Abstract
The University of Pittsburgh Molecular Library Screening Center (Pittsburgh, PA) conducted a screen with the National Institutes of Health compound library for inhibitors of in vitro cell division cycle 25 protein (Cdc25) B activity during the pilot phase of the Molecular Library Screening Center Network. Seventy-nine (0.12%) of the 65,239 compounds screened at 10 μM met the active criterion of ≥50% inhibition of Cdc25B activity, and 25 (31.6%) of these were confirmed as Cdc25B inhibitors with 50% inhibitory concentration (IC50) values <50 μM. Thirteen of the Cdc25B inhibitors were represented by singleton chemical structures, and 12 were divided among four clusters of related structures. Thirteen (52%) of the Cdc25B inhibitor hits were quinone-based structures. The Cdc25B inhibitors were further characterized in a series of in vitro secondary assays to confirm their activity, to determine their phosphatase selectivity against two other dual-specificity phosphatases, mitogen-activated protein kinase phosphatase (MKP)-1 and MKP-3, and to examine if the mechanism of Cdc25B inhibition involved oxidation and inactivation. Nine Cdc25B inhibitors did not appear to affect Cdc25B through a mechanism involving oxidation because they did not generate detectable amounts of H2O2 in the presence of dithiothreitol, and their Cdc25B IC50 values were not significantly affected by exchanging the dithiothreitol for β-mercaptoethanol or reduced glutathione or by adding catalase to the assay. Six of the nonoxidative hits were selective for Cdc25B inhibition versus MKP-1 and MKP-3, but only the two bisfuran-containing hits, PubChem substance identifiers 4258795 and 4260465, significantly inhibited the growth of human MBA-MD-435 breast and PC-3 prostate cancer cell lines. To confirm the structure and biological activity of 4260465, the compound was resynthesized along with two analogs. Neither of the substitutions to the two analogs was tolerated, and only the resynthesized hit 26683752 inhibited Cdc25B activity in vitro (IC50 = 13.83 ± 1.0 μM) and significantly inhibited the growth of the MBA-MD-435 breast and PC-3 prostate cancer cell lines (IC50 = 20.16 ± 2.0 μM and 24.87 ± 2.25 μM, respectively). The two bis-furan-containing hits identified in the screen represent novel nonoxidative Cdc25B inhibitor chemotypes that block tumor cell proliferation. The availability of non-redox active Cdc25B inhibitors should provide valuable tools to explore the inhibition of the Cdc25 phosphatases as potential mono- or combination therapies for cancer.
Introduction
The critical roles that the three human cell division cycle 25 proteins (Cdc25s) (A, B, and C) play in the regulation of the cell cycle and DNA damage-activated checkpoints make them attractive targets for the development of antiproliferative cancer drugs.1–5 The Cdc25s are dual-specificity phosphatases that dephosphorylate threonine and tyrosine residues (T14 and Y15) on cyclin-dependent kinases (CDKs), thereby activating the CDK-cyclin complexes, which regulate progression through the cell cycle.1,6 Historically, specific Cdc25 isoforms have been ascribed to the regulation of distinct transition phases of the mammalian cell cycle: Cdc25A was thought to primarily activate CDK1-cyclin E and CDK2-cyclin A complexes during the G1-S transition, while Cdc25B and Cdc25C were believed to preferentially activate CDK1-cyclin B complexes during the G2-M transition.1–3,5,6 It is now believed that all three Cdc25 isoforms cooperate to play essential roles in the temporal and spatial regulation of the CDKs during all stages of the cell cycle.1–3,6 In response to DNA damage cells activate checkpoint signaling pathways that lead to a transient cell cycle arrest, which is maintained until either the DNA is repaired or the cell undergoes apoptosis.1–3,5,6 The proximal ataxia telangiestasia mutated and ataxia-telangiestasia-and-RAD3-related kinases recognize DNA damage and trigger signaling cascades that activate the checkpoint effector kinases (CHKs) 1 and 2, which in turn modulate the activity of other cellular proteins including inactivation of the Cdc25 phosphatases.1–3,5,7,8 Inactivation of the Cdc25s is achieved through CHK1/CHK2 phosphorylation of specific residues that regulate the proteasome-mediated degradation of Cdc25s, Cdc25 catalytic activity, or interactions between Cdc25s and 14-3-3 family members that sequester the Cdc25s in the cytoplasm away from their substrates.1–3,5,7,8 Cdc25 inactivation produces either a G1 or G2 arrest that is dictated by the nature of the stressor, the type of DNA damage induced, and the signaling pathway that is activated.1–3,5,7,8 DNA damage checkpoints serve to maintain genetic integrity by preventing cells with damaged DNA from dividing and allowing DNA repair to occur.1–3,5,7,8 In noncancerous cells, irreparably damaged cells undergo apoptosis, thereby eliminating them from the pool of replicating cells.1–3,5,7,8
Cdc25s have been linked to oncogenic transformation in mice, and elevated expression of Cdc25A and Cdc25B at the mRNA and/or protein levels has been reported in a wide variety of primary human tumor samples.1,2,5 Enhanced expression of Cdc25A and Cdc25B in tumors correlates with specific clinical and pathological features: higher-grade or later-stage tumors, more aggressive tumors, and a shorter disease-free survival.1,2,5 Increased phosphatase activity resulting from the higher expression of Cdc25A and Cdc25B in tumor cells may contribute to unregulated proliferation through the overactivation of CDK-cyclin complexes that inappropriately drive cells through the cell cycle.1,2,5 Inappropriate and accelerated entry into S phase and/or the initiation of mitosis with incompletely replicated DNA may also lead to the accumulation of genetic aberrations and tumorigenesis.1,2,5 In normal cells DNA damage checkpoint activation induces degradation and/or inactivation of Cdc25s and cell cycle arrest.1,2,5,7,8 However, the increased expression of Cdc25A and Cdc25B in tumor cells may overwhelm the inactivation machinery, and the continued activation of CDK-cyclin complexes could conceivably push through the checkpoint barrier before the damaged DNA has been repaired, again contributing to genetic instability and malignant transformation.1,2,8
Entry into mitosis is initiated by the activation of the CDK1-cyclin B complex, which is maintained in its inactive form by Wee1 and Myt1 kinase-mediated phosphorylation of the Thr14 and Tyr15 residues of CDK1.1,2,5,8–10 During prophase, active CDK1-cyclin B complexes first appear on centrosomes and subsequently translocate into the nucleus.7,8 Cdc25B accumulates from late S phase through early G2, and its activity peaks during the G2-M transition when it functions to initiate mitosis by dephosphorylating and activating CDK1-cyclin B complexes on centrosomes, thus triggering spindle formation.7,8 The nuclear import of CDK1-cyclin B complexes and the subsequent stabilization of Cdc25C lead to chromatin condensation and the onset of mitosis.7,8 Ectopic overexpression of Cdc25B produced both chromatin aberrations and multipolar spindle phenotypes.7–9 Once DNA damage has been repaired, the DNA damage checkpoint is silenced so that cell cycle progression may resume.7–9 The resumption of the cell cycle after G2-M arrest has been shown to depend upon both Cdc25B and polo-like-kinase activities.7–12 Mechanistically distinct DNA-damaging agents induce a rapid increase in Cdc25B levels, and the extent of protein expression influence the kinetics of cell cycle resumption after DNA damage-induced arrest.11,12 Increased levels of Cdc25B, produced by ectopic overexpression or by constitutive expression in cancer cells, permit cells to bypass the DNA damaging agent-induced G2-M checkpoint arrest and prematurely enter mitosis.9 The inappropriate re-initiation of mitosis after genotoxic-induced G2-M checkpoint arrest in tumor cells with increased Cdc25B expression could reduce the therapeutic efficacy of DNA-damaging agents and further the propagation of genetic defects to daughter cells that may contribute to tumor growth.1,2,5,6 Cdc25B inhibitors therefore have potential as combination anticancer therapy because they should impair checkpoint recovery and likely produce a more pronounced G2 arrest in response to DNA-damaging drugs that could lead to increased apoptosis and better efficacy.1,2,5,6
The University of Pittsburgh Molecular Library Screening Center (PMLSC) (Pittsburgh, PA) accepted a project to screen the National Institutes of Health (NIH) compound library for inhibitors of in vitro Cdc25B activity during the pilot phase of the Molecular Library Screening Center Network (MLSCN).13–18 We present here the results of that screening campaign and the subsequent follow-up hit characterization of the Cdc25B inhibitors that were identified.
Materials and Methods
Reagents and Supplies
Trizma, dithiothreitol (DTT), β-mercaptoethanol (BME), reduced glutathione (GSH), tris(2-carboxyethyl)phosphine (TCEP), H2O2 (30% wt/wt), phenol red, horseradish peroxidase (HRP), catalase (CAT), and 3-O-methylfluorescein phosphate (OMFP) were purchased from Sigma-Aldrich Co. (St. Louis, MO). Sodium chloride was from VWR (West Chester, PA). EDTA and sodium hydroxide were from EMD (Gibbstown, NJ). Bovine serum albumin was from Invitrogen (Carlsbad, CA). Dimethyl sulfoxide (DMSO) (99.9% high-performance liquid chromatography-grade, under argon) was from Alfa Aesar (Ward Hill, MA). All other materials were reagent grade.
Cdc25B, Mitogen-Activated Protein Kinase Phosphatase (MKP)-1, and MKP-3 Protein Expression and Purification
The expression and purification of an epitope-tagged (hexahistidine) recombinant Cdc25B catalytic domain, MKP-1, and MKP-3 have been described previously.19 In brief, His-tagged Cdc25B, MKP-1, and MKP-1 were expressed in BL21(DE3) competent bacterial cells transformed with the appropriate pET expression vectors encoding the required phosphatase and induced with isopropyl-β-d-thiogalactopyranoside. Biologically active phosphatase was partially purified from isopropyl-β-d-thiogalactopyranoside-induced bacterial cell lysates by cobalt-column chromatography eluted with imidazole, frozen, aliquoted, and stored at −80°C. The relative purity of the phosphatase enzyme preparation was visualized by separating the protein components by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie Brilliant Blue. The identity of the phosphatase enzyme was confirmed by the presence of a protein band with the anticipated relative mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels together with immunoreactivity with antibodies against the specific phosphatase and the His-tag epitope on western blots.
Compound Library
The NIH library of 65,239 compounds was formatted at 10 mM concentration in DMSO, arrayed into 384-well microtiter master plates, and distributed to the PMLSC by the small molecule repository Biofocus-DPI (A Galapagos Company, San Francisco, CA).13,14,16,17,20 Compounds were identified by their PubChem substance identity numbers (SIDs). Daughter plates containing 2 μl of 1 mM compounds in DMSO were prepared and replicated from the MLSCN master plates using the Velocity11 (Menlo Park, CA) Vprep® outfitted with a 384-well transfer head. Aluminum adhesive plate seals were applied with an ABgene (Rochester, NY) plate sealer, and plates were stored at −20°C in a Matrical (Spokane, WA) MatriMinistore™ automated compound storage and retrieval system. Immediately prior to use daughter plates were withdrawn from −20°C storage, thawed at ambient temperature, and centrifuged 1–2 min at 50 g, and the plate seals were removed prior to the transfer of the appropriate volume of deionized H2O to ensure a concentration of the library compounds of 30 μM (in 3% DMSO) using the Velocity11 Vprep outfitted with a 384-well transfer head. The diluted compounds were mixed by repeated aspiration and dispensing using the 384-well transfer head of the Velocity11 Vprep, and 5 μl was transferred to the compound wells of assay plates.
Cdc25B, MKP-1, and MKP-3 Phosphatase Assays
The development and optimization of 384-well-format low-volume homogeneous fluorescence intensity assays for Cdc25B, MKP-1, and MKP-3 have been described previously.16,19 In brief, the assay involved three consecutive 5-μl additions to low-volume microtiter plates (catalog number 784076, Greiner BioOne, (Monroe, NC) performed on either the Velocity11 Vprep or the Evolution P3™ (PerkinElmer, Waltham, MA) automated liquid handler outfitted with a 384-well transfer head, plate controls and compounds, phosphatase enzyme, and OMFP substrate. Compounds were individually tested at 10 μM in the Cdc25B primary screen in an assay buffer comprising 30 mM Tris (pH 8.0), 75 mM NaCl, and 1.0 mM EDTA, at a final DMSO concentration of 2%, with 1% each contributed by the diluted compounds and OMFP substrate. For the MKP-1 and MKP-3 assays the pH of the assay buffer was 7.0 rather than 8.0 to ensure optimal enzyme activity.16,19 The phosphatase reactions were terminated after a 60-min incubation at ambient temperature by a 5-μl addition of either 2 mM Na3VO4 in deionized H2O for Cdc25B or 500 mM NaOH in deionized H2O for MKP-1 and MKP-3,16,19 performed on the Velocity11 Vprep outfitted with a 384-well transfer head. The fluorescence intensity of OMF product was measured on a Molecular Devices (Sunnyvale, CA) SpectraMax M5 plate reader (excitation filter, 485 nm; emission filter, 525 nm; auto cutoff, 515 nm).
For concentration-response confirmation and hit characterization assays, compounds were tested in singleton 10-point twofold dilution series concentration-response assays, starting at a maximum final concentration of 50 μM (2% DMSO). Compounds were diluted to 150 μM in deionized H2O (3% DMSO final concentration) and serially diluted in deionized H2O and 3% DMSO to provide the desired concentration range. Five microliters of the compound dilutions was transferred to a total assay volume of 15 μl (threefold dilution) performed on the Velocity11 Vprep outfitted with a 384-well transfer head.
Cdc25B Inhibitor Hit Characterization
Cdc25B inhibitors that were confirmed in concentration-response assays were tested for phosphatase selectivity against MKP-1 and MKP-3 in singleton 10-point twofold dilution series concentration-response assays, starting at a maximum final concentration of 50 μM as described above. To identify Cdc25B inhibitors that might be inactivating the enzyme through a mechanism involving oxidation and inactivation,21 we modified various components of the primary Cdc25B assay described above and determined if they either abolished the inhibition of Cdc25B or significantly shifted the potencies of the Cdc25B 50% inhibitory concentration (IC50) values higher (Table 1). In two of the Cdc25B hit characterization assays the 1 mM DTT reducing agent used in the HTS was replaced with either 1 mM BME or 1 mM GSH. In other Cdc25B formats, 100 U of CAT was added to the assay to degrade H2O2, the DTT concentration was increased from 1 mM to 25 mM, or reactions were conducted in the presence of both 100 U of CAT and 25 mM DTT.
Table 1.
Cdc25B Inhibitor Hit Characterization
| |
|
IC50 (μM) for AID in assay |
AC50 (μM) for AID in assay |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SID | AID 368 HTS (% inhibition) | 569 CFM | 650 HCF | 674 MKP-1 | 673 MKP-3 | 670 BME | 671 GSH | 668 CAT | 649 25D | 669 CAT 25D | 682 H2O2 0.5D | 683 H2O2 1.0D |
| 850758 | 100.6 | 0.542 | 0.421 | 0.333 | 0.629 | 41.845 | >50 | >50 | 17.227 | >50 | 0.646 | 1.046 |
| 845167 | 100.9 | 0.400 | 0.493 | 0.485 | 0.615 | >50 | >50 | 20.022 | 21.125 | >50 | 0.731 | 1.605 |
| 4251194 | 101.8 | 0.349 | 0.551 | 0.423 | 0.433 | 32.365 | 35.510 | 23.000 | 34.529 | >50 | 1.166 | 3.158 |
| 847359 | 100.4 | 13.538 | 20.375 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 3.888 | 13.622 |
| 3712327 | 65.2 | 15.495 | 14.711 | >50 | >50 | 23.011 | 24.189 | 23.582 | >50 | >50 | 7.905 | 24.962 |
| 845964 | 101.7 | 0.949 | 0.502 | 2.379 | 2.739 | 33.080 | 42.970 | 14.055 | 49.271 | >50 | 12.880 | 23.940 |
| 7973677 | 98.4 | 3.627 | 5.117 | 43.772 | 19.774 | >50 | >50 | >50 | >50 | >50 | 13.158 | 41.370 |
| 3712249 | 101.2 | 5.252 | 1.410 | 14.647 | 3.707 | > 50 | > 50 | > 50 | > 50 | > 50 | 13.172 | 27.490 |
| 856108 | 66.0 | 9.528 | 9.535 | >50 | >50 | 26.310 | 31.737 | 47.092 | >50 | >50 | 24.117 | >50 |
| 8139964 | 99.0 | 13.113 | 12.171 | >50 | >50 | 20.380 | 24.974 | 25.398 | >50 | >50 | 25.648 | > 50 |
| 857882 | 62.0 | 49.563 | 25.491 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 26.104 | 45.673 |
| 4242461 | 96.4 | 4.306 | 4.934 | > 50 | 48.661 | >50 | >50 | >50 | >50 | >50 | 28.608 | >50 |
| 4251929 | 85.4 | 3.276 | 2.220 | 10.678 | 12.599 | >50 | >50 | 45.752 | 47.983 | >50 | >50 | >50 |
| 4249736 | 80.9 | 4.229 | 2.238 | 8.746 | 12.079 | >50 | >50 | 45.033 | 42.269 | >50 | >50 | >50 |
| 4249621 | 63.4 | 9.099 | 3.117 | 4.606 | >50 | 3.802 | 2.787 | 3.804 | 7.420 | 8.693 | >50 | >50 |
| 4241893 | 61.3 | 4.695 | 3.495 | 7.018 | >50 | 1.783 | 2.133 | 4.197 | 14.673 | 16.784 | >50 | >50 |
| 844096 | 52.6 | 30.223 | 5.429 | 42.517 | >50 | 4.198 | 4.729 | 7.115 | 17.712 | 20.486 | >50 | >50 |
| 850390 | 50.9 | 8.056 | 5.730 | >50 | >50 | 8.114 | 8.229 | 6.177 | 11.702 | 13.238 | >50 | >50 |
| 843791 | 51.4 | 5.852 | 5.984 | >50 | >50 | 5.197 | 5.891 | 7.351 | 11.390 | 12.062 | >50 | >50 |
| 7964733 | 58.6 | 24.131 | 6.275 | >50 | 19.763 | >50 | >50 | 48.860 | >50 | >50 | >50 | >50 |
| 851514 | 55.6 | 13.939 | 7.097 | >50 | >50 | 7.420 | 6.761 | 11.000 | 12.813 | 16.013 | >50 | >50 |
| 847214 | 50.5 | 21.954 | 7.162 | >50 | >50 | 6.351 | 6.798 | 7.109 | 10.909 | 11.399 | >50 | >50 |
| 4260465 | 50.3 | 15.534 | 9.213 | >50 | >50 | 10.328 | 11.124 | 15.655 | >50 | >50 | >50 | >50 |
| 4258795 | 51.0 | 11.637 | 15.349 | >50 | >50 | 19.221 | 23.651 | 22.375 | 39.842 | 40.874 | >50 | >50 |
| 4248661 | 81.3 | 26.368 | 24.251 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
CFM, confirmatory concentration-response assay; HCF, hit confirmation concentration-response assay; MKP-1, MKP-1 concentration-response assay; MKP-3, MKP-3 concentration-response assay; BME, Cdc25B concentration-response assay conducted with BME; GSH, Cdc25B concentration-response assay conducted with GSH; CAT, Cdc25B concentration-response assay conducted in the presence of CAT; 25D, Cdc25B concentration-response assay conducted in the presence of 25 mM DTT; CAT 25D, Cdc25B concentration-response assay conducted in the presence of CAT and DTT; H2O2 0.5D, redox cycling H2O2 generation assay conducted in 0.5 mM DTT; H2O2 1.0D, redox cycling H2O2 generation assay conducted in the presence of 1.0 mM DTT.
384-Well Redox Cycling H2O2 Generation Assay
The H2O2 generation assay was performed in 384-well flat-bottomed clear polystyrene microtiter plates with a well volume of 120 μl (catalog number 781101, Greiner BioOne) as described previously.15 Compounds and plate controls were diluted to the required concentrations in Hanks' Balanced Salt Solution. Plate controls were represented by 100 μM H2O2 (maximum, n = 32) or 1% DMSO (minimum, n = 24) (final concentrations) in the well. DTT was diluted in Hanks' Balanced Salt Solution from 200 mM stocks to the required concentrations (typically 0.5–1.0 mM final concentration in the well). The phenol red-HRP detection reagent contains 300 μg/ml phenol red and 180 μg/ml HRP in Hanks' Balanced Salt Solution. The assay involved three liquid transfer steps into the 384-well plate performed on the Evolution P3 liquid handling platform using a 384-well P30 dispensing head and 20 μl each of compounds/controls, DTT, and the phenol red-HRP detection reagent to give a final assay volume of 60 μl. Compounds and DTT were incubated together at ambient temperature for a minimum of 15 min before addition of the phenol red-HRP detection reagent (100 μg/ml phenol red and 60 μg/ml HRP final concentration in the well). After an additional incubation period at ambient temperature, minimally 5 min, the assay was terminated by addition of 10 μl of 1 N NaOH, and the absorbance of the phenol red was measured at 610 nm in a SpectraMax M5 microtiter plate reader.
MDA-MB-435 and PC-3 Growth Inhibition Assays
The proliferation of the human MBA-MD-435 breast and PC-3 prostate cancer cell lines was measured using a previously described high-content imaging assay.22 MBA-MD-435 and PC-3 cells were seeded at 2,000 cells per well in collagen-coated 384-well black-walled clear-bottom plates (catalog number 781091, Greiner BioOne) and cultured for 2–72 h at 37°C in a 5% CO2 atmosphere and 95% humidity. After a 2-h cell attachment period, one plate of cells was fixed, stained, and analyzed as described below to establish the cell density at the time of treatment; the other plates were treated with the indicated compound concentrations transferred to the 384-well plate on the Evolution P3 liquid handling platform using a 384-well P30 dispensing head and cultured for an additional 24–72 h. At the indicated time points, plates were fixed with 3.7% formaldehyde containing 2 μg/ml Hoechst 33342 at 37°C for 10 min, the fixed cells were then washed twice with Dulbecco's phosphate-buffered saline, and aluminum adhesive plate seals were applied with the ABgene plate sealer. Images of the Hoechst-stained nuclei from nine fields of view per well were acquired on an ArrayScan VTI imaging platform (Thermo Fisher Scientific, Pittsburgh, PA) using a 10× 0.3NA objective and the XF100 filter set. Hoechst-stained nuclei were quantified using an image analysis algorithm that segmented the images and identified nuclear objects on the basis of total and average fluorescent intensity and size (area) and shape (width and length) thresholds. The total number of selected objects (nuclei) from nine fields of view covering the whole well provided the total cell count per well. The extent of growth inhibition observed in compound-treated wells is expressed as a percentage of the total cell counts in these wells relative to the total cell counts in DMSO control wells: % growth inhibition = ([mean total cell count in DMSO control wells – total cell count in treated wells] × 100)/mean total cell count in DMSO control wells.
Data Analysis: HTS and IC50 Values
We utilized GraphPad Software (La Jolla, CA) Prism® version 5.0 software to plot and analyze assay development data and perform linear regression analysis, four-parametric nonlinear regression curve fitting, and enzyme kinetics analysis. To analyze the percentage inhibition of Cdc25B at 10 μM in the primary screen, we constructed an ActivityBase™ (IDBS, Guilford, UK) HTS template to calculate the percentage inhibition, based on the mean maximum (n = 32) and mean minimum (n = 24) plate controls, plate signal-to-background ratios (mean maximum signal/mean minimum signal), and Z-factors.23 The data were exported to Spotfire® (TIBCO, Somerville, MA) for visualization to help identify process errors and facilitate quality control analysis. To analyze the Cdc25B inhibition, we constructed an ActivityBase concentration-response template to calculate percentage inhibition together with plate control signal-to-noise ratio and Z-factors for quality control purposes. IC50 values were calculated using an XLfit four-parameter logistic model, also called the Sigmoidal Dose-Response Model: y = (A + ((B − A)/(1 + ((C/x)^D)))), where y is the percentage inhibition and x is the corresponding compound concentration. The fitted C parameter is the IC50, defined as the concentration giving a response halfway between the fitted top (B) and bottom (A) of the curve. The fitted D parameter is called the “Slope” and equals the Hill coefficient. The top parameter B was locked at 100%, and the bottom parameter A was set at 0%.
Compound Structure Clustering Analysis
Cdc25B inhibitors that were identified in the HTS and confirmed in 10-point concentration-response IC50 assays were analyzed using Leadscope® (Columbus, OH) Enterprise version 2.4.6–1 software. The active compounds were subjected to structure-based clustering and classification techniques based on recursive partitioning24 as described previously.16,17
Results
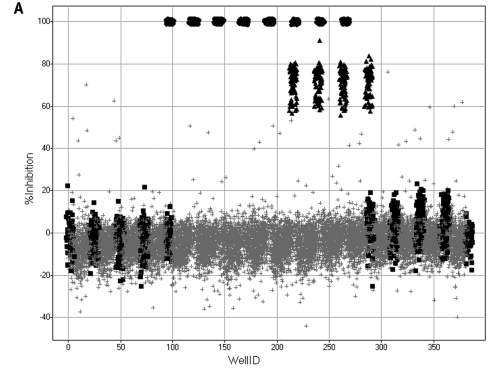
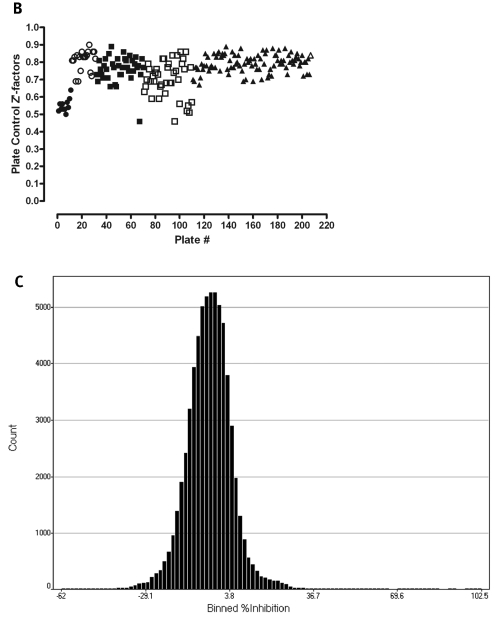
The PMLSC screened the NIH compound library of 65,239 compounds against the Cdc25B assay in 6 days, and the Cdc25B primary HTS data from a single 40-plate day of operations together with the plate Z′-factors, signal-to-background ratios, and percentage inhibition frequency distribution from the entire HTS campaign are presented in Fig. 1. The 40-plate overlay of the percentage inhibition of Cdc25B data from a single day of screening operations (Fig. 1A) demonstrates that the assay plate controls and corresponding assay signal window performed consistently and reproducibly, that the majority of compounds did not inhibit Cdc25B, and that an acceptable number of compounds (0.12% overall for the HTS) met the active criterion of ≥50% inhibition of Cdc25B activity (Fig. 1A). Overall, the Cdc25B HTS of the 65,239 compound library performed extremely well and exhibited a mean Z′-factor of 0.76 ± 0.09 with only two of the 207 plates producing Z′-factors below 0.5 (Fig. 1B). The frequency distribution of the percentage inhibition of Cdc25B for the screen approximates a normal distribution in which the majority of compounds were inactive (Fig. 1C), and only 79 (0.12%) of the 65,239 compounds screened achieved the active criterion of ≥50% inhibition of Cdc25B activity at 10 μM.
Fig. 1.
Cdc25B high-throughput screen performance data. (A) Forty HTS plate overlay scatter plot of the percentage inhibition Cdc25B data from the third day of screening operations. An ActivityBase HTS template was written to calculate the percentage inhibition based on the maximum (n = 32) and minimum (n = 24) plate controls for the Cdc25B HTS campaign. A Spotfire visualization of the percentage inhibition data from 40 384-well plates of the Cdc25B HTS conducted on the third day of screening operations overlaid in a single scatter plot is presented: maximum (2% DMSO, n = 32) plate controls (▪), minimum (500 μM Na3 VO4 and 2% DMSO, n = 24) plate controls (•), 50% (500 μM Na3VO4 and 2% DMSO, n = 8) plate controls (▴), and 10 μM compounds (2% DMSO, n = 320) (+). (B) Plate controls Z′-factors for the Cdc25B HTS campaign. An ActivityBase HTS template was written to calculate Z′-factors for each plate based on the maximum (2% DMSO, n = 32) and minimum (500 μM Na3VO4 and 2% DMSO) plate controls. The Cdc25B HTS campaign was conducted over 6 days of screening operations, and the associated plate Z′-factors for all 207 plates are plotted for each day: (○) day 1, (•) day 2, (□) day 3, (▪) day 4, (▴) day 5, (▵) day 6. (C) Compound percentage inhibition frequency distribution for the Cdc25B HTS campaign. An ActivityBase HTS template was written to calculate the percentage inhibition, based on the mean maximum (2% DMSO, n = 32) and mean minimum (500 μM Na3VO4 and 2% DMSO, n = 24) plate controls, and the data were uploaded to an Oracle database. The percentage inhibition of Cdc25B for all compounds run in the screen was queried from the database and exported to Spotfire for visualization of the frequency distribution of the calculated results.
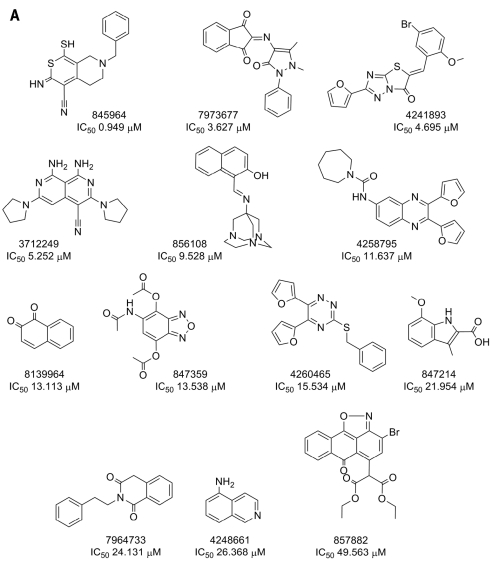
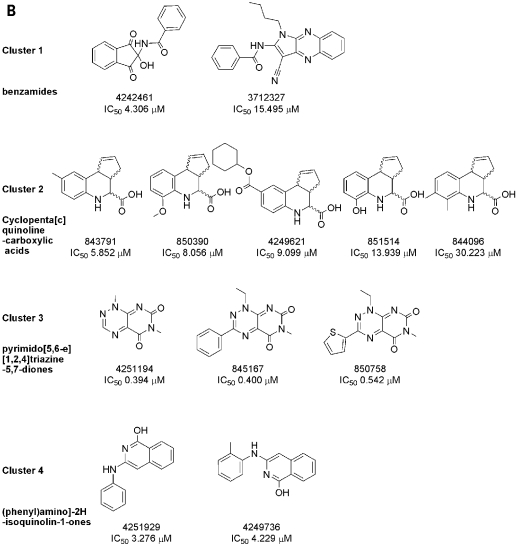
The active compounds identified in the Cdc25B primary HTS were cherry-picked by the NIH small molecule repository, and 25 (31.6%) of the 79 active compounds were confirmed as concentration-dependent Cdc25B inhibitors (Fig. 2 and Table 1, PubChem assay identifier [AID] 569, confirmatory concentration–response assay). The 25 confirmed Cdc25B inhibitors exhibited IC50 values in the following ranges: four were <1.0 μM, 10 were between 1.0 and 10.0 μM, and 11 were between 10.0 and 50.0 μM (Table 1, AID 569). Thirteen of the confirmed Cdc25B inhibitors were represented by singleton chemical structures, and their IC50 values ranged between 1.0 and 50.0 μM (Fig. 2A). The remaining 12 confirmed Cdc25B inhibitors were divided among four clusters with related structures with IC50 values ranging between 400 nM and 50.0 μM: two benzamides, five cyclopenta[c]quinoline-carboxylic acids, three pyrimido[5,6-e][1,2,4]triazine-5,7-diones, and two phenyl-amino-2H-isoquinolin-1-ones (Fig. 2B).
Fig. 2.
Compound structures and IC50 values for Cdc25B inhibitors confirmed in 10-point concentration dependence assays: (A) singletons and (B) clusters. Cdc25B inhibitors that were identified in the HTS and confirmed in 10-point dose–response IC5o assays were analyzed using the Leadscope Enterprise version 2.4.6-1 software. The active compounds were subjected to structure-based clustering and classification techniques based on recursive partitioning that produced singleton structures and clusters of related structures. The PubChem SIDs, Cdc25B IC5o values (in μM), chemical structures, and names of the chemical clusters are presented.
The Cdc25B inhibitor hits were then further characterized in a series of in vitro secondary assays to confirm their Cdc25B activity (AID 650, hit confirmation concentration-response assay), to determine their phosphatase selectivity against MKP-1 (AID 674) and MKP-3 (AID 673), to examine whether they might be inhibiting Cdc25B through a mechanism involving oxidation and inactivation21 (AIDs 670, 671, 668, 649, and 669), and to test their activity in the redox cycling H2O2 generation assay15 (AIDs 682 and 683) (Table 1). The 25 confirmed Cdc25B inhibitors (AID 569) consistently inhibited Cdc25B with IC50 values <26 μM (AID 650); as shown under AIDs 674 and 673, 12 appeared to be selective for Cdc25B, three also inhibited MKP-1, two also inhibited MKP-3, and eight inhibited all three dual-specificity phosphatases (Table 1). Twelve of the 25 Cdc25B inhibitors gave 50% activation concentration (AC50) values for H2O2 generation in the presence of 0.5 mM DTT (AID 682), and nine of these also produced AC50 values in the presence of 1.0 mM DTT (AID 683) (Table 1). Two additional compounds, the phenylamino-2H-isoquinolin-1-ones, SIDs 4251929 and 4249736, did not produce an AC50 in either of these redox cycling assays but reproducibly achieved between 10% to 20% activation at 50 μM (data not shown) and should therefore be considered capable of redox cycling. For the 14 compounds that exhibited activity in one or both of the redox cycling assays, their Cdc25B IC50 values were either abolished or significantly increased by exchanging the DTT for BME (AID 670) or GSH (AID 671), by adding CAT to the assay (AID 668), and/or by raising the DTT concentration to 25 mM (AIDs 649 and 669) (Table 1). Although two quinone-based compounds (SIDs 7964733 and 4248661) failed to produce detectable amounts of H2O2 in the presence of DTT, their Cdc25B IC50 values were either abolished or significantly increased by the indicated modifications to the Cdc25B assay (Table 1). Six of the nine non-redox active compounds that were active in the Cdc25B primary HTS (AID 368) and both the confirmation (AID 569) and hit characterization (AID 650) concentration-response assays appeared to be selective Cdc25B inhibitors since they did not significantly inhibit either MKP-1 or MKP-3 at ≤50 μM (Table 1). Three of the non-redox active Cdc25B inhibitors are from the cyclopenta[c]quinoline-carboxylic acid cluster of hits (SIDs 843791, 850390, and 851514), and the other three (SIDs 4258795, 4260465, and 847214) were represented by singleton chemical structures (Fig. 2).
A cross-target query of the PubChem database revealed that the six Cdc25B inhibitors had been tested in between 192 and 227 assays uploaded to PubChem and on average had produced active flags in 10.7% of those assays (Table 2). SID 4258795 exhibited confirmed activity against 13 different targets, whereas the other five Cdc25B inhibitors have only been confirmed active against a relatively small number of other targets (Table 2). To date, SID 847214 has only been confirmed as an inhibitor of Cdc25B and the antigen receptor-induced activation of nuclear factor κB (Table 2). However, the other four Cdc25B inhibitors have exhibited confirmed activity against four to seven other targets, including three common targets: hydroxysteroid 17β-dehydrogenase 4, hydroxyacyl-coenzyme A dehydrogenase type II, and a protein tyrosine phosphatase (PTP) expressed in hematopoietic cells (Table 2).
Table 2.
Cdc25B Inhibitor Specificity: Cross-Target Query of PubChem
| |
|
|
Type of flag |
|
|
|---|---|---|---|---|---|
| PubChem SID | Number of assays | Number of active flags | HTS | Confirmation | Confirmed targets |
| 847214 | 192 | 13 | 5 | 2 | Cdc25B, NFκB |
| 843791 | 193 | 19 | 8 | 5 | Cdc25B, HADH2, HSD17B4, EukTI, HePTP |
| 850390 | 192 | 18 | 6 | 6 | Cdc25B, HADH2, HSD17B4, EukTI, DNA Pol III, HePTP |
| 851514 | 206 | 23 | 9 | 8 | Cdc25B, HADH2, HSD17B4, EukTI, DNA Pol III, Tau, Hsp90, HePTP |
| 4258795 | 227 | 37 | 17 | 13 | Cdc25B, HADH2, HSD17B4, EukTI, DNA Pol III, Tau, Hsp90, Cruzain, 15-hLO, MLVN, Bim-Bfl-1, RT-RNH, MKP-1 |
| 4260465 | 211 | 19 | 8 | 7 | Cdc25B, HADH2, HSD17B4, Tau, Cruzain, 15-hLO, E. coli, HePTP |
NFκB, antigen receptor-induced NF-κB activation; HSD17B4, hydroxysteroid 17β-dehydrogenase 4; HADH2, hydroxyacyl-coenzyme A dehydrogenase, type II; EukTI, eukaryotic translation initiation; DNA Pol III, DNA polymerase III holoenzyme; Tau, tau filament binding; Hsp90, heat shock protein 90 co-chaperone interaction; Cruzain, inhibition of cruzain (the major cysteine protease of Trypanosoma cruzi); 15-hLO, human 15-lipoxygenase; MLVN, mevalonate pathway in Streptococcus pneumoniae; E. coli, antimicrobial Escherichia coli BW25113 ΔtolC::kan; HePTP, a PTP expressed in hematopoietic cells; Bim-Bfl-1, regulators of Bcl-2 family protein interactions, specifically Bim-Bfl-1; RT-RNH, human immunodeficiency virus type 1 reverse transcriptase associated ribonuclease H.
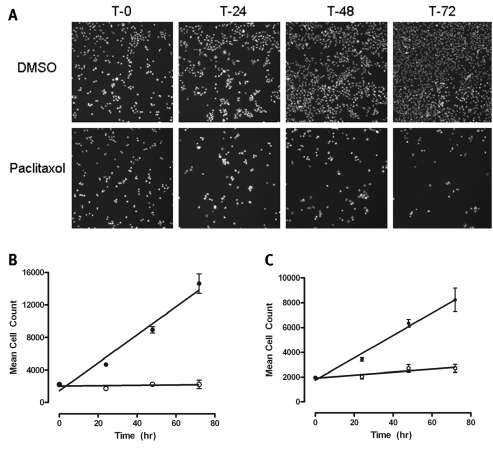
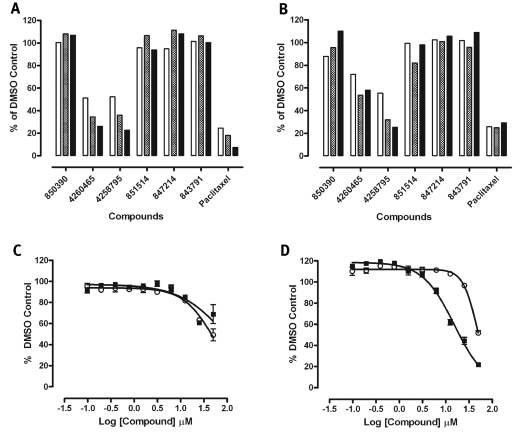
Because Cdc25B plays a key role(s) in controlling cell cycle progression, we next examined the ability of the six Cdc25B hits to inhibit the proliferation of the human MBA-MD-435 breast and PC-3 prostate cancer cell lines, both of which express Cdc25B22 (Figs. 3 and 4). Under the conditions utilized for the assay, both the MBA-MD-435 breast and PC-3 prostate cancer cell lines are actively proliferating as evidenced by the linear increase in cell counts throughout the 72-h time course of the assay (Fig. 3). Consistent with previous reports,22 the three cyclopenta[c]quinoline-carboxylic acid Cdc25B inhibitors at 50 μM failed to inhibit the proliferation of either cell line (Fig. 4A and B), and the 7-methoxy-3-methyl-1H-indole-2-carboxylic acid hit (SID 847214) at 50 μM also failed to inhibit the growth of either cell line. However, both of the bis-furan compounds, N-[2,3-di(furan-2-yl)quinoxalin-6-yl]azepane-1-carboxamide (SID 4258795) and 5,6-di(furan-2-yl)-3-(phenylmethylsulfanyl)-1,2,4-triazine (SID 4260465), inhibited the growth of both cell lines in a time- and concentration-dependent manner (Fig. 4). SID 4258795 exhibited IC50 values of 96.2 ± 68 μM and 15.3 ± 1.36 μM against the MBA-MD-435 and PC-3 cell lines, respectively, while 4260465 exhibited IC50 values of 50.4 ± 8.6 μM and 47.5 ± 2.75 μM against the MBA-MD-435 and PC-3 cell lines, respectively.
Fig. 3.
Human MBA-MD-435 breast and PC-3 prostate cancer cell proliferation assay. (A) Representative images of MBA-MD-435 cells cultured for 0, 24, 48, and 72 h with and without 100 nM paclitaxel. MBA-MD-435 and PC-3 cells were cultured, harvested, seeded, and treated in culture as described in Materials and Methods. After a 2-h cell attachment period, one plate of cells was fixed, stained, and analyzed as described below to establish the cell density at the time of treatment (To). The other plates were cultured for an additional 24 h, 48 h, or 72 h with or without 100 nM paclitaxel. At the indicated time points, plates were fixed and stained with formaldehyde containing 2 μg/ml Hoechst 33342 as described in Material and Methods. Images of the Hoechst-stained nuclei from nine fields of view per well were acquired on the ArrayScan VTI imaging platform using a 10× 0.3NA objective and the XF100 filter set. Representative images of a single field of view of Hoechst-stained nuclei from MBA-MD-435 cells cultured for the indicated times with or without 100 nM paclitaxel are presented. (B and C) Proliferation of (B) MBA-MD-435 and (C) PC-3 cells in culture over a 72-h period with or without 100 nM paclitaxel. Hoechst-stained nuclei from replicate wells (n = 16) were quantified using an image analysis algorithm that segmented the images and identified nuclear objects on the basis of total and average fluorescent intensity and size (area) and shape (width and length) thresholds. The total number of selected objects (nuclei) from nine fields of view covering the whole well provided the total cell count per well. The data are presented as the mean total cell counts per well ± SD of the mean (n = 16) for control (•) and 25 nM paclitaxel-treated (○) cells at the indicated time points together with a linear regression analysis of the data using Graphpad Prism software version 4.03. The r2 correlation coefficients of the linear analysis exceeded 0.969 for actively proliferating cells from both cell lines.
Fig. 4.
Time- and concentration-dependent inhibition of (A) MBA-MD-435 and (B) PC-3 cancer cell growth by Cdc25B inhibitors. MBA-MD-435 and PC-3 cells were cultured, harvested, seeded, and treated in culture as described in Materials and Methods. After a 2-h cell attachment period, cells were treated with DMSO, the indicated Cdc25B inhibitor compounds at 50 μM, or 25 nM paclitaxel, and plates were cultured for an additional 24–72 h. At the indicated time points, plates were fixed and stained with formaldehyde containing 2 μg/ml Hoechst 33342 as described in Materials and Methods. Images of the Hoechst-stained nuclei from nine fields of view per well were acquired on the ArrayScan VTI imaging platform using a 10× 0.3NA objective and the XF100 filter set. Hoechst-stained nuclei were quantified using an image analysis algorithm to segment the images as described in Materials and Methods. The total number of selected objects (nuclei) from nine fields of view covering the whole well provided the total cell count per well. The total cell counts observed in compound-treated wells (n = 1) are expressed as a percentage of the mean total cell counts in DMSO control wells (n = 16): % of DMSO controls = (total cell count in compound-treated wells × 100)/mean total cell count of DMSO control wells. A bar graph of the total cell counts per well of compound-treated wells at 24 h (empty bars), 48 h (cross-hatched bars), and 72 h (solid filled bars) expressed as a percentage of DMSO controls is presented. (C and D) Concentration-dependent inhibition of (C) MBA-MD-435 and (D) PC-3 cancer cell growth by Cdc25B inhibitors 4258795 and 4260465. MBA-MD-435 and PC-3 cells were cultured, harvested, seeded, and treated in culture as described in Materials and Methods. After a 2-h cell attachment period, cells in triplicate wells were treated with the indicated concentrations Cdc25B inhibitors 4258795 and 4260465, and plates were cultured for an additional 72 h. Plates were then fixed and stained, and images of the Hoechst stained nuclei were acquired and analyzed as described in (A) and (B). The total cell counts observed in compound-treated wells (n = 3) are expressed as a percentage of the mean total cell counts in DMSO control wells (n = 16): % of DMSO controls = (total cell count in compound-treated wells × 100)/mean total cell count of DMSO control wells. The data are presented as the mean percentage of DMSO control cell counts ± SD of the mean (n = 3), and the concentration–response data for the effects of the Cdc25B inhibitors 4258795 (▪) and 4260465 (○) on MBA-MD-435 (C) and PC-3 (D) cell proliferation were fit to curves using the sigmoidal dose–response variable slope equation y = Bottom + (Top – Bottom)/(1 + 10^((LogEC50 −x) × Hill Slope)), where EC50 is the 50% effective concentration, using Graphpad Prism software version 4.03.
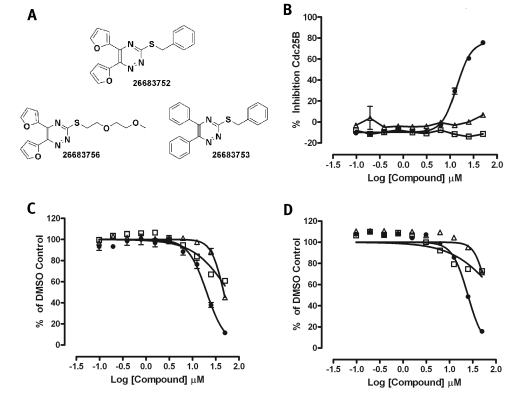
To confirm the structure and activity of N-[2,3-di(furan-2-yl) quinoxalin-6-yl]azepane-1-carboxamide (SID 4260465), the PMLSC chemistry core resynthesized the compound (SID 26683752) and two analogs (Fig. 5A). In the 5,6-di(phenyl)-3-(phenylmethylsulfanyl)-1,2,4-triazine analog (SID 26683753), two phenyl rings were substituted for the two furan rings in 26683752, and in the 5,6-di(furan-2-yl)-3-[2-(2-methoxyethoxy)ethylsulfanyl]-1,2,4-triazine analog (SID 26683756), a 3-(methoxyethoxy)ethylsulfanyl side chain was substituted for the 3-phenylmethylsulfanyl side chain in SID 26683752. Only the resynthesized hit SID 26683752 exhibited inhibition in the in vitro Cdc25B assay, exhibiting an IC50 of 13.83 ± 1.0 μM (Fig. 5B). Although both of the analogs (SIDs 26683753 and 26683756) exhibit partial growth inhibition of both the MBA-MD-435 breast and PC-3 prostate cancer cell lines at 25 and 50 μM, the resynthesized hit (SID 26683752) was significantly more potent and produced IC50 values of 20.16 ± 2.0 μM and 24.87 ± 2.25 μM against the MBA-MD-435 and PC-3 cell lines, respectively (Fig. 5C and D).
Fig. 5.
Chemical structures, in vitro Cdc25B inhibition, and inhibition of MBA-MD-435 and PC-3 cancer cell growth by Cdc25B inhibitor analogs. (A) Chemical structures of the resynthesized Cdc25B inhibitor 4260465 and analogs. The Cdc25B inhibitor hit from the screen SID 4260465 was resynthesized by the PMLSC chemistry core along with two analogs. The PubChem SIDs and chemical structures of the resynthesized compound SID 26683752 and two analogs, 26683753 (□) and 26683756 (▵), are presented. (B) Inhibition of the in vitro Cdc25B activity assay by the resynthesized Cdc25B inhibitor hit 26683752 (•) and two analogs. Recombinant Cdc25B enzyme was incubated with 40 μM OMFP in assay buffer together with the indicated concentrations of compounds in triplicate wells of a 384-well low-volume microtiter plate for 60 min, and the resulting fluorescent intensity at 485 nm excitation/525 nm emission was measured on a SpectraMaxM5 plate reader. An ActivityBase HTS template was written to calculate percentage inhibition for each well based on the maximum (2% DMSO, n = 32) and minimum (500 μM Na3 VO4 and 2% DMSO) plate controls. The mean percentage inhibition ± SD of triplicate values produced at each compound concentration after the 60-min incubation are plotted together with the resulting nonlinear regression curves plotted using the sigmoidal dose–response variable slope equation y = Bottom + (Top – Bottom)/(1 + 10^((LogEC50 – x) × Hill Slope)) using Graphpad Prism software version 4.03. (C and D) Concentration-dependent inhibition of (C) MBA-MD-435 and (D) PC-3 cancer cell growth by the resynthesized hit 26683752 and two analogs, 26683753 and 26683756. MBA-MD-435 and PC-3 cells were seeded in 384-well plates as described in Fig. 4. After a 2-h cell attachment period, cells in triplicate wells were treated with the indicated concentrations of compounds, and plates were cultured for an additional 72 h. Images of the fixed and stained cells were acquired and analyzed as described in Fig. 4. The total cell counts observed in compound-treated wells (n = 3) are expressed as a percentage of the mean total cell counts in DMSO control wells (n = 16): % of DMSO controls = (total cell count in compound-treated wells × 100)/mean total cell count of DMSO control wells. The data are presented as the mean percentage of DMSO control cell counts ± SD of the mean (n = 3), and the concentration–response data for the effects of the resynthesized hit 26683752 (•) and two analogs, 26683753 (□) and 26683756 (▵), on (C) MBA-MD-435 and (D) PC-3 cell proliferation were fit to curves using the sigmoidal dose–response variable slope equation y = Bottom + (Top – Bottom)/(1+10^((LogEC50 – x) × Hill Slope)), where EC50 is the 50% effective concentration, using Graphpad Prism software version 4.03.
Discussion
A significant drug discovery effort has been mounted to find Cdc25 inhibitors for cancer therapy because of the critical roles that these phosphatases have in regulating cell cycle progression and DNA damage-induced checkpoint arrest and recovery.1–5 The N-terminal regulatory domains of the Cdc25s are highly divergent but contain multiple sequence motifs that modulate their catalytic activity, stability, subcellular localization, and protein–protein interactions with partners, regulators, and substrates. These motifs include nuclear localization and nuclear export sequences, 14-3-3 binding sites, and multiple sites for phosphorylation or ubiquitination.2,5,6 The N-terminal regulatory domains of the Cdc25s are also subject to alternate splicing that generate at least two variants for Cdc25A and five each for Cdc25B and Cdc25C.2,6 In contrast, the C-terminal catalytic domains of the Cdc25s are highly conserved (~60% identity) and share a common C(X)5R(S/T) active site sequence motif with the 107 PTPs of the human genome, with X being any amino acid, R an invariant arginine, and C the active site cysteine required for catalytic activity.2,6,25 The PTP active site cysteine performs a nucleophilic attack on the phosphotyrosine residues of substrates to form covalent thiol–phosphate intermediates that are then hydrolyzed to release free phosphate.2,6,25 The cysteines of PTP active sites have a low pKa of 4.7–5.4 and exist as thiolate anions at neutral pH, which enhances their catalytic activity as a nucleophile but also makes them susceptible to oxidation and inactivation.2,6,25 PTP cysteine thiolates react with reactive oxygen species (ROS) such as H2O2 to produce a sulfenic acid form of the enzyme that is inactive.15,25–35 PTPs oxidized to the sulfenic acid form may be reduced back to the active thiolate species by cellular thiol donors such as GSH, but sulfenic acids are highly reactive and may readily undergo further oxidation to sulfinic and sulfonic forms that are irreversibly oxidized and inactive.15,25–35 The inactivation of PTPs by the specific and reversible oxidation of the active site cysteine by H2O2 has been demonstrated for numerous key PTPs that regulate the signaling pathways and networks involved in normal physiological processes and disease: PTP1B, PTPα, LAR, VHR, PTEN, SHP-2, Cdc25B, MKP-3, and MKP-1.15,26–35
The crystal structures of the catalytic domains of Cdc25A and Cdc25B revealed them to be small globular domains with a shallow solvent-exposed active site and no obvious sites for substrate recognition or inhibitor binding.6,36,37 Cdc25–protein substrate recognition appears to occur through a broad protein–protein interaction surface,6,36,37 making the molecular modeling of inhibitors exceedingly challenging and mandating an HTS approach. Almost all of the in vitro Cdc25 inhibitors that have been described to date were identified in assays that employed the catalytic domains of the Cdc25s and small molecule substrates.3,19,21,22,38–44 A number of potent Cdc25 inhibitors have now been synthesized or isolated from natural product extracts, and although many of these exhibit selectivity against other dual-specificity phosphatases or PTPs, most inhibit all three Cdc25 isoforms.3,4,10,21,22,38–44 The majority of the known Cdc25 inhibitors are quinone-based structures, phosphate surrogates, or electrophiles.3,4,10,21,22,38–44 The mechanism of action of many quinoid compounds involves the oxidation and inactivation of the Cdc25 active site cysteine by ROS either generated through in vitro redox cycling of these compounds in the presence of DTT or induced within cells by exposure to these compounds.21,38,45 We have previously shown that the in vitro inhibition of Cdc25B by cell-active quinolinediones was irreversible, time dependent, and sensitive to pH, CAT, and reducing agents.46,47 The inhibition of Cdc25B in vitro by the quinolinediones required the presence of reducing agents and was abolished by the addition of CAT, implicating the redox cycling generation of H2O2 and oxidation of the active site cysteine as the mode of inhibition.46,47 Exposure of cells to quinolinediones directly triggered the production of intracellular ROS.46 o-Quinone-based inhibition of PTPα was shown to be time dependent, sensitive to superoxide dismutase and CAT, and mediated by the generation of H2O2 through a cyclic redox mechanism that required both the compound and DTT.26 o-Quinones produced H2O2 in the presence of strong reducing agents such as DTT or TCEP but not with weaker reducing agents such as GSH or Cys.26 Alternatively, some quinone compounds have been shown to covalently modify the Cdc25 active site cysteines or vicinal serines resulting in inhibition.3,4
The current study describes the first systematic interrogation of a large publicly sponsored compound library for inhibitors of in vitro Cdc25B activity. A recently described assay that measures the generation of H2O2 by redox active compounds in DTT15 was utilized to identify and filter out compounds acting through the oxidation and inactivation of the Cdc25 active site cysteine. Twenty-five compounds were subsequently confirmed as concentration-dependent inhibitors of Cdc25B activity with IC50 values <50 μM; 13 were represented by singleton chemical structures, and 12 were divided among four clusters of related structures (Fig. 2). Thirteen (52%) of the 25 Cdc25B inhibitor hits have quinone-based structures similar to those previously shown to generate ROS (see above): five cyclopenta[c]quinoline-carboxylic acids (SIDs 843791, 850390, 844096, 851514, and 4249621), three pyrimidotriazine-5,7-diones (SIDs 4251194, 850758, and 845167), two isoquinolin-1-ones (SIDs 4251929 and 4249736), one naphthoquinone (SID 8139964), one isoquinoline-1,3-dione (SID 7964733), and one isoquinolin-5-amine (SID 4248661) (Fig. 2).
The Cdc25B inhibitors were further characterized in a series of in vitro secondary assays to identify compounds that might inactivate the active site cysteine through oxidation and to evaluate their selectivity for Cdc25B over MKP-1 and MKP-3 dual-specificity phosphatases (Table 1). We incorporated an assay to directly measure H2O2 generated by redox cycling compounds in reducing environments.15 This assay has been used to profile the redox potential of MKP-1 inhibitors and a series of p-quinone-based Cdc25B inhibitors.15,45 The conditions of the primary Cdc25B assay were altered to determine if the inhibition of Cdc25B would be significantly affected by any or all of the following modifications: the strong nonphysiological reducing agent DTT was replaced with the weaker BME or by the physiologically relevant GSH; CAT was added to the assay to degrade any H2O2 produced; the DTT concentration was increased from 1 to 25 mM; or reactions were conducted in the presence of both CAT and high levels of DTT. To facilitate the evaluation and analysis of the Cdc25B inhibitor hit characteristics we have simplified and summarized the data in Table 1 into an activity profile table (Table 3). At least 16 of the hit compounds (SIDs 4242461, 8139964, 856108, 3712249, 7973677, 845964, 3712327, 847359, 4251194, 845167, 850758, 857882, 4251929, 4249736, 7964733, and 4248661) inhibit Cdc25B activity via the redox cycling generation of ROS, which illustrates the importance of incorporating the assay to measure redox cycling H2O2 generation early in the hit characterization process for targets susceptible to oxidation. This conclusion was further supported by the observation that eight (50%) of the redox active compounds were promiscuous inhibitors of all three phosphatases (Tables 1 and 3). The redox cycling generation of H2O2 by the three pyrimido-triazine-diones (SIDs 850758, 845167, and 4251194) has been reported previously,15,48 and a cross-target query of the PubChem database revealed that these compounds had produced active flags in 36–39% of the assays against which they have been screened, and their activity has been confirmed for a number of targets with active site cysteines including several PTPs, cathepsins, and caspases. The redox cycling activity and promiscuous inhibition of PTPs by the 3-amino-7-(phenylmethyl)-1-sulfanylidene-6,8-dihydro-5H-thiopyrano[5,4-c]pyridine-4-carbonitrile (SID 845964) have also been reported previously.15
Table 3.
Cdc25B Inhibitor Activity Profile
| SID | Cdc25B HTS % inhibition >50% | Cdc25B IC50 <50 μM | Selective vs. MKP-1/3 | Redox H2O2 generator | Cdc25B IC50 modulated | Inhibition of tumor cell proliferation |
|---|---|---|---|---|---|---|
| 850758 | + | + | − | + | + | NT |
| 845167 | + | + | − | + | + | NT |
| 4251194 | + | + | − | + | + | NT |
| 847359 | + | + | + | + | + | NT |
| 3712327 | + | + | + | + | + | NT |
| 845964 | + | + | − | + | + | NT |
| 7973677 | + | + | − | + | + | NT |
| 3712249 | + | + | − | + | + | NT |
| 856108 | + | + | + | + | + | NT |
| 8139964 | + | + | + | + | + | NT |
| 857882 | + | + | + | + | + | NT |
| 4242461 | + | + | − | + | + | NT |
| 4251929 | + | + | − | − | + | NT |
| 4249736 | + | + | − | − | + | NT |
| 4249621 | + | + | − | − | − | NT |
| 4241893 | + | + | − | − | − | NT |
| 844096 | + | + | − | − | − | NT |
| 850390 | + | + | + | − | − | − |
| 843791 | + | + | + | − | − | − |
| 7964733 | + | + | − | − | + | NT |
| 851514 | + | + | + | − | − | − |
| 847214 | + | + | + | − | − | − |
| 4260465 | + | + | + | − | − | + |
| 4258795 | + | + | + | − | − | + |
| 4248661 | + | + | + | − | + | NT |
Cdc25B HTS % inhibition >50%, inhibition >50% in the primary Cdc25B screen; Selective vs. MKP-1/3, selectivity for Cdc25B versus MKP-1 and/or MKP-3; Redox H2O2 generator, evidence of redox cycling H2O2 generation; Cdc25B IC50 modulated, the inhibition of Cdc25B was sensitive to the modifications in the assay conditions described in Table 1; Inhibition of tumor cell proliferation, the ability to inhibit the proliferation of the human MBA-MD-435 breast and PC-3 prostate cancer cell lines. NT = not tested. + indicates that the compound met this criterion; – indicates that the compound did not meet this criterion.
Importantly, nine of the Cdc25B hits exhibited a secondary assay activity profile consistent with a nonoxidative mechanism of action because their inhibition of Cdc25B was resistant to the indicated modifications to the activity assay (Tables 1 and 3). Six of the nine compounds selectively inhibited Cdc25B over both MKP-1 and MKP-3, and a cross-target query of the PubChem database revealed that each of the six hits had been tested in ~200 bioassays and that their activity had been confirmed against several other targets, including a PTP expressed in hematopoietic cells (Table 2). We next examined the ability of the six Cdc25B hits to inhibit the proliferation of the human MBA-MD-435 breast and PC-3 prostate cancer cell lines (Figs. 3 and 4). Two of the six hits, N-[2,3-di(furan-2-yl)quinoxalin-6-yl]azepane-1-carboxamide (SID 4258795) and 5,6-di(furan-2-yl)-3-(phenylmethylsulfanyl)-1,2,4-triazine (SID 4260465), significantly inhibited the growth of both tumor cell lines in a time- and concentration-dependent manner (Figs. 3 and 4).
Interestingly, five of the nine non-redox active Cdc25B hits are cyclopentaquinoline carboxylic acid analogs, and three of these compounds appear to selectively inhibit Cdc25B (SIDs 843791, 850390, and 851514) over MKP-1 and MKP-3, whereas two inhibit both Cdc25B and MKP-1 but not MKP-3 (SIDs 844096 and 4249621) (Tables 1 and 3). Cyclopentaquinoline carboxylic acid compounds were previously shown to inhibit Cdc25B activity in vitro22 and have more recently been shown to inhibit the activity of other PTPs, including PTP1B, PTPN5, PTPN7, PTPRR,49 and MKP-1.16 Cyclopentaquinoline carboxylic acid analogs have also been reported to inhibit both MKP-3 (AID 425) and hematopoietic cell PTP (AID 521) in data uploaded to the PubChem database. However, consistent with previously published data,22 none of the three selective cyclopentaquinoline carboxylic acid Cdc25B hits produced significant growth inhibition in either tumor cell line (Fig. 4A and B). The 7-methoxy-3-methyl-1H-indole-2-carboxylic acid (SID 847214) Cdc25B hit also failed to inhibit the proliferation of either cell line (Fig. 4A and B).
The cell-active Cdc25B inhibitor 4260465 was resynthesized (SID 26683752) along with two analogs (SIDs 26683753 and 26683756) (Fig. 5A). Neither of the substitutions in the two analogs was tolerated, and only the resynthesized hit SID 26683752 inhibited Cdc25B activity in vitro (IC50 = 13.83 ± 1.0 μM) and significantly inhibited the growth of the MBA-MD-435 breast and PC-3 prostate cancer cell lines (IC50 = 20.16 ± 2.0 μM and 24.87 ± 2.25 μM, respectively) (Fig. 5). The two bis-furan-containing hits identified in the screen represent novel nonoxidative Cdc25B inhibitor chemotypes that inhibit the proliferation of cultured tumor cells. In this context, it is noteworthy to mention the known toxicity of furans, which is believed to be due in vivo (mainly) or in vitro (less frequently) to the P450-catalyzed oxidation of furan to a highly reactive 2-butene-1,4-dial metabolite.50,51 This electrophilic metabolite could readily deactivate catalytic or structural sulfhydryl groups in Cdc25B or deplete cellular GSH, and the bis-furans may be a likely source of reactive metabolites.
Studies have been initiated to further evaluate the cell-based activity of these two novel non-redox cycling bis-furan-containing Cdc25B inhibitors to determine whether they induce readily detectable effects on the cell cycle such as a G1/S or G2/M arrest and if they induce hyperphosphorylation of Cdc25B substrates such as CDK1. Positive results will trigger a more extensive chemistry analog synthesis effort to try to enhance the modest potency of these hits and establish a structure-activity relationship. Despite the large number of potent quinoid Cdc25 inhibitors with antiproliferative activity against tumor cell lines that are currently available, very few of these have demonstrated in vivo activity in human tumor cell xenograft animal models.2–4 There are also concerns about the potential toxicity of quinonoid compounds in vivo due to the redox cycling generation of ROS.2–4 The availability of non-redox active Cdc25B inhibitors should provide valuable tools to explore the inhibition of the Cdc25 phosphatases as potential mono- or combination therapies for cancer.
Abbreviations
- AC50
50% activation concentration
- AID
PubChem assay identifier
- BME
β-mercaptoethanol
- CAT
catalase
- Cdc25
cell division cycle 25 protein
- CDK
cyclin-dependent kinase
- CHK
checkpoint effector kinase
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- GSH
reduced glutathione
- HRP
horseradish peroxidase
- IC50
50% inhibitory concentration
- MKP
mitogen-activated protein kinase phosphatase
- MLSCN
Molecular Library Screening Center Network
- NIH
National Institutes of Health
- OMFP
3-O-methylfluorescein phosphate
- PMLSC
Pittsburgh Molecular Library Screening Center
- PTP
protein tyrosine phosphatase
- ROS
reactive oxygen species
- SID
PubChem substance identifier
- TCEP
tris(2-carboxyethyl)phosphine.
Acknowledgments
The authors would like to thank Harold Takyi for information technology and data analysis support. This work was supported by grants from the National Institutes of Health: MLSCN (U54MH074411), RO3 (MH78959), and PO1 (CA78093).
Disclosure Statement
No competing financial interests exist.
References
- 1.Boutros R. Dozier C. Ducommun B. The when, wheres of CDC25 phosphatases. Curr Opin Cell Biol. 2006;18:185–191. doi: 10.1016/j.ceb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Boutros R. Lobjois V. Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 3.Contour-Galcera MO. Sidhu A. Prévost G. Bigg D. Ducommun B. What's new on CDC25 phosphatase inhibitors. Pharmacol Ther. 2006;115:1–12. doi: 10.1016/j.pharmthera.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Jiang ZX. Zhang ZY. Targeting PTPs with small molecule inhibitors in cancer treatment. Cancer Metastasis Rev. 2008;27:263–272. doi: 10.1007/s10555-008-9113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristjánsdóttir K. Rudolph J. Cdc25 phosphatases and cancer. Chem Biol. 2004;11:1043–1051. doi: 10.1016/j.chembiol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Rudolph J. Cdc25 phosphatases: structure, specificity, and mechanism. Biochemistry. 2007;46:3595–3604. doi: 10.1021/bi700026j. [DOI] [PubMed] [Google Scholar]
- 7.Löffler H. Rebacz B. Ho AD. Lukas J. Bartek J. Krämer A. Chk1-dependent regulation of Cdc25B functions to coordinate mitotic events. Cell Cycle. 2006;5:2543–2547. doi: 10.4161/cc.5.21.3435. [DOI] [PubMed] [Google Scholar]
- 8.van Vugt MATM. Bras A. Medema RH. Restarting the cell cycle when the checkpoint comes to a halt. Cancer Res. 2005;65:7037–7040. doi: 10.1158/0008-5472.CAN-05-1054. [DOI] [PubMed] [Google Scholar]
- 9.Bugler B. Quaranta M. Aressy B. Brezak MC. Prevost G. Ducommun B. Genotoxic-activated G2-M checkpoint exit is dependent on CDC25B phosphatase expression. Mol Cancer Ther. 2006;5:1446–1451. doi: 10.1158/1535-7163.MCT-06-0099. [DOI] [PubMed] [Google Scholar]
- 10.Cazales M. Boutros R. Brezak MC. Chaumeron S. Prevost G. Ducommun B. Pharmacologic inhibition of CDC25 phosphatases impairs interphase microtubule dynamics and mitotic spindle assembly. Mol Cancer Ther. 2007;6:318–325. doi: 10.1158/1535-7163.MCT-06-0299. [DOI] [PubMed] [Google Scholar]
- 11.Aressy B. Bugler B. Valette A. Biard D. Ducommun B. Moderate variations in CDC25B protein levels modulate the response to DNA damaging agents. Cell Cycle. 2008;7:2234–2240. doi: 10.4161/cc.7.14.6305. [DOI] [PubMed] [Google Scholar]
- 12.Bansal P. Lazo JS. Induction of Cdc25B regulates cell cycle resumption after genotoxic stress. Cancer Res. 2007;67:3356–3363. doi: 10.1158/0008-5472.CAN-06-3685. [DOI] [PubMed] [Google Scholar]
- 13.Austin CP. Brady LS. Insel TR. Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 14.Inglese J. Auld DS. Jadhav A. Johnson RL. Simeonov A. Yasgar A, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston PA. Soares KM. Shinde SN. Foster CA. Shun TY. Takyi HK, et al. Development of a simple 384-well colorimetric assay to quantify hydrogen peroxide generated by the redox cycling of compounds in the presence of reducing agents. Assay Drug Dev Technol. 2008;6:505–518. doi: 10.1089/adt.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston PA. Foster CA. Shun TY. Skoko JJ. Shinde S. Wipf P, et al. Development, implementation of a 384-well homogeneous fluorescence intensity high-throughput screening assay to identify mitogen-activated protein kinase phosphatase-1 dual-specificity protein phosphatase inhibitors. Assay Drug Dev Technol. 2007;5:319–332. doi: 10.1089/adt.2007.066. [DOI] [PubMed] [Google Scholar]
- 17.Johnston PA. Phillips J. Shun TY. Shinde S. Lazo JS. Huryn DM, et al. HTS identifies novel, specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 18.Lazo JS. Roadmap or roadkill: a pharmacologist's analysis of the NIH Molecular Libraries Initiative. Mol Interv. 2006;6:240–243. doi: 10.1124/mi.6.5.1. [DOI] [PubMed] [Google Scholar]
- 19.Tierno MB. Johnston PA. Foster C. Skoko JJ. Shinde SN. Shun TY, et al. Development, optimization of high-throughput in vitro protein phosphatase screening assays. Nat Protoc. 2007;2:1134–1144. doi: 10.1038/nprot.2007.155. [DOI] [PubMed] [Google Scholar]
- 20.Lazo JS. Nemoto K. Pestell KE. Cooley K. Southwick EC. Mitchell DA, et al. Identification of a potent, selective pharmacophore for Cdc25 dual specificity phosphatase inhibitors. Mol Pharmacol. 2002;61:720–728. doi: 10.1124/mol.61.4.720. [DOI] [PubMed] [Google Scholar]
- 21.Brisson M. Nguyen T. Wipf P. Joo B. Day BW. Skoko JS, et al. Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol. 2005;68:1810–1820. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- 22.Brisson M. Nguyen T. Vogt A. Yalowich J. Giorgianni A. Tobi D, et al. Discovery, characterization of novel small molecule inhibitors of human Cdc25B dual specificity phosphatase. Mol Pharmacol. 2004;66:824–833. doi: 10.1124/mol.104.001784. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Blower PE., Jr Cross KP. Fligner MA. Myatt GJ. Verducci JS. Yang C. Systematic analysis of large screening sets in drug discovery. Curr Drug Discov Technol. 2004;1:37–47. doi: 10.2174/1570163043484879. [DOI] [PubMed] [Google Scholar]
- 25.Alonso A. Sasin J. Bottini N. Friedberg I. Friedberg I. Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Bova MP. Mattson MN. Vasile S. Tam D. Holsinger L. Bremer M, et al. The oxidative mechanism of action of ortho-quinone inhibitors of protein-tyrosine phosphatase alpha is mediated by hydrogen peroxide. Arch Biochem Biophys. 2004;429:30–41. doi: 10.1016/j.abb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Denu JM. Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 28.Kwon J. Lee SR. Yang KS. Ahn Y. Kim YJ. Stadtman ER, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci U S A. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SR. Yang KS. Kwon J. Lee C. Jeong W. Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 30.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 31.Rhee SG. Bae YS. Lee SR. Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000(53):PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 32.Rhee SG. Chang TS. Bae YS. Lee SR. Kang SW. Cellular regulation by hydrogen peroxide. J Am Soc Nephrol. 2003;14(8 Suppl 3):S211–S215. doi: 10.1097/01.asn.0000077404.45564.7e. [DOI] [PubMed] [Google Scholar]
- 33.Rhee SG. Kang SW. Jeong W. Chang TS. Yang KS. Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Salmeen A. Andersen JN. Myers MP. Meng TC. Hinks JA. Tonks NK, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenylamide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 35.Seth D. Rudolph J. Redox regulation of MAP kinase phosphatase 3. Biochemistry. 2006;45:8476–8487. doi: 10.1021/bi060157p. [DOI] [PubMed] [Google Scholar]
- 36.Sohn J. Buhrman G. Rudolph J. Kinetic and structural studies of specific protein-protein interactions in substrate catalysis by Cdc25B phosphatase. Biochemistry. 2007;46:807–818. doi: 10.1021/bi061257y. [DOI] [PubMed] [Google Scholar]
- 37.Sohn J. Parks JM. Buhrman G. Brown P. Kristjánsdóttir K. Safi A, et al. Experimental validation of the docking orientation of Cdc25 with its Cdk2-CycA protein substrate. Biochemistry. 2005;44:16563–16573. doi: 10.1021/bi0516879. [DOI] [PubMed] [Google Scholar]
- 38.Brisson M. Foster C. Wipf P. Joo B. Tomko RJ., Jr Nguyen T, et al. Independent mechanistic inhibition of cdc25 phosphatases by a natural product caulibugulone. Mol Pharmacol. 2007;71:184–192. doi: 10.1124/mol.106.028589. [DOI] [PubMed] [Google Scholar]
- 39.Cao S. Foster C. Lazo JS. Kingston DG. Four diterpenoid inhibitors of Cdc25B phosphatase from a marine anemone. Bioorg Med Chem. 2005;13:5830–5834. doi: 10.1016/j.bmc.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 40.Cao S. Foster C. Lazo JS. Kingston DG. Sesterterpenoids and an alkaloid from a Thorectandra sp. as inhibitors of the phosphatase Cdc25B. Bioorg Med Chem. 2005;13:5094–5098. doi: 10.1016/j.bmc.2005.04.070. [DOI] [PubMed] [Google Scholar]
- 41.Cao S. Foster C. Brisson M. Lazo JS. Kingston DG. Halenaquinone and xestoquinone derivatives, inhibitors of Cdc25B phosphatase from a Xestospongia sp. Bioorg Med Chem. 2005;13:999–1003. doi: 10.1016/j.bmc.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 42.Kar S. Lefterov IM. Wang M. Lazo JS. Scott CN. Wilcox CS, et al. Binding and inhibition of Cdc25 phosphatases by vitamin K analogues. Biochemistry. 2003;42:10490–10497. doi: 10.1021/bi027418p. [DOI] [PubMed] [Google Scholar]
- 43.Peyregne VP. Kar S. Ham SW. Wang M. Wang Z. Carr BI. Novel hydroxyl naphthoquinones with potent Cdc25 antagonizing and growth inhibitory properties. Mol Cancer Ther. 2005;4:595–602. doi: 10.1158/1535-7163.MCT-04-0274. [DOI] [PubMed] [Google Scholar]
- 44.Wipf P. Joo B. Nguyen T. Lazo JS. Synthesis and biological evaluation of caulibugulones A-E. Org Biomol Chem. 2004;2:2173–2174. doi: 10.1039/b408184f. [DOI] [PubMed] [Google Scholar]
- 45.Keinan S. Paquette WD. Skoko JJ. Beratan DN. Yang W. Shinde S, et al. Computational design, synthesis and biological evaluation of para-quinone-based inhibitors for redox regulation of the dual-specificity phosphatase Cdc25B. Org Biomol Chem. 2008;6:3256–3263. doi: 10.1039/b806712k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brisson M. Nguyen T. Wipf P. Joo B. Day BW. Skoko JS, et al. Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol. 2005;68:1810–1820. doi: 10.1124/mol.105.016360. [DOI] [PubMed] [Google Scholar]
- 47.Guo J. Parise RA. Joseph E. Lan J. Pan SS. Joo B, et al. Pharmacology and antitumor activity of a quinolinedione Cdc25 phosphatase inhibitor DA3003-1 (NSC 663284) Anticancer Res. 2007;27:3067–3073. [PubMed] [Google Scholar]
- 48.Lor LA. Schneck J. McNulty DE. Diaz E. Brandt M. Thrall SH, et al. A simple assay for detection of small-molecule redox activity. J Biomol Screen. 2007;12:881–890. doi: 10.1177/1087057107304113. [DOI] [PubMed] [Google Scholar]
- 49.Eswaran J. von Kries JP. Marsden B. Longman E. Debreczeni JE. Ugochukwu E, et al. Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases. Biochem J. 2006;395:483–491. doi: 10.1042/BJ20051931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen LJ. Hecht SS. Peterson LA. Characterization of amino acid and glutathione adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 1997;10:866–874. doi: 10.1021/tx9700174. [DOI] [PubMed] [Google Scholar]
- 51.Peterson L. Electrophilic intermediates produced by bioactivation of furan. Drug Metab Rev. 2006;38:615–626. doi: 10.1080/03602530600959417. [DOI] [PubMed] [Google Scholar]