL-12/IL-18-stimulated PECs from mice lacking the common γ chain, which is shared by both IL-2 and IL-15 receptors. Another additional signal that could be implicated in macrophage differentiation to IFN-γ production is the one provided by the CD40–CD40L costimulatory pathway, which results from interaction with lymphocytes and other cell types. This possibility is suggested by data showing that i.p. injection of anti-CD40 antibodies elicits intracellular IFN-γ expression by 55%–75% of F4/80+ peritoneal macrophages.54
The results presented here reinforce the notion that IL-12, in synergism with IL-18, can program macrophages through an autocrine pathway,14–24 corroborating our previous observation that IL-12 or IL-23 or both directly influence the macrophage activation profile.32,33 In addition, the present study suggests that at least in some circumstances, macrophages can be an alternative source of IFN-γ. The biologic relevance of intracellular IFN-γ in macrophages is still under discussion.41 Our results indicate that by producing minute quantities of IFN-γ, which could be insufficient for directly activating effector functions, macrophages can be programmed for an optimized response to a future encounter with pathogens. Based on these findings, we consider the elucidation of the molecular basis involved in macrophage differentiation an exciting field for the near future.
Acknowledgments
This study was supported by grants from FAPESP and CNPq (Brazil). We thank Rogério Nascimento and Eliane Costa for technical assistance.
References
- 1.Kobayashi M. Fitz L. Ryan M. Hewick RM. Clark SC. Chan S. Loudon R. Sherman F. Perussia B. Trinchieri G. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 1989;170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heufler C. Koch F. Stanzl U. Topar G. Wysocka M. Trinchieri G. Enk A. Steinman RM. Romani N. Schuler G. Interleukin-12 is produced by dendritic cells and mediates T helper 1 development as well as interferon-γ production by T helper 1 cells. Eur. J. Immunol. 1996;26:659–668. doi: 10.1002/eji.1830260323. [DOI] [PubMed] [Google Scholar]
- 3.D'Andrea A. Rengaraju M. Valiante NM. Chehimi J. Kubin M. Aste M. Chan SH. Kobayashi M. Young D. Nickbarg E. Production of natural killer cell stimulatory factor (interleukin 12) by peripheral blood mononuclear cells. J. Exp. Med. 1992;176:1387–1398. doi: 10.1084/jem.176.5.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stobie L. Gurunathan S. Prussin C. Sacks DL. Glaichenhaus N. Wu CY. Seder RA. The role of antigen and IL-12 in sustaining Th1 memory cells in vivo: IL-12 is required to maintain memory/effector Th1 cells sufficient to mediate protection to an infectious parasite challenge. Proc. Natl. Acad. Sci. USA. 2000;97:8427–8432. doi: 10.1073/pnas.160197797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germann T. Bongartz M. Dlugonska H. Hess H. Schmitt E. Kolbe L. Kolsch E. Podlaski FJ. Gately MK. Rude E. Interleukin-12 profoundly up-regulates the synthesis of antigen-specific complement-fixing IgG2a, IgG2b and IgG3 antibody subclasses in vivo. Eur. J. Immunol. 1995;25:823–829. doi: 10.1002/eji.1830250329. [DOI] [PubMed] [Google Scholar]
- 6.Meko JB. Yim JH. Tsung K. Norton JA. High cytokine production and effective antitumor activity of a recombinant vaccinia virus encoding murine interleukin 12. Cancer Res. 1995;55:4765–4770. [PubMed] [Google Scholar]
- 7.Heinzel FP. Schoenhaut DS. Rerko RM. Rosser LE. Gately MK. Recombinant interleukin 12 cures mice infected with Leishmania major. J. Exp. Med. 1993;177:1505–1509. doi: 10.1084/jem.177.5.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magram J. Connaughton SE. Warrier RR. Carvajal DM. Wu CY. Ferrante J. Stewart C. Sarmiento U. Faherty DA. Gately MK. IL-12-deficient mice are defective in IFN gamma production and type 1 cytokine responses. Immunity. 1996;4:471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- 9.Mattner F. Magram J. Ferrante J. Launois P. Di Padova K. Behin R. Gately MK. Louis JA. Alber G. Genetically resistant mice lacking interleukin-12 are susceptible to infection with Leishmania major and mount a polarized Th2 cell response. Eur. J. Immunol. 1996;26:1553–1559. doi: 10.1002/eji.1830260722. [DOI] [PubMed] [Google Scholar]
- 10.Aliberti JCS. Cardoso MA. Martins GA. Gazzinelli RT. Vieira LQ. Silva JS. Interleukin-12 mediates resistance to Trypanosoma cruzi in mice and is produced by murine macrophages in response to live trypomastigotes. Infect. Immun. 1996;64:1961–1967. doi: 10.1128/iai.64.6.1961-1967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper AM. Magram J. Ferrante J. Orme IM. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with Mycobacterium tuberculosis. J. Exp. Med. 1997;186:39–45. doi: 10.1084/jem.186.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchieri G. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu. Rev. Immunol. 1995;13:251–276. doi: 10.1146/annurev.iy.13.040195.001343. [DOI] [PubMed] [Google Scholar]
- 13.Trinchieri G. Cytokines acting on or secreted by macrophages during intracellular infection (IL-10, IL-12, IFN-gamma) Curr. Opin. Immunol. 1997;9:17–23. doi: 10.1016/s0952-7915(97)80154-9. [DOI] [PubMed] [Google Scholar]
- 14.Puddu P. Fantuzzi L. Borghi P. Varano B. Rainaldi G. Guillemard E. Malorni W. Nicaise P. Wolf SF. Belardelli F. Gessani S. IL-12 induces IFN-gamma expression and secretion in mouse peritoneal macrophages. J. Immunol. 1997;159:3490–3497. [PubMed] [Google Scholar]
- 15.Fenton M. Vermeulen MW. Kim S. Burdick M. Strieter RM. Kornfield H. Induction of interferon production in human alveolar macrophages by Mycobacterium tuberculosis. Infect. Immun. 1997;65:5149–5156. doi: 10.1128/iai.65.12.5149-5156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munder M. Mallo M. Eichmann K. Modolell M. Murine macrophages secrete interferon gamma upon combined stimulation with interleukin (IL)-12 and IL-18: a novel pathway of autocrine macrophage activation. J. Exp. Med. 1998;187:2103–2108. doi: 10.1084/jem.187.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salkowski CA. Kopydlowski K. Blanco J. Cody MJ. McNally R. Vogel SN. IL-12 is dysregulated in macrophages from IRF-1 and IRF-2 knockout mice. J. Immunol. 1999;163:1529–1536. [PubMed] [Google Scholar]
- 18.Schindler H. Lutz MB. Rollinghoff M. Bogdan C. The production of IFN-gamma by IL-12/IL-18-activated macrophages requires Stat4 signaling and is inhibited by IL-4. J. Immunol. 2001;166:3075–3082. doi: 10.4049/jimmunol.166.5.3075. [DOI] [PubMed] [Google Scholar]
- 19.Wang J. Wakeham J. Harkness R. Xing Z. Macrophages are a significant source of type 1 cytokines during mycobacterial infection. J. Clin. Invest. 1999;103:1023–1029. doi: 10.1172/JCI6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami K. Qureshi MH. Koguchi Y. Zhang T. Okamura H. Kurimoto M. Saito A. Role of TNF-alpha in the induction of fungicidal activity of mouse peritoneal exudate cells against Cryptococcus neoformans by IL-12 and IL-18. Cell. Immunol. 1999;193:9–16. doi: 10.1006/cimm.1999.1460. [DOI] [PubMed] [Google Scholar]
- 21.Fantuzzi L. Puddu P. Varano B. Del Corno M. Belardelli F. Gessani S. IFN-alpha and IL-18 exert opposite regulatory effects on the IL-12 receptor expression and IL-12-induced IFN-gamma production in mouse macrophages: novel pathways in the regulation of the inflammatory response of macrophages. J. Leukoc. Biol. 2000;68:707–714. [PubMed] [Google Scholar]
- 22.Xing Z. Zganiacz A. Santosuosso M. Role of IL-12 in macrophage activation during intracellular infection: IL-12 and mycobacteria synergistically release TNF-alpha and nitric oxide from macrophages via IFN-gamma induction. J. Leukoc. Biol. 2000;68:897–902. [PubMed] [Google Scholar]
- 23.Grohmann U. Belladonna ML. Vacca C. Bianchi R. Fallarino F. Orabona C. Fioretti M. Puccetti P. Positive regulatory role of IL-12 in macrophages and modulation by IFN-γ. J. Immunol. 2001;167:221–227. doi: 10.4049/jimmunol.167.1.221. [DOI] [PubMed] [Google Scholar]
- 24.Puddu P. Carollo M. Pietraforte I. Spadaro F. Tombesi M. Ramoni C. Belardelli F. Gessani S. IL-2 induces expression and secretion of IFN-gamma in murine peritoneal macrophages. J. Leukoc. Biol. 2005;78:686–695. doi: 10.1189/jlb.0105035. [DOI] [PubMed] [Google Scholar]
- 25.Shnyra A. Brewington R. Alipio A. Amura C. Morrison DC. Reprogramming of lipopolysaccharide-primed macrophages is controlled by a counterbalanced production of IL-10 and IL-12. J. Immunol. 1998;160:3729–3736. [PubMed] [Google Scholar]
- 26.Okamura H. Tsutsi H. Komatsu T. Yutsudo M. Hakura A. Tanimoto T. Torigoe K. Okura T. Nukada Y. Hattori K. Akita K. Namba M. Tanabe F. Konishi K. Fukuda S. Kurimoto M. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimoto T. Takeda K. Tanaka T. Ohkusu K. Kashiwamura S. Okamura H. Akira S. Nakanishi K. Tsutsui H. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J. Immunol. 1998;161:3400–3407. [PubMed] [Google Scholar]
- 28.Takeda K. Tsutsui H. Yoshimoto T. Adachi O. Yoshida N. Kishimoto T. Okamura H. Nakanishi K. Akira S. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 29.Fukao T. Matsuda S. Koyasu S. Synergistic effects of IL-4 and IL-18 on IL-12-dependent IFN-gamma production by dendritic cells. J. Immunol. 2000;164:64–71. doi: 10.4049/jimmunol.164.1.64. [DOI] [PubMed] [Google Scholar]
- 30.Stober D. Schirmbeck R. Reimann J. IL-12/IL-18-dependent IFN-gamma release by murine dendritic cells. J. Immunol. 2001;167:957–965. doi: 10.4049/jimmunol.167.2.957. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson KA. Aung H. Wu M. Toossi Z. Modulation of transforming growth factor beta-1 gene expression by interleukin-12. Scand. J. Immunol. 2000;52:271–277. doi: 10.1046/j.1365-3083.2000.00772.x. [DOI] [PubMed] [Google Scholar]
- 32.Bastos KR. Alvarez JM. Marinho CR. Rizzo LV. Lima MR. Macrophages from IL-12p40-deficient mice have a bias toward the M2 activation profile. J. Leukoc. Biol. 2002;71:271–278. [PubMed] [Google Scholar]
- 33.Bastos KR. Barboza R. Elias RM. Sardinha LR. Grisotto MG. Marinho CR. Amarante-Mendes GP. Alvarez JM. Lima MR. Impaired macrophage responses may contribute to exacerbation of blood-stage Plasmodium chabaudi chabaudi malaria in interleukin-12-deficient mice. J. Interferon Cytokine Res. 2002;22:1191–1199. doi: 10.1089/10799900260475713. [DOI] [PubMed] [Google Scholar]
- 34.Kawai T. Adachi O. Ogawa T. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:15–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 35.Reis LFL. Ruffner H. Stark G. Aguet M. Weissmann C. Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EMBO J. 1994;13:4798–4806. doi: 10.1002/j.1460-2075.1994.tb06805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flick DA. Gifford GE. Comparison of in vitro cell cytotoxic assays for tumor necrosis factor. J. Immunol. Methods. 1984;68:167–175. doi: 10.1016/0022-1759(84)90147-9. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi T. Ogasawara K. Takaoka A. Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 38.Bafica A. Santiago HC. Goldszmid R. Ropert C. Gazzinelli RT. Sher A. Cutting edge: TLR9 and TLR2 signaling together account for MyD88-dependent control of parasitemia in Trypanosoma cruzi infection. J. Immunol. 2006;177:3515–3519. doi: 10.4049/jimmunol.177.6.3515. [DOI] [PubMed] [Google Scholar]
- 39.Gazzinelli RT. Oswald IP. Hieny S. James SL. Sher A. The microbicidal activity of interferon-gamma-treated macrophages against Trypanosoma cruzi involves an l-arginine-dependent, nitrogen oxide-mediated mechanism inhibitable by interleukin-10 and transforming growth factor-beta. Eur. J. Immunol. 1992;22:2501–2506. doi: 10.1002/eji.1830221006. [DOI] [PubMed] [Google Scholar]
- 40.Schleicher U. Hesse A. Bogdan C. Minute numbers of contaminant CD8+ T cells or CD11b+CD11c+ NK cells are the source of IFN-gamma in IL-12/IL-18-stimulated mouse macrophage populations. Blood. 2005;105:1319–1328. doi: 10.1182/blood-2004-05-1749. [DOI] [PubMed] [Google Scholar]
- 41.Bogdan C. Schleicher U. Production of interferon-gamma by myeloid cells—fact or fancy? Trends Immunol. 2006;27:282–290. doi: 10.1016/j.it.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Golab J. Zagozdzon R. Stoklosal T. Kaminski R. Kozar K. Jakobisiak M. Direct stimulation of macrophages by IL-12 and IL-18—a bridge too far? Immunol. Lett. 2000;72:153–157. doi: 10.1016/s0165-2478(00)00178-4. [DOI] [PubMed] [Google Scholar]
- 43.Matsuura M. Saito S. Hirai Y. Okamura H. A pathway through interferon-γ is the main pathway of nitric oxide upon stimulation with bacterial lipopolysaccharide in mouse peritoneal cells. Eur. J. Biochem. 2003;270:4016–4025. doi: 10.1046/j.1432-1033.2003.03792.x. [DOI] [PubMed] [Google Scholar]
- 44.Lowenstein CJ. Alley EW. Raval P. Snowman AM. Snyder SH. Russell SW. Murphy WJ. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon γ and lipopolysaccharide. Proc. Natl. Acad. Sci. USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamijo R. Harada H. Matsuyama T. Bosland M. Gerecitano J. Shapiro D. Le J. Koh SI. Kimura T. Green SJ. Mak TW. Taniguchi T. Vilcek J. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 46.Martin E. Nathan C. Xie QW. Role of interferon regulatory factor 1 in induction of nitric oxide synthase. J. Exp. Med. 1994;180:977–984. doi: 10.1084/jem.180.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sariban E. Imamura K. Luebbers R. Kufe DJ. Transcriptional and posttranscriptional regulation of tumor necrosis factor gene expression in human monocytes. J. Clin. Invest. 1998;81:1506–1510. doi: 10.1172/JCI113482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill LA. How toll-like receptors signal: what we know and what we don't know. Curr. Opin. Immunol. 2006;18:3–9. doi: 10.1016/j.coi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 49.Hoebe K. Du X. Georgel P. Janssen E. Tabeta K. Kim SO. Goode J. Lin P. Mann N. Mudd S. Crozat K. Sovath S. Han J. Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TLR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 50.Takaoka A. Yanai H. Interferon signalling networks in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 51.Salins S. Newton C. Widen R. Klein TW. Friedman H. Differential induction of gamma interferon in Legionella pneumophila-in-fected macrophages from BALB/c and A/J mice. Infect. Immun. 2001;69:3605–3610. doi: 10.1128/IAI.69.6.3605-3610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothfuchs AG. Gigliotti D. Palmblad K. Andersson U. Wigzell H. Rottenberg ME. IFN-alpha beta-dependent, IFN-gamma secretion by bone marrow-derived macrophages controls an intracellular bacterial infection. J. Immunol. 2001;167:6453–6461. doi: 10.4049/jimmunol.167.11.6453. [DOI] [PubMed] [Google Scholar]
- 53.Chan CW. Crafton E. Fan HN. Flook J. Yoshimura K. Skarica M. Brockstedt D. Dubensky TW. Stins MF. Lanier LL. Pardoll DM. Housseau F. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat. Med. 2006;12:207–213. doi: 10.1038/nm1352. [DOI] [PubMed] [Google Scholar]
- 54.Buhtoiarov IN. Lum H. Berke G. Paulnock DM. Sondel PM. Rakhmilevich AL. CD40 ligation activates murine macrophages via an IFN-gamma-dependent mechanism resulting in tumor cell destruction in vitro. J. Immunol. 2005;174:6013–6022. doi: 10.4049/jimmunol.174.10.6013. [DOI] [PubMed] [Google Scholar]