Abstract
Hepatitis C virus (HCV) is a global public health problem that mediates a persistent infection in nearly 200 million people. HCV is efficient in establishing chronicity due in part to the inefficiency of the host immune system in controlling and counteracting HCV-mediated evasion strategies. HCV persistence is linked to the ability of the virus to suppress the RIG-I pathway and interferon production from infected hepatocytes, thus evading innate immune defenses within the infected cell. This review describes the virus and host processes that regulate the RIG-I pathway during HCV infection. An understanding of these HCV–host interactions could lead to more effective therapies for HCV designed to reactivate the host immune response following HCV infection.
Introduction
Hepatitis C virus (HCV), a single-stranded hepatotropic RNA virus, continues to be a major public health problem, persistently infecting nearly 200 million people worldwide, with 3–4 million new infections annually. While 20%–30% of those acutely infected with HCV may clear the virus, the majority of those infected develop chronic infection, leading to liver inflammation, fibrosis, and cirrhosis. HCV is a primary indicator for liver transplantation and a leading etiology for hepatocellular carcinoma, the third leading cause of cancer death worldwide (Bosch and others 1999; Sharma and Lok 2006). The disease associated with HCV places a substantial global burden on the health care system (Lavanchy 2009). There is no vaccine for HCV, and the current HCV therapy of pegylated interferon (IFN)-α in combination with ribavirin leads to a sustained virological response (SVR) in only half of those chronically infected (Soriano and others 2009). In recent years, HCV-related liver disease has become the leading cause of non-AIDS death in those coinfected with HIV (Pineda and others 2007).
The innate immune response to virus infection is activated when conserved pattern-associated molecular patterns (PAMPs) generated during infection are recognized in cells by proteins known as pattern recognition receptors (PRRs). The 3 major classes of PRRs are Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and nucleotide oligomerization domain (NOD)-like receptors (NLRs). Viral engagement of TLRs and RLRs leads to downstream signaling that results in the activation of latent transcription factors, including the IFN regulatory factors (IRFs) and nuclear factor-κB (NF-κB), and culminates in the induction of IRF3 target genes, type I IFN, and proinflammatory cytokines. Type I IFN coordinates immunity to prevent virus infection of new cells and limit virus spread. Importantly, type I IFN signals in an autocrine and paracrine manner through the type I IFN receptor to induce hundreds of IFN-stimulated genes (ISGs) that establish a tissue-wide antiviral state, and it sensitizes infected cells to apoptosis (de Veer and others 2001; Stetson and Medzhitov 2006). Furthermore, type I IFN modulates several aspects of adaptive immunity, including establishment of cytotoxic T-cell responses, generation of natural killer cells, and B-cell differentiation, for the elimination of virus-infected cells (Stetson and Medzhitov 2006; Purtha and others 2008). NLR-mediated PAMP detection activates caspase-1 leading to the secretion of proinflammatory cytokines, such as interleukin-1β (Petrilli and others 2007), which directs an antiviral inflammatory response.
Recent studies have made progress toward our understanding of how HCV is detected by the host cell for the activation of innate immune response. In spite of this detection and ability to stimulate innate immune defenses and IFN production, HCV is successful in mediating a chronic course of infection in the majority of those infected. This review will discuss how HCV is sensed by the innate immune response and then how the virus subsequently antagonizes the antiviral effects of type I IFN.
Hepatocytes Detect HCV Infection Through RIG-I
The robust replication of HCV produces an estimated 1012 virions per day (Neumann and others 1998), and this high level of HCV transiently activates the innate immune system early during infection. Acute HCV infection in chimpanzees induces an ISG expression profile in the liver consistent with a type I IFN response (Su and others 2002). Furthermore, HCV RNA is potent inducer of type I IFN (Sumpter and others 2005; Saito and others 2008), mediated by HCV engagement of PRRs.
Pattern recognition receptors
Viral PAMPs in the host are recognized as nonself by 3 distinct classes of sensors that initiate intracellular signaling, the TLRs, NLRs, and RLRs (Saito and Gale 2007). The function of these antiviral sensors is regulated in part by their compartmentalization within the cell. RLRs and NLRs sense PAMPs in the cytoplasm of most cells, including hepatocytes, while TLRs are expressed in endosomes and on the cell surface and primarily detect viral PAMPs in various immune cells, such as macrophages, dendritic cells, B cells, and some types of T cells (Eisenacher and others 2007). The role of NLRs in sensing RNA viruses is still unclear, and they are primarily thought to be activated by bacterial PAMPs or intracellular stress signals (Kanneganti and others 2006; Petrilli and others 2007).
The TLR family members that detect viral PAMPs include TLR2, TLR3, TLR4, TLR7/8, and TLR9, and their activation leads to production of proinflammatory cytokines and IFN-α. TLR2 and TLR4 are expressed on the plasma membrane and are involved in detecting viral proteins on the cell surface. Both the HCV core and NS3 proteins have been shown to activate TLR2 signaling leading to host inflammation (Dolganiuc and others 2004; Chang and others 2007). TLR3, TLR7/8, and TLR9 detect viral nucleic acids. TLR7/8 and TLR9 are primarily expressed in endosomes, while TLR3 can be found in endosomes and at the cell surface, depending on the cell type (Eisenacher and others 2007). TLR9 senses unmethylated CpG motifs in DNA, and therefore its role in the detection of HCV, an RNA virus, would be expected to be minor. TLR7/8 sense uridine- and guanosine-rich single-stranded RNA (ssRNA) (Diebold and others 2004; Heil and others 2004) in conventional dendritic cells and plasmacytoid dendritic cells, the major producers of IFN-α in response to virus infection. TLR3 is expressed in conventional dendritic cells, macrophages, and several nonimmune cell types, such as fibroblasts and epithelial cells, and has been proposed to sense double-stranded RNA (dsRNA). The physiological viral ligand and antiviral function of TLR3 and TLR7/8 are undefined (Schroder and Bowie 2005; Vercammen and others 2008), although it appears that TLR3 is involved in the induction of type II IFN following picornavirus infection (Negishi and others 2008). Because TLR3 and TLR7/8, which are transmembrane proteins, primarily sense nucleic acids in an intracellular location, detection of HCV by these TLRs would likely require phagocytosis of virus-infected cells. Hepatocytes, the primary site of HCV infection, are deficient in TLR signaling (Seki and Brenner 2008). To date, no HCV RNA ligand has been shown to directly activate TLR pathways. Therefore, TLRs likely mediate a secondary response to HCV infection, perhaps sensing viral products in phagocytosed cells (Schulz and others 2005), resulting in amplification of intrahepatic inflammatory signals.
The RLR family consists of retinoic acid-inducible gene-I, RIG-I; melanoma differentiation-associated gene 5, MDA5; and laboratory of genetics and physiology 2, LGP2. Because these RLRs are expressed in the cytoplasm of most cells, including hepatocytes, they are candidates to be the primary intracellular sensors of HCV infection (Kato and others 2006). Both RIG-I and MDA5 contain 2 N-terminal caspase activation and recruitment domains (CARD). All the RLRs have a DExD/H RNA helicase domain and bind to RNA ligands. Studies using RIG-I−/− and MDA5−/− mice revealed that RIG-I and MDA5 recognize distinct classes of viruses (Kato and others 2006). RIG-I has been shown to be essential for detection of specific set of ssRNA viruses, including paramyxoviruses, flaviviruses, orthomyxoviruses, and rhabdoviruses, while MDA5 is essential for detection of picornaviruses, such as encephalomyocarditis virus (Yoneyama and others 2004; Sumpter and others 2005; Kato and others 2006; Loo and others 2008). These studies also showed that production of IFN-β following stimulation with poly I:C, the synthetic double-stranded (ds) RNA analog, was mediated by MDA5, and not RIG-I (Gitlin and others 2006; Kato and others 2006). The proposed ligand for MDA5 is long dsRNA greater than 3 kb (Kato and others 2008).
Transfection of HCV RNA into human hepatoma cells leads to transient activation of IRF3 and IFN-β (Sumpter and others 2005; Saito and others 2007). This activation is defective in cells that lack functional RIG-I, but can be complemented with wild-type RIG-I, demonstrating that RIG-I is required for activation of IRF3 by HCV RNA (Sumpter and others 2005). HCV infection is also more efficient in cells that lack functional RIG-I signaling (Loo and others 2006). Taken together, these studies indicate that RIG-I is the primary intracellular sensor of HCV. Biochemical studies revealed that RIG-I, but not MDA5, recognizes the 5′ and 3′ noncoding regions (NCRs) of HCV RNA (Saito and others 2007). The HCV NCRs are highly structured and contain partial double-stranded regions (Fig. 1). Recent work has defined the detailed mechanism of HCV RNA ligand recognition by RIG-I for the induction of antiviral signaling (Saito and others 2008; Uzri and Gehrke 2009).
FIG. 1.
RNA genome structure of hepatitis C virus (HCV). The 9.6 kb positive strand RNA genome of HCV is illustrated. The RNA secondary structure of the 5′ noncoding region (NCR) and 3′ NCR are depicted. The HCV 3′ NCR contains 3 domains: a variable region with 2 RNA stem loops, a single-stranded poly U/UC region, and a conserved “X” region with 3 RNA stem loops. The minimal HCV pattern-associated molecular pattern (PAMP) that activates RIG-I signaling is indicated by the arrow.
How is HCV recognized by RIG-I?
Cytoplasmic ssRNA containing a 5′ triphosphate (ppp), short dsRNA, and uridine- or adenosine-rich viral RNA motifs are recognized by RIG-I (Hornung and others 2006; Pichlmair and others 2006; Kato and others 2008; Saito and others 2008; Takahasi and others 2008; Uzri and Gehrke 2009). The RNA ligand base composition, sugar backbone, and length of the RNA sequence also provide important signals required for RIG-I activation (Saito and others 2008; Uzri and Gehrke 2009). The HCV genome contains several motifs that facilitate its recognition by RIG-I. HCV RNA is not capped and therefore its RNA contains the 5′ ppp required for detection by RIG-I. The 3′ NCR of the HCV genome is the primary HCV PAMP that activates RIG-I signaling (Sumpter and others 2005; Saito and others 2007). Importantly, this region is highly conserved among HCV genotypes and is critical for HCV replication (You and Rice 2008). The 3′ NCR consists of 3 parts: a variable region containing 2 stem loops, a poly U-rich region that is single-stranded, and a conserved “X” region, which contains 3 stem loops (Fig. 1). It was expected that dsRNA or RNA with secondary structure located within the 3′ NCR would be the primary HCV PAMP for RIG-I through interactions with the RIG-I helicase domain. In fact, the highly structured X region of the HCV genome does not activate RIG-I signaling, but surprisingly, the poly U/UC region is a potent activator of RIG-I signaling. A 5′ ppp is necessary for this activation, but it is not sufficient, as the X region containing a 5′ ppp does not activate RIG-I (Saito and others 2008).
How does RIG-I detect these 2 distinct signatures (5′ ppp and poly U/UC sequence) in 1 RNA molecule, when they are separated by over 9,600 nucleotides in the HCV genome? There are 3 possibilities. First, well-established long-range RNA interactions between the 5′ and 3′ ends of the HCV genome that are important for efficient HCV translation could bring the 5′ ppp and poly U/UC into close proximity for stable interaction with and activation of RIG-I (Ito and others 1998; Song and others 2006). Alternatively, it was recently shown that the ability of HCV poly U/UC RNA to activate RIG-I does not require immediate adjacency of the 5′ ppp, suggesting that RIG-I recognizes 2 distinct features of the PAMP RNA, both the 5′ ppp and the poly U/UC sequence (Uzri and Gehrke 2009). In fact, structural studies suggest that RIG-I has 2 distinct RNA sensing domains (Cui and others 2008; Takahasi and others 2008). RIG-I binds 5′ ppp ssRNA through a positively charged groove in its C-terminal domain, which includes the RD. As this groove can only accommodate 3 nucleotides, RIG-I must use an additional domain, most likely the helicase domain, to discriminate specific RNA sequences, such as the poly U/UC present in the HCV PAMP. Finally, the corresponding replication intermediate sequence of the poly U/UC region, poly A/AG, also activates RIG-I signaling. In this case, during HCV replication, the 5′ ppp of the negative strand would be in close proximity to the poly A/AG region, and together these motifs could bind to RIG-I. In fact, it has been proposed that RIG-I may bind to substrate RNA and then translocate on the RNA to scan for activating sequences (Myong and others 2009).
It is not known where in the cell and when in the HCV replication program the PAMP that activates RIG-I is generated. HCV replicates in association with cellular membranes and induces weblike membranes structures, which compartmentalize HCV replication complexes and probably protect HCV RNA from RIG-I detection (Egger and others 2002; Gosert and others 2003). However, this does not exclude the possibility of some nonmembranous HCV RNA present in the cytoplasm. In addition to these large perinuclear replication complexes on weblike cellular membranes, smaller HCV replication complexes are distributed on membranes located throughout the cytoplasm (Targett-Adams and others 2008; Wolk and others 2008). These smaller replication complexes, distinct from the large perinuclear replication complexes, appear to be formed during early times after infection (<24 h) (Targett-Adams and others 2008; Wolk and others 2008), and therefore could be initial detection sites of the HCV genome by RIG-I. In support of this idea, HCV infection activates IRF3 at early times during infection before extensive viral protein synthesis has occurred (Loo and others 2006; Lau and others 2008). Further studies are needed to determine the intracellular localization and activation of RIG-I during HCV infection.
The HCV PAMP (5′ ppp poly U/UC ssRNA) triggers hepatic immune responses in vivo, and these responses are mediated by RIG-I (Saito and others 2008). The PAMPs in the 3′ NCR of 3 different HCV strains (Con1, J4L6, JFH-1) have been tested so far, and all 3 activate RIG-I signaling (Saito and others 2008; Uzri and Gehrke 2009). The PAMP from the HCV genotype 2a strain JFH-1, which replicates and produces infectious particles in cell culture, is a relatively weak activator of RIG-I signaling, suggesting that the ability of the JFH-1 strain to replicate in cell culture could be due in part to its low immunostimulatory activity (Uzri and Gehrke 2009). Other viruses that activate RIG-I, including rabies virus, ebola virus, and measles virus, all contain PAMP regions with similar sequences to the HCV PAMP that potently activate RIG-I signaling thereby demonstrating a conserved mechanism of PAMP recognition by RIG-I (Saito and others 2008).
The features present in the HCV PAMP distinguish it from cellular RNAs and illustrate the exquisite discrimination that RIG-I uses to distinguish “self” (endogenous RNA) from “nonself” (viral RNA). While HCV RNA is uncapped and contains a 5′ ppp, endogenous RNA has 5′ end modifications, such as a methylguanosine cap or a monophosphate, interacts with ribonucleoproteins, or has extensive base modifications, all of which would mask detection by RIG-I. In fact, some viruses have evolved mechanisms to avoid detection by RIG-I, such as removal of the 5′ ppp from their viral genomes or attachment of viral proteins to the 5′ ends of their genomes (Ferrer-Orta and others 2006; Habjan and others 2008). The poly U/UN (where N denotes an interspersed non-U nucleotide) sequence seems to be a general immune mechanism for nonself recognition, as TLR7/8 are also activated by ssRNA PAMPs containing this sequence (Diebold and others 2004; Heil and others 2004).
Following RNA virus infection or dsRNA treatment of cells, the antiviral endoribonuclease RNaseL is required for the production of IFN-β. Upon activation by 2′,5′-linked oligoadenylate, which is produced by the 2′–5′ oligoadenylate synthetase (2–5 OAS), RNaseL cleaves RNA to generate small RNA cleavage products that signal through the RIG-I/MDA5 pathway (Malathi and others 2007). RNaseL can cleave HCV RNA at single-stranded UA and UU dinucleotides in vitro (Han and Barton 2002; Washenberger and others 2007); however, it is unclear how RNaseL-cleaved RNA is recognized by RIG-I. It is possible that cleavage of HCV RNA by RNaseL could generate higher levels of HCV PAMP leading to amplification of RIG-I signaling during virus infection.
Mechanism of RIG-I activation
In the absence of ligand, RIG-I is held in an inactive confirmation by intramolecular interactions between the N-terminal CARD and the C-terminal RD, which suppress ATPase activity of RIG-I (Saito and others 2007; Gee and others 2008). Structural studies revealed that both short dsRNA and 5′ ppp RNA bind to the C-terminal domain of RIG-I (Takahasi and others 2008). RIG-I interaction with these RNA ligands, including the HCV PAMP, induces a conformational change in RIG-I that facilitates oligomerization through the RD, activation of the ATPase domain, and exposure of the CARD for interaction with the CARD domain of the adaptor protein IFN promoter-stimulating factor-1 (IPS-1, also known as Cardif, MAVS, and VISA) and activation of downstream signaling molecules (see below) (Saito and others 2007; Cui and others 2008; Saito and Gale 2008; Takahasi and others 2008). The details of RIG-I activation, as well as the role of the RIG-I helicase domain in RIG-I activation, are still unclear. Structural studies of full-length RIG-I in the presence and absence of PAMP are needed to understand the exact mechanism of RIG-I activation after engagement of the PAMP.
In addition to internal regulation, RIG-I activation is subject to external regulation. Ubiquitination of RIG-I on the CARD by TRIM25 or on the C-terminal domain by Riplet/ RNF135, promotes the interaction of RIG-I with IPS-1 and activation of downstream signaling (Gack and others 2007; Oshiumi and others 2009). The RLR LGP2 also plays a role in regulating RIG-I signaling and controlling virus infection. LGP2, which lacks a CARD, has been reported to be a negative regulator of the RIG-I pathway (Yoneyama and others 2005; Saito and others 2007). Several mechanisms for this negative regulation have been suggested, including sequestration of RNA ligands, heterodimerization with RIG-I, and preventing the kinase IκB kinase-ɛ (IKK-ɛ) from binding to IPS-1 (Yoneyama and others 2005; Komuro and Horvath 2006; Saito and others 2007). While the C-terminal domain of LGP2 can bind to dsRNA, including HCV RNA, this binding activity does not appear to be required for its ability to regulate RIG-I signaling (Saito and others 2007; Bamming and Horvath 2009; Li and others 2009). Studies in LGP2−/− mice demonstrate that depending on the type of virus, LGP2 is either a positive or negative regulator of antiviral signaling (Venkataraman and others 2007). Future studies are needed to define the mechanism of LGP2 regulation of RIG-I signaling during HCV infection.
IFN Signaling During HCV Infection
During HCV infection, RIG-I detects the 3′ NCR of HCV leading to activation of IRF3 (Sumpter and others 2005; Saito and others 2007). Activated IRF3 directly stimulates the expression of specific set of genes within the infected cell, including type I IFN (Grandvaux and others 2002; Elco and others 2005). Secreted IFN-β transmits local and tissue-wide signals for the induction of hundreds of ISGs by interacting in an autocrine and paracrine manner with the IFN-α/β receptor located on the cell surface. This leads to activation of the Janus kinase (Jak) signal transducer and activator of transcription (STAT) pathway. STAT1 and STAT2 are phosphorylated by JAK-1 and protein tyrosine kinase-2 and, together with IRF9, form the ISG factor-3 (ISGF3) transcription factor complex. ISGF3 binds to IFN-stimulated response elements within the promoters of ISGs to activate gene transcription.
Type I IFN successfully blocks HCV replication in cell culture (Guo and others 2001; Pawlotsky 2003) and induces expression of many ISGs (de Veer and others 2001). In addition to having roles in antiviral activities, ISGs are involved in such processes as lipid metabolism, apoptosis, protein degradation, and inflammatory responses (de Veer and others 2001). Microarray analysis of liver samples from HCV-infected chimpanzees or chronically infected humans demonstrates that many ISGs are expressed in the HCV-infected liver (Bigger and others 2001; Su and others 2002; Smith and others 2003; Bigger and others 2004; Helbig and others 2005; Lanford and others 2006; Lau and others 2008). Characterization of these ISGs has revealed that several have anti-HCV activity, including protein kinase R (PKR), ISG56, ISG20, ADAR1, and viperin. Both PKR and ISG56 block HCV replication at the level of translational inhibition. PKR phosphorylates the α-subunit of the eukaryotic initiation factor (eIF) 2 and the IRF3-activated gene ISG56 binds to eIF3; both of these processes prevent initiation of viral protein translation (Wang and others 2003). ADAR1 is an RNA-editing enzyme that deaminates adenosines in dsRNA, resulting in destabilization of RNA and accumulation of mutations (Taylor and others 2005). Viperin has also been characterized as an anti-HCV effector. Its expression induces the formation of crystalloid ER and interferes with protein secretion (Hinson and Cresswell 2009). Deletion of the ER localization domain of viperin abrogates its anti-HCV activity (Jiang and others 2008). Therefore, it is possible that viperin inhibits HCV replication by interfering with the formation or activity of the HCV replication complex on ER membranes.
The current HCV therapy of pegylated IFN-α and ribavirin is only successful for ∼50% of those treated, despite the fact that type I IFN can block HCV replication and stimulate the expression of ISGs that have anti-HCV activity. HCV has several mechanisms for disrupting the IFN pathway (see below) that may contribute to the low success rate of HCV therapy. This disruption of the IFN pathway by HCV may prevent the activation of a specific subset of ISGs important for HCV clearance. Examination of paired liver biopsies from patients with chronic HCV before and after pegylated IFN-α therapy found that nonresponders to therapy had similarly high levels ISGs before and after therapy (Sarasin-Filipowicz and others 2008). Additionally, this preactivation of ISGs was higher in patients infected with the more difficult to treat HCV genotypes 1 and 4 than in genotypes 2 and 3, which are easier to treat. While the exact mechanism for this preactivation of ISGs and non-response to therapy is not known, this study, along with similar studies in chimpanzees and human patients (Bigger and others 2001; Bigger and others 2004; Chen and others 2005), suggests that nonresponders may not be activating a specific subset of ISGs that are important for viral clearance or that negative feedback loops, such as those mediated by ISG15 protease USP18 (Randall and others 2006), due to the high levels of IFN signaling in these patients prevent the induction of anti-HCV ISGs following pegylated IFN-α therapy. Indeed, consensus IFN, which has increased anti-HCV potency over pegylated IFN-α, induces a subset of ISGs distinct from those induced by pegylated IFN-α (Erickson and others 2008). Perhaps one of these ISGs or a specific IFN-induced micro-RNA will be implicated as a factor required for successful HCV clearance (Pedersen and others 2007).
Control of Innate Immune Signaling by HCV
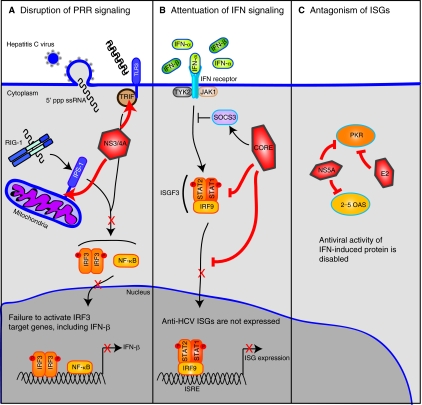
Despite the fact that RIG-I signaling and ISGs are induced by HCV, 80% of those infected with HCV become chronically infected. HCV successfully evades several aspects of the host immune response, including expression of IFN-β, IFN signaling, and antiviral activities of IFN-induced proteins (Fig. 2). These activities blunt the intracellular innate immune response and also most likely cause defects in the adaptive immune response, contributing to HCV persistence.
FIG. 2.
Hepatitis C virus (HCV) evades the innate immune response of the host cell. The intracellular innate immune response to HCV is activated when RIG-I (retinoic inducible gene-1) binds to the HCV pattern-associated molecular pattern, uridine-rich single-stranded RNA (ssRNA) with a 5′ triphosphate (5′ ppp). HCV proteins ablate the signaling pathways that activate this immune response. (A) The HCV NS3/4A protease cleaves the RIG-I and TLR3 (Toll-like receptor-3) signaling adaptor proteins, IFN-β promoter stimulator-1 (IPS-1), and Toll/interleukin-1 receptor/resistance domain-containing adaptor-inducing IFN (TRIF), respectively. Cleavage of these proteins by NS3/4A prevents IRF3 (interferon regulatory factor-3) and NF-κB (nuclear factor-κB) signaling and IFN-β activation in the infected cell. (B) The HCV core protein blocks several aspects of Janus kinase (Jak) signal transducer or activator of transcription (STAT) signaling to prevent expression of IFN-stimulated genes (ISGs). HCV core protein induces suppressor of cytokine signaling-3 (SOCS3), a negative regulator of Jak/ STAT signaling. HCV core also prevents STAT1 phosphorylation and nuclear import. (C) HCV proteins antagonize the antiviral activity of IFN-induced proteins. Both the HCV E2 and NS5A proteins bind to protein kinase R (PKR) and inhibit its activation. NS5A also blocks the antiviral activity of 2′–5′ oligoadenylate synthetase (2–5 OAS). Abbreviations: ISRE, interferon stimulated response element; TYK2, protein tyrosine kinase-2; PRR, pattern recognition receptor.
HCV disrupts signaling to IFN-β
RIG-I signaling is activated upon interaction with the HCV PAMP that promotes a conformational change in RIG-I to facilitate its interaction with IPS-1. This interaction and subsequent activation of IPS-1 leads to assembly of a complex on the mitochondria that signals downstream to activate NF-κB and the kinases IKK-ɛ and Tank-binding kinase 1 (TBK1), which phosphorylate the transcription factors IRF3 and IRF7. STING (also called MITA (Zhong and others 2008) and MPYS (Jin and others 2008)) has been identified as a new member of this pathway (Ishikawa and Barber 2008). It appears to be localized in both the ER and the mitochondria, and upon virus infection, it recruits TBK1 to the mitochondria for the phosphorylation and activation of IRF3. The exact mechanism by which interaction of RIG-I with IPS-1 activates signal transduction through IPS-1 remains to be determined, especially since overexpression of IPS-1 alone leads to downstream signaling. Both NLRX1 and the C1q receptor gC1qR have been identified as negative regulators of IPS-1 signaling (Moore and others 2008; Tattoli and others 2008; Xu and others 2009). When these proteins are overexpressed during virus infection, they interact with IPS-1 on the mitochondria and prevent IPS-1 signaling. Therefore, activation of RIG-I by PAMP binding and subsequent ubiquitination of RIG-I may promote strong interactions with IPS-1 to displace these negative regulators from IPS-1 for the activation of downstream antiviral signaling. Interestingly, extracellular HCV core protein interacts with gC1qR on the surface of T cells and inhibits T-cell activation (Kittlesen and others 2000; Yao and others 2004). It will be of interest to determine if HCV core protein interacts with intracellular gC1qR or if any of the HCV proteins antagonize the function of any of these new members in the RIG-I pathway.
HCV has several mechanisms to disrupt the RIG-I signaling pathway that it triggers to prevent and control activation of type I IFN (Gale and Foy 2005). The NS3/4A protein is the primary HCV protein responsible for evasion of these antiviral signaling pathways. NS3/4A is the major HCV protease and is essential for viral replication. NS3 interacts with its cofactor, the NS4A peptide, to anchor the NS3/4A complex to intracellular membranes and to facilitate complete activation of the NS3 protease domain (Wolk and others 2000; Brass and others 2008). During HCV replication, NS3/4A uses its serine protease domain to cleave the HCV polyprotein and release the mature nonstructural viral proteins. To block signaling by RIG-I, NS3/4A also cleaves the innate immune adaptor proteins IPS-1. Cleavage of IPS-1 by NS3/4A releases it from the mitochondrial membrane thereby abolishing RIG-I-mediated signal transduction, including IRF3 activation, in the infected cell (Foy and others 2003; Li and others 2005; Meylan and others 2005; Loo and others 2006). Recent studies have demonstrated that IPS-1 oligomerization is required for activation of antiviral signaling, and it appears that NS3/4A cleavage of IPS-1 also prevents signaling by disrupting IPS-1 oligomerization (Baril and others 2009; Tang and Wang 2009). NS3/4A has also been shown to cleave TRIF (Toll/interleukin-1 receptor/resistance domain-containing adaptor-inducing IFN), the signaling adaptor molecule for TLR3, to prevent TLR3-mediated antiviral signaling (Li and others 2005). However, the role of TLR3/TRIF in innate immunity to HCV has not been defined and other studies have not reproduced this finding (Dansako and others 2007; Dansako and others 2009). NS3/4A protease inhibitors are currently under development as antiviral therapies for HCV (Soriano and others 2009). These protease inhibitors, which block the ability of NS3/4A to cleave the HCV polyprotein and IPS-1, restore innate immune signaling in HCV-infected cells (Johnson and others 2007; Liang and others 2008). In addition to NS3/4A-mediated cleavage of IPS-1 and TRIF, NS3 has also been reported to physically bind to TBK1 to block the association of TBK1 with IRF3 and the activation of IRF3 (Otsuka and others 2005).
The HCV core protein interacts with the DEAD box protein-3 (DDX3), a member of the DEAD box helicase family (Mamiya and Worman 1999; Owsianka and Patel 1999; You and others 1999). It has recently been shown that during virus infection, DDX3 interacts with IKK-ɛ to prevent IRF activation (Schroder and others 2008). Further studies are required to determine if the interaction of HCV core with DDX3, which is required for HCV replication (Ariumi and others 2007), has an effect on IRF activation during HCV infection. In 1 study, HCV NS4B was shown to block RIG-I- and IPS-1-mediated signal transduction, although other studies have not observed an effect of NS4B expression on IRF3 activation (Foy and others 2003; Tasaka and others 2007).
HCV regulates IFN signaling and antagonizes antiviral activities of IFN-induced proteins
The Jak/STAT pathway is another target of HCV. Expression of the HCV polyprotein may attenuate Jak/ STAT signal transduction by inducing expression of protein phosphatase 2A (PP2A) (Heim and others 1999) in chronic HCV infection, resulting in a decreased activity of STAT1 and loss of ISG activation by the transcription factor ISGF3 (Blindenbacher and others 2003; Duong and others 2004). The HCV core protein expressed alone can block Jak/STAT signal transduction. It directly binds to STAT1 to prevent its phosphorylation and subsequent activation of downstream anti-HCV ISGs, including PKR, MxA, and 2–5 OAS (Melen and others 2004; de Lucas and others 2005; Lin and others 2006). Additionally the HCV core protein induces expression of the suppressor of cytokine signaling-3 (SOCS3), a negative regulator of the Jak/STAT signaling pathway, resulting in decreased STAT1 activation in response to type I IFN (Bode and others 2003). Therefore, antagonism of the Jak/STAT pathway by HCV core protein prevents ISG induction in response to virus infection.
The HCV NS5A and E2 proteins directly interfere with the antiviral actions of ISGs, including PKR. The HCV glycoprotein E2 acts as competitive substrate with eIF2α for PKR binding resulting in inhibition of PKR kinase activity (Taylor and others 1999). The HCV NS5A protein, which is involved in RNA replication and HCV particle assembly (Huang and others 2007; Appel and others 2008; Tellinghuisen and others 2008), prevents PKR activation through direct interactions that prevents PKR-mediated translation control resulting in increased HCV replication and subversion of the IFN response (Gale and others 1998; Pflugheber and others 2002). NS5A also blocks the antiviral function of 2–5 OAS through a direct interaction (Taguchi and others 2004). Expression of NS5A seems to induce secretion of IL-8 resulting in a reduced expression of ISGs (Polyak and others 2001a). In fact, IL-8 levels are increased in HCV-infected patients who do not respond to therapy, as compared to those that do respond to therapy, suggesting that NS5A may have direct effects on responses to IFN therapy (Polyak and others 2001b). Finally, NS5A binds to MyD88, the major signaling adaptor protein for TLRs (except for TLR3), in macrophages and prevents cytokine induction in response to TLR ligands, although the relevance of this is unclear because HCV has not been reported to infect macrophages (Abe and others 2007).
Conclusion
The recent discovery of the HCV PAMP has provided us with a more complete understanding of how RIG-I engages its ligands for activation of downstream signaling pathways that mediate the production of type I IFN and ISGs. In spite of the fact that HCV induces RIG-I signaling, 80% of those infected do not clear the virus and become chronically infected. HCV has a diverse set of strategies to evade the actions of RIG-I signaling, type I IFN, and antiviral ISGs, creating an environment suitable for HCV persistence. These innate immune evasion mechanisms most likely prevent a functional adaptive immune response to HCV and contribute to the ability of HCV to mediate a chronic course of infection. A detailed understanding of the interactions of HCV with the innate immune response is necessary to develop novel therapeutics for HCV and to design strategies to improve the outcome of current HCV therapy.
Acknowledgments
The authors would like to thank Takeshi Saito and Mehul S. Suthar for helpful discussion and critical reading of the manuscript. S.M.H. is a fellow of the Irvington Institute Fellowship Program of the Cancer Research Institute. Work in the Gale laboratory is supported by funds from the State of Washington, National Institutes of Health (AI060389, AI40035, DA024563, and AI057568), and by the Burroughs-Wellcome Fund.
References
- Abe T. Kaname Y. Hamamoto I. Tsuda Y. Wen X. Taguwa S. Moriishi K. Takeuchi O. Kawai T. Kanto T. Hayashi N. Akira S. Matsuura Y. Hepatitis C virus nonstructural protein 5A modulates the toll-like receptor-MyD88-dependent signaling pathway in macrophage cell lines. J Virol. 2007;81(17):8953–8966. doi: 10.1128/JVI.00649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N. Zayas M. Miller S. Krijnse-Locker J. Schaller T. Friebe P. Kallis S. Engel U. Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4(3):e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y. Kuroki M. Abe K. Dansako H. Ikeda M. Wakita T. Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81(24):13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamming D. Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I and LGP2. J Biol Chem. 2009;284(15):9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril M. Racine ME. Penin F. Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83(3):1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB. Brasky KM. Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75(15):7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB. Guerra B. Brasky KM. Hubbard G. Beard MR. Luxon BA. Lemon SM. Lanford RE. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78(24):13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blindenbacher A. Duong FH. Hunziker L. Stutvoet ST. Wang X. Terracciano L. Moradpour D. Blum HE. Alonzi T. Tripodi M. La Monica N. Heim MH. Expression of hepatitis c virus proteins inhibits interferon alpha signaling in the liver of transgenic mice. Gastroenterology. 2003;124(5):1465–1475. doi: 10.1016/s0016-5085(03)00290-7. [DOI] [PubMed] [Google Scholar]
- Bode JG. Ludwig S. Ehrhardt C. Albrecht U. Erhardt A. Schaper F. Heinrich PC. Haussinger D. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17(3):488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- Bosch FX. Ribes J. Borras J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19(3):271–285. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- Brass V. Berke JM. Montserret R. Blum HE. Penin F. Moradpour D. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc Natl Acad Sci USA. 2008;105(38):14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S. Dolganiuc A. Szabo G. Toll-like receptors 1 and 6 are involved in TLR2-mediated macrophage activation by hepatitis C virus core and NS3 proteins. J Leukoc Biol. 2007;82(3):479–487. doi: 10.1189/jlb.0207128. [DOI] [PubMed] [Google Scholar]
- Chen L. Borozan I. Feld J. Sun J. Tannis LL. Coltescu C. Heathcote J. Edwards AM. McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128(5):1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- Cui S. Eisenacher K. Kirchhofer A. Brzozka K. Lammens A. Lammens K. Fujita T. Conzelmann KK. Krug A. Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29(2):169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Dansako H. Ikeda M. Ariumi Y. Wakita T. Kato N. Double-stranded RNA-induced interferon-beta and inflammatory cytokine production modulated by hepatitis C virus serine proteases derived from patients with hepatic diseases. Arch Virol. 2009;154(5):801–810. doi: 10.1007/s00705-009-0375-z. [DOI] [PubMed] [Google Scholar]
- Dansako H. Ikeda M. Kato N. Limited suppression of the interferon-beta production by hepatitis C virus serine protease in cultured human hepatocytes. FEBS J. 2007;274(16):4161–4176. doi: 10.1111/j.1742-4658.2007.05942.x. [DOI] [PubMed] [Google Scholar]
- de Lucas S. Bartolome J. Carreno V. Hepatitis C virus core protein down-regulates transcription of interferon-induced antiviral genes. J Infect Dis. 2005;191(1):93–99. doi: 10.1086/426509. [DOI] [PubMed] [Google Scholar]
- de Veer MJ. Holko M. Frevel M. Walker E. Der S. Paranjape JM. Silverman RH. Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
- Diebold SS. Kaisho T. Hemmi H. Akira S. Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Dolganiuc A. Oak S. Kodys K. Golenbock DT. Finberg RW. Kurt-Jones E. Szabo G. Hepatitis C core and nonstructural 3 proteins trigger toll-like receptor 2-mediated pathways and inflammatory activation. Gastroenterology. 2004;127(5):1513–1524. doi: 10.1053/j.gastro.2004.08.067. [DOI] [PubMed] [Google Scholar]
- Duong FH. Filipowicz M. Tripodi M. La Monica N. Heim MH. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology. 2004;126(1):263–277. doi: 10.1053/j.gastro.2003.10.076. [DOI] [PubMed] [Google Scholar]
- Egger D. Wolk B. Gosert R. Bianchi L. Blum HE. Moradpour D. Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76(12):5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenacher K. Steinberg C. Reindl W. Krug A. The role of viral nucleic acid recognition in dendritic cells for innate and adaptive antiviral immunity. Immunobiology. 2007;212(9–10):701–714. doi: 10.1016/j.imbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elco CP. Guenther JM. Williams BR. Sen GC. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol. 2005;79(7):3920–3929. doi: 10.1128/JVI.79.7.3920-3929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AK. Seiwert S. Gale M., Jr Antiviral potency analysis and functional comparison of consensus interferon, interferon-alpha2a and pegylated interferon-alpha2b against hepatitis C virus infection. Antivir Ther. 2008;13(7):851–862. [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta C. Arias A. Escarmis C. Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16(1):27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Foy E. Li K. Wang C. Sumpter R., Jr Ikeda M. Lemon SM. Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300(5622):1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Gack MU. Shin YC. Joo CH. Urano T. Liang C. Sun L. Takeuchi O. Akira S. Chen Z. Inoue S. Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gale M., Jr Blakely CM. Kwieciszewski B. Tan SL. Dossett M. Tang NM. Korth MJ. Polyak SJ. Gretch DR. Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18(9):5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M., Jr Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436(7053):939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Gee P. Chua PK. Gevorkyan J. Klumpp K. Najera I. Swinney DC. Deval J. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J Biol Chem. 2008;283(14):9488–9496. doi: 10.1074/jbc.M706777200. [DOI] [PubMed] [Google Scholar]
- Gitlin L. Barchet W. Gilfillan S. Cella M. Beutler B. Flavell RA. Diamond MS. Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103(22):8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R. Egger D. Lohmann V. Bartenschlager R. Blum HE. Bienz K. Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77(9):5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N. Servant MJ. tenOever B. Sen GC. Balachandran S. Barber GN. Lin R. Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76(11):5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT. Bichko VV. Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75(18):8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M. Andersson I. Klingstrom J. Schumann M. Martin A. Zimmermann P. Wagner V. Pichlmair A. Schneider U. Muhlberger E. Mirazimi A. Weber F. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3(4):e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ. Barton DJ. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA. 2002;8(4):512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F. Hemmi H. Hochrein H. Ampenberger F. Kirschning C. Akira S. Lipford G. Wagner H. Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Heim MH. Moradpour D. Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73(10):8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig KJ. Lau DT. Semendric L. Harley HA. Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42(3):702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- Hinson ER. Cresswell P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009;284(7):4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V. Ellegast J. Kim S. Brzozka K. Jung A. Kato H. Poeck H. Akira S. Conzelmann KK. Schlee M. Endres S. Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–947. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Huang Y. Staschke K. De Francesco R. Tan SL. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology. 2007;364(1):1–9. doi: 10.1016/j.virol.2007.01.042. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T. Tahara SM. Lai MM. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72(11):8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D. Guo H. Xu C. Chang J. Gu B. Wang L. Block TM. Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82(4):1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L. Waterman PM. Jonscher KR. Short CM. Reisdorph NA. Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28(16):5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL. Owen DM. Gale M., Jr Functional and therapeutic analysis of hepatitis C virus NS3.4A protease control of antiviral immune defense. J Biol Chem. 2007;282(14):10792–10803. doi: 10.1074/jbc.M610361200. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD. Body-Malapel M. Amer A. Park JH. Whitfield J. Franchi L. Taraporewala ZF. Miller D. Patton JT. Inohara N. Nunez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281(48):36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Mikamo-Satoh E. Hirai R. Kawai T. Matsushita K. Hiiragi A. Dermody TS. Fujita T. Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Sato S. Yoneyama M. Yamamoto M. Matsui K. Uematsu S. Jung A. Kawai T. Ishii KJ. Yamaguchi O. Otsu K. Tsujimura T. Koh CS. Reis e Sousa C. Matsuura Y. Fujita T. Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kittlesen DJ. Chianese-Bullock KA. Yao ZQ. Braciale TJ. Hahn YS. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000;106(10):1239–1249. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A. Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80(24):12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE. Guerra B. Lee H. Chavez D. Brasky KM. Bigger CB. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43(5):961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- Lau DT. Fish PM. Sinha M. Owen DM. Lemon SM. Gale M., Jr Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008;47(3):799–809. doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- Li X. Ranjith-Kumar CT. Brooks MT. Bharmaiah S. Herr AB. Kao C. Li P. The RIG-I like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284(20):13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD. Sun L. Seth RB. Pineda G. Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102(49):17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. Ishida H. Lenz O. Lin TI. Nyanguile O. Simmen K. Pyles RB. Bourne N. Yi M. Li K. Lemon SM. Antiviral suppression vs restoration of RIG-I signaling by hepatitis C protease and polymerase inhibitors. Gastroenterology. 2008;135(5):1710–1718.e2. doi: 10.1053/j.gastro.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Lin W. Kim SS. Yeung E. Kamegaya Y. Blackard JT. Kim KA. Holtzman MJ. Chung RT. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol. 2006;80(18):9226–9235. doi: 10.1128/JVI.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM. Fornek J. Crochet N. Bajwa G. Perwitasari O. Martinez-Sobrido L. Akira S. Gill MA. Garcia-Sastre A. Katze MG. Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM. Owen DM. Li K. Erickson AK. Johnson CL. Fish PM. Carney DS. Wang T. Ishida H. Yoneyama M. Fujita T. Saito T. Lee WM. Hagedorn CH. Lau DT. Weinman SA. Lemon SM. Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103(15):6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K. Dong B. Gale M., Jr Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448(7155):816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N. Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274(22):15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- Melen K. Fagerlund R. Nyqvist M. Keskinen P. Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73(4):536–547. doi: 10.1002/jmv.20123. [DOI] [PubMed] [Google Scholar]
- Meylan E. Curran J. Hofmann K. Moradpour D. Binder M. Bartenschlager R. Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Moore CB. Bergstralh DT. Duncan JA. Lei Y. Morrison TE. Zimmermann AG. Accavitti-Loper MA. Madden VJ. Sun L. Ye Z. Lich JD. Heise MT. Chen Z. Ting JP. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451(7178):573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Myong S. Cui S. Cornish PV. Kirchhofer A. Gack MU. Jung JU. Hopfner KP. Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi H. Osawa T. Ogami K. Ouyang X. Sakaguchi S. Koshiba R. Yanai H. Seko Y. Shitara H. Bishop K. Yonekawa H. Tamura T. Kaisho T. Taya C. Taniguchi T. Honda K. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci USA. 2008;105(51):20446–20451. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann AU. Lam NP. Dahari H. Gretch DR. Wiley TE. Layden TJ. Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282(5386):103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- Oshiumi H. Matsumoto M. Hatakeyama S. Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284(2):807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- Otsuka M. Kato N. Moriyama M. Taniguchi H. Wang Y. Dharel N. Kawabe T. Omata M. Interaction between the HCV NS3 protein and the host TBK1 protein leads to inhibition of cellular antiviral responses. Hepatology. 2005;41(5):1004–1012. doi: 10.1002/hep.20666. [DOI] [PubMed] [Google Scholar]
- Owsianka AM. Patel AH. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology. 1999;257(2):330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- Pawlotsky JM. The nature of interferon-alpha resistance in hepatitis C virus infection. Curr Opin Infect Dis. 2003;16(6):587–592. doi: 10.1097/00001432-200312000-00012. [DOI] [PubMed] [Google Scholar]
- Pedersen IM. Cheng G. Wieland S. Volinia S. Croce CM. Chisari FV. David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli V. Dostert C. Muruve DA. Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19(6):615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Pflugheber J. Fredericksen B. Sumpter R., Jr Wang C. Ware F. Sodora DL. Gale M., Jr Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc Natl Acad Sci USA. 2002;99(7):4650–4655. doi: 10.1073/pnas.062055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A. Schulz O. Tan CP. Naslund TI. Liljestrom P. Weber F. Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pineda JA. Garcia-Garcia JA. Aguilar-Guisado M. Rios-Villegas MJ. Ruiz-Morales J. Rivero A. del Valle J. Luque R. Rodriguez-Bano J. Gonzalez-Serrano M. Camacho A. Macias J. Grilo I. Gomez-Mateos JM. Clinical progression of hepatitis C virus-related chronic liver disease in human immunodeficiency virus-infected patients undergoing highly active antiretroviral therapy. Hepatology. 2007;46(3):622–630. doi: 10.1002/hep.21757. [DOI] [PubMed] [Google Scholar]
- Polyak SJ. Khabar KS. Paschal DM. Ezelle HJ. Duverlie G. Barber GN. Levy DE. Mukaida N. Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol. 2001a;75(13):6095–6106. doi: 10.1128/JVI.75.13.6095-6106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak SJ. Khabar KS. Rezeiq M. Gretch DR. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J Virol. 2001b;75(13):6209–6211. doi: 10.1128/JVI.75.13.6209-6211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtha WE. Chachu KA. Virgin HWT. Diamond MS. Early B-cell activation after West Nile virus infection requires alpha/beta interferon but not antigen receptor signaling. J Virol. 2008;82(22):10964–10974. doi: 10.1128/JVI.01646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G. Chen L. Panis M. Fischer AK. Lindenbach BD. Sun J. Heathcote J. Rice CM. Edwards AM. McGilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131(5):1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- Saito T. Gale M., Jr Principles of intracellular viral recognition. Curr Opin Immunol. 2007;19(1):17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Saito T. Gale M., Jr Regulation of innate immunity against hepatitis C virus infection. Hepatol Res. 2008;38(2):115–122. doi: 10.1111/j.1872-034X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Saito T. Hirai R. Loo YM. Owen D. Johnson CL. Sinha SC. Akira S. Fujita T. Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104(2):582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. Owen DM. Jiang F. Marcotrigiano J. Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M. Oakeley EJ. Duong FH. Christen V. Terracciano L. Filipowicz W. Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci USA. 2008;105(19):7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M. Baran M. Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27(15):2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M. Bowie AG. TLR3 in antiviral immunity: key player or bystander? Trends Immunol. 2005;26(9):462–468. doi: 10.1016/j.it.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Schulz O. Diebold SS. Chen M. Naslund TI. Nolte MA. Alexopoulou L. Azuma YT. Flavell RA. Liljestrom P. Reis e Sousa C. Tolllike receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433(7028):887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- Seki E. Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48(1):322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- Sharma P. Lok A. Viral hepatitis and liver transplantation. Semin Liver Dis. 2006;26(3):285–297. doi: 10.1055/s-2006-947298. [DOI] [PubMed] [Google Scholar]
- Smith MW. Yue ZN. Korth MJ. Do HA. Boix L. Fausto N. Bruix J. Carithers RL., Jr Katze MG. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology. 2003;38(6):1458–1467. doi: 10.1016/j.hep.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Song Y. Friebe P. Tzima E. Junemann C. Bartenschlager R. Niepmann M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80(23):11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano V. Peters MG. Zeuzem S. New therapies for hepatitis C virus infection. Clin Infect Dis. 2009;48(3):313–320. doi: 10.1086/595848. [DOI] [PubMed] [Google Scholar]
- Stetson DB. Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Su AI. Pezacki JP. Wodicka L. Brideau AD. Supekova L. Thimme R. Wieland S. Bukh J. Purcell RH. Schultz PG. Chisari FV. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci USA. 2002;99(24):15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R., Jr Loo YM. Foy E. Li K. Yoneyama M. Fujita T. Lemon SM. Gale M., Jr Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79(5):2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi T. Nagano-Fujii M. Akutsu M. Kadoya H. Ohgimoto S. Ishido S. Hotta H. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol. 2004;85(Pt 4):959–969. doi: 10.1099/vir.0.19513-0. [DOI] [PubMed] [Google Scholar]
- Takahasi K. Yoneyama M. Nishihori T. Hirai R. Kumeta H. Narita R. Gale M., Jr Inagaki F. Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Tang ED. Wang CY. MAVS self-association mediates antiviral innate immune signaling. J Virol. 2009;83(8):3420–3428. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targett-Adams P. Boulant S. McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82(5):2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka M. Sakamoto N. Itakura Y. Nakagawa M. Itsui Y. Sekine-Osajima Y. Nishimura-Sakurai Y. Chen CH. Yoneyama M. Fujita T. Wakita T. Maekawa S. Enomoto N. Watanabe M. Hepatitis C virus non-structural proteins responsible for suppression of the RIG-I/Cardif-induced interferon response. J Gen Virol. 2007;88(Pt 12):3323–3333. doi: 10.1099/vir.0.83056-0. [DOI] [PubMed] [Google Scholar]
- Tattoli I. Carneiro LA. Jehanno M. Magalhaes JG. Shu Y. Philpott DJ. Arnoult D. Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9(3):293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR. Puig M. Darnell ME. Mihalik K. Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79(10):6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR. Shi ST. Romano PR. Barber GN. Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285(5424):107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL. Foss KL. Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4(3):e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D. Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83(9):4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T. Valdes M. Elsby R. Kakuta S. Caceres G. Saijo S. Iwakura Y. Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178(10):6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Vercammen E. Staal J. Beyaert R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev. 2008;21(1):13–25. doi: 10.1128/CMR.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Pflugheber J. Sumpter R., Jr Sodora DL. Hui D. Sen GC. Gale M., Jr Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77(7):3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washenberger CL. Han JQ. Kechris KJ. Jha BK. Silverman RH. Barton DJ. Hepatitis C virus RNA: dinucleotide frequencies and cleavage by RNase L. Virus Res. 2007;130(1–2):85–95. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk B. Buchele B. Moradpour D. Rice CM. A dynamic view of hepatitis C virus replication complexes. J Virol. 2008;82(21):10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk B. Sansonno D. Krausslich HG. Dammacco F. Rice CM. Blum HE. Moradpour D. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74(5):2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. Xiao N. Liu F. Ren H. Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc Natl Acad Sci USA. 2009;106(5):1530–1535. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao ZQ. Eisen-Vandervelde A. Waggoner SN. Cale EM. Hahn YS. Direct binding of hepatitis C virus core to gC1qR on CD4+ and CD8+ T cells leads to impaired activation of Lck and Akt. J Virol. 2004;78(12):6409–6419. doi: 10.1128/JVI.78.12.6409-6419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M. Kikuchi M. Matsumoto K. Imaizumi T. Miyagishi M. Taira K. Foy E. Loo YM. Gale M., Jr Akira S. Yonehara S. Kato A. Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M. Kikuchi M. Natsukawa T. Shinobu N. Imaizumi T. Miyagishi M. Taira K. Akira S. Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- You LR. Chen CM. Yeh TS. Tsai TY. Mai RT. Lin CH. Lee YH. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. 1999;73(4):2841–2853. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S. Rice CM. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J Virol. 2008;82(1):184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B. Yang Y. Li S. Wang YY. Li Y. Diao F. Lei C. He X. Zhang L. Tien P. Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]