Abstract
Activation of chemokine genes in response to interferon (IFN)-γ or NF-κB is an important aspect of inflammation. Using the chemokine gene ip-10 in mouse embryonic fibroblast cells as an example, we show that the response to IFN-γ is long lasting but secondary: initial STAT1 activation drives IRF1 synthesis, and IRF1 then binds to IFN-stimulated regulatory elements (ISREs) in the ip-10 promoter. The promoters of most IKK-β-dependent IFN-stimulated genes (ISGs) also contain ISREs. In response to IFN-γ, inhibitor of NF-κB (IκB) kinase β (IKK-β) is required to activate both newly synthesized IRF1 and the p65 subunit of NF-κB, which contributes to ip-10 expression by binding to κB sites in the ip-10 promoter, with little or no activation of classical NF-κB. In contrast to IFN-γ, IL-1β induces ip-10 expression rapidly but transiently, by activating classical NF-κB and increasing the synthesis of IRF1. Together, IL-1β and IFN-γ induce ip-10 synergistically. IFN-γ does not affect the transient activation of classical NF-κB by IL-1β and synergistic induction of ip-10 expression by IFN-γ and IL-1β occurs even after the activation of classical NF-κB has returned to basal levels. Therefore, IKK-β has a novel role in IFN-γ-dependent activation of chemokine gene expression through its activation of IRF1 and p65.
Introduction
Interferon-γ (IFN-γ) drives the formation of Stat1 homodimers, which in turn activate the transcription of most IFN-stimulated genes (ISGs) directly, by binding to gamma-activated sequence (GAS) elements in their promoters (Stark and others 1998). However, several IFN-γ-stimulated genes are regulated by IFN-stimulated regulatory elements (ISREs) instead of GAS elements. ISREs can be activated either by ISGF3, composed of IRF9 and a Stat1–Stat2 dimer, formed in response to type I IFN-dependent signaling, or by IRF proteins, activated by IFNs or Toll-like receptor (TLR) signaling pathways. Several mechanisms of ISRE activation by IFN-γ have been described, including activation of variants of ISGF3 and the induction of IRF1 (Bluyssen and others 1995; Matsumoto and others 1999; Schroder and others 2004).
Previously, we identified several ISGs that require IKK-β in a novel role for their induction by IFN-γ in both MEFs and RAW 264.7 macrophages. Most of these IKK-β-dependent genes have κB elements as well as ISREs in their promoters instead of GAS elements. Furthermore, many IKK-β-dependent genes are activated synergistically by the combination of IFN-γ-dependent signaling and NF-κB activation. The super-repressor mutant of IκB inhibits the induction by IFN-γ of the IKK-β-dependent ISG ip-10, and p65 is also required for induction of this gene. In the same cells, however, IFN-γ does not activate NF-κB. From this information, we concluded that IKK-β is required for IFN-γ-dependent gene induction, either because it supports a low level of basal NF-κB activation necessary for the induction of some ISGs (Shi and others 2003; Hurgin and others 2007) or because IFN-γ activates a novel IKK-β- and p65-dependent pathway that was not detected by the methods employed previously (Sizemore and others 2004; Shultz and others 2007).
Here, we show that ip-10 is induced by IFN-γ with delayed kinetics due to the need to synthesize the ISRE-binding factor IRF1. Furthermore, the expression and nuclear localization of IRF1 alone are insufficient to stimulate ip-10 transcription; an additional IFN-γ-mediated signal is required. Both IRF1 and, surprisingly, p65 bind to the ip-10 promoter in response to IFN-γ via a mechanism that requires IKK-β. Together, IFN-γ and IL-1β induce ip-10 expression synergistically.
Materials and Methods
Constructs
The promoter region from the original construct was cloned into pGL3. The construct used to put IKK-β back into null cells was described previously (Shultz and others 2007). The retroviral construct expressing IRF1-ER was generated by cloning mouse IRF1 cDNA in frame with the 5′ end of a modified version of the human estrogen receptor gene, within a pBabepuro-based vector.
Biological reagents and cell culture
Recombinant murine IFN-γ and IL-1β from Peprotech, Inc. (Rocky Hill, NJ) were used at 1,000 U/mL and 10 ng/mL, respectively. 4-Hydroxytamoxifen was from Sigma-Aldrich (St. Louis, MO). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum. IKK-β-null cells were a gift from Dr. Michael Karin. IRF1-null MEFs and their wild-type counterparts were a gift from Dr. Tadatsugu Taniguchi.
Retroviral transduction
Bosc packaging cells were transfected with pBabeHygro-IKK-β using Lipofectamine Plus (Invitrogen, Carlsbad, CA). Virus-containing supernatant medium, collected 24 and 48 h later, was passed through a 0.2 μM filter and combined with 5 μg/mL of polybrene. Equal parts of filtered virus and fresh medium were then used to infect cells. Twenty-four hours after the final round of virus treatment, IKK-β-null cells were seeded at 20% confluence and grown in antibiotic-containing medium for selection.
Western analysis
After treatment, cells at 90% confluence in 100-mm dishes were washed twice with phosphate-buffered saline (PBS), scraped into Eppendorf tubes, and lysed for 10 min in a buffer containing 1% Triton X-100, 50 mM Tris HCl, pH 8, 150 mM NaCl, 10 mM sodium fluoride, 5 mM sodium pyrophosphate, 10 mM orthovanadate, 1 mM leupeptin, 10 mM aprotonin, and 1 mM phenylmethanesulfonyl fluoride. Cellular debris was removed by centrifugation at 16,000g for 10 min. Cell extracts were fractionated by electrophoresis in 10% SDS-PAGE gels and transferred to PVDF membranes. Anti-mouse IRF1 (M-20), p65, and anti-histone antibodies were from Santa-Cruz (Santa Cruz, CA).
Electrophoretic mobility shift assays (EMSAs)
Cells at 90% confluence were stimulated and then washed twice with PBS. Nuclear extracts (10 μg), prepared as described previously (Qing and Stark 2004), were incubated for 20 min at room temperature in 20 μL of binding buffer (10 mM Hepes, pH 7.9, 50 mM KCl, 1 mM EDTA, 5% glycerol) with 1 μg of poly(dI–dC) with a labeled ds-DNA oligonucleotide probe. κB, ISRE, and mutant ISRE probes specific for the ip-10 minimal essential promoter were described previously (Ohmori and Hamilton 1993). Protein–DNA complexes were separated in a 6% native polyacrylamide gel in 45 mM Tris, 32 mM boric acid, and 1.25 mM EDTA. The gels were dried and the complexes were visualized by autoradiography.
Northern analysis
Total RNA, isolated by using Trizol (Invitrogen, Carlsbad, CA), was denatured, separated by electrophoresis in a formaldehyde–1% agarose gel, and transferred to Hybond N+ membrane (Amersham Biosciences, Buckinghamshire, UK). mRNA was detected with cDNA probes labeled by using the Megaprime DNA Labeling Kit (Amersham Biosciences, Buckinghamshire, UK).
Quantitative real-time PCR
Total RNA was isolated from cells by using Trizol (Invitrogen, Carlsbad, CA). Two micrograms of RNA was treated in a 20 μL reaction mixture containing two units of DNAse I (Invitrogen) for 15 min at 25°C; 2 μL of 25 mM EDTA was added and the reaction was placed at 65°C for 10 min. cDNA was synthesized from 8 μL of the DNAse-treated RNA reaction mixture (0.75 μg) by using the random priming procedure with the SuperScript II First Strand Synthesis Kit (Invitrogen). The final cDNA reaction mixture (20 μL) was diluted 10-fold and 5 μL was used as a template for PCR. Primer sequences were developed using the PerlPrimer program. The forward primer, gagagtcaggagaaagggcga, anneals within the first intron of ip-10. The reverse primer, gcaacttgtcagttacgaaatcct, anneals within the first exon. Amplicons were designed to be 100 to 200 base pairs long and to overlap a splice site. Primers were then tested in serial dilutions of cDNA to assure 90%–100% efficiency.
Real-time PCR was performed by using 500 nM of primers, 5 μL of diluted cDNA sample, and 10 μL of iQ SYBER Green Supermix (BioRad, Hercules, CA). All samples were amplified in triplicate and normalized against gapdh. Amplification was performed by using a BioRad iCycler IQ real-time PCR instrument. Parameters for amplification were: 95°C for 3 min, followed by 40 cycles at 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s. The initial amplicon for each primer was assayed by melt curve analysis and agarose gel electrophoresis and all subsequent amplifications with that primer set were assayed by melt curve analysis after 40 cycles.
Chromatin immunoprecipitation (ChiP) assays
These assays were performed as described (Sakamoto and others 2004). In brief, MEFs were untreated or treated as described, fixed in 1% formaldehyde, resuspended in SDS-containing lysis buffer, and sonicated. Soluble chromatin was collected by centrifugation, precleared with protein G-agarose, and immunoprecipitated with anti-IRF1 or anti-p65 or an isotype control antibody overnight before extensive washing, elution, reversal of cross-linking, and digestion with RNAse A and proteinase K. The DNA was then amplified by PCR, using primers specific to the ip-10 promoter. The forward primer was caaggcactcatctgatttctcaaacagct. The reverse primer was ggagggaaactctttgcagata. The PCR program used was 94°C × 5′ (94°C × 1′, 55° × 1′, 72° × 2′) × 30 cycles, 72° × 5′.
Results
IFN-γ stimulates p65 binding to the ip-10 promoter in a process that requires IKK-β
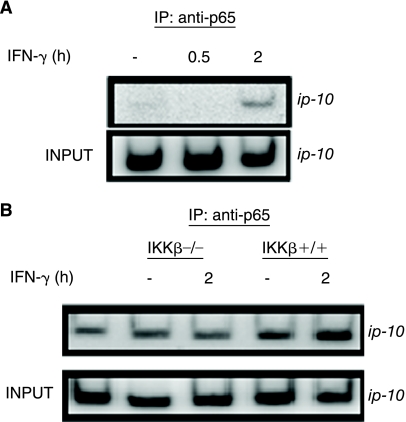
Previously we showed that the induction of ip-10 by IFN-γ requires p65 expression, although NF-κB did not appear to be activated by IFN-γ. To detect binding of p65 to the ip-10 promoter, we treated wild-type MEFs with IFN-γ, followed by analysis by ChIP. IFN-γ promotes the binding of p65 to the ip-10 promoter after 2 h (Fig. 1A). The process requires IKK-β, since p65 does not bind to this promoter in response to IFN-γ in IKK-β-null cells (Fig. 1B).
FIG. 1.
Interferon (IFN)-γ stimulates p65 binding to the ip-10 promoter in an IKK-β-dependent fashion. (A) Wild-type MEFs or (B) IKK-β-null MEFs or a pool of the same cells in which IKK-β was stably restored were treated with 1,000 IU/mL of murine IFN-γ. Cross-linked chromatin fragments were immunoprecipitated with anti-p65. The recovered DNA was amplified by PCR with 32P-labeled α-dCTP, using primers specific for the promoter region of ip-10. The products were separated in an 8% polyacrylamide gel, dried, and analyzed by autoradiography. An example representative of three experiments is shown. The lane at the far left of B represents a sample immunoprecipitated with an isotype-matched control antibody.
The expression of ip-10 is induced by IFN-γ with kinetics that match the binding of a complex to the ip-10 ISRE
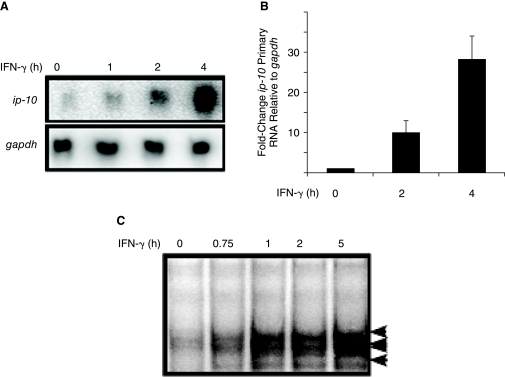
Wild-type MEFs were treated for various times with IFN-γ and ip-10 mRNA expression was measured (Fig. 2A). Robust induction did not occur until 2–4 h after IFN-γ stimulation. The pattern of ip-10 induction by IFN-γ suggests that a secondary signal is required. We tried to use cycloheximide to evaluate the requirement for new protein synthesis (data not shown) but, unfortunately, cycloheximide increases both basal and IFN-γ-induced levels of ip-10 (Ohmori and Hamilton 1995), making it difficult to obtain this information. Next, to determine if the increase in ip-10 expression between 2 and 4 h was due to an increased rate of transcription, we measured the amounts of unspliced transcript (Levy and others 1986; Chang and others 2004; Ramsauer and others 2007) induced by IFN-γ at different times (Fig. 2B). For this assay, primers were designed to amplify a region of RNA spanning an intron–exon junction, thus specifically measuring unspliced RNA. The rate of transcription increased between 2 and 4 h, consistent with a requirement for intermediate protein synthesis. Control experiments confirmed that contaminating genomic DNA or mature mRNA transcripts did not contribute to the signal observed (data not shown). We observed previously that most IKK-β-dependent ISGs, including ip-10, contain ISREs in their promoters instead of GAS elements, and that activation of the ip-10 promoter by IFN-γ requires the binding of proteins to the ISRE (Ohmori and Hamilton 1993). Therefore, we analyzed the kinetics of protein binding to the ip-10 promoter by electrophoretic mobility shift assay (EMSA) (Fig. 2C). Several complexes bound to the ISRE probe with intensities that increased similarly to the increase in gene induction. From this result, we conclude that the factor(s) binding to this ISRE are likely to be required for gene induction.
FIG. 2.
Ip-10 is transcribed with delayed kinetics due to synthesis of an interferon (IFN)-stimulated regulatory element (ISRE)-binding factor. (A) Wild-type MEFs were treated with 1,000 IU/mL of murine IFN-γ for various times and ip-10 and gapdh mRNAs were measured by the Northern method. (B) The rate of ip-10 transcription in wild-type MEFs in response to IFN-γ measured by qPCR, using primers that specifically amplify primary ip-10 RNA transcripts. (C) Electrophoretic mobility shift assay (EMSA) analysis of nuclear lysates from IFN-γ-treated wild-type MEFs, using an oligonucleotide probe representing the ISRE from the ip-10 promoter.
IRF1 binds to the ip-10 ISRE
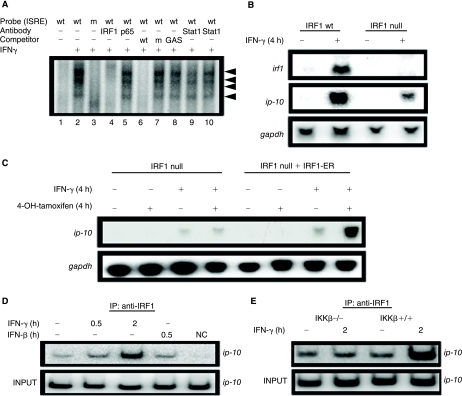
IRF1 is an ISRE-binding protein whose synthesis is induced in response to IFN-γ (Taniguchi and others 2001). We tested its binding to the ISRE in the ip-10 promoter by supershift analysis (Fig. 3A). Anti-IRF1 supershifted the ISRE-binding complex formed in response to a 4-h treatment with IFN-γ (lane 4). Anti-p65 (an isotype control), on the other hand, did not (lane 5), as expected since p65 binds to κB elements, which were not present in the probe. Furthermore, a version of the ISRE probe carrying mutations in the core ISRE-binding domain (GGAAAGTGAAACC→ GGACAGTGACACC) did not bind at all (lane 3). In the same gel, we also ran samples from cells treated with IFN-γ for 4 h and then preincubated with a 50-fold excess of unlabeled wild-type ISRE probe, mutant ISRE probe, or GAS probe (lanes 6, 7, and 8, respectively). Only the unlabeled wild-type ISRE probe competed successfully, showing that the complex is specific for that sequence. Finally, two different antibodies that shift Stat1 bound to a GAS probe (data not shown) did not shift the ISRE-bound complex (lanes 9 and 10).
FIG. 3.
IRF1 recruitment to the ip-10 promoter by interferon (IFN)-γ requires IKK-β. (A) Wild-type MEFs were untreated (lane 1) or treated for 4 h with 1,000 IU/mL of murine IFN-γ (lanes 2–10) and nuclear lysates were analyzed by electrophoretic mobility shift assay (EMSA) using the IFN-stimulated regulatory element (ISRE) from the ip-10 promoter. A mutant version of the ISRE probe did not produce a band shift (lane 3). In addition, adding anti-IRF1 (lane 4) supershifted the ISRE-bound complex, while anti-p65 did not (lane 5), nor did 2 different antibodies to Stat1 (lanes 9 and 10). Finally, competition with a 50-fold excess of unlabeled wild-type ISRE probe competed the labeled probe while a 50-fold excess of mutant ISRE and wild-type GAS probe did not (lanes 6, 7, and 8, respectively). (B) IRF1-null MEFs and wild-type MEFs derived from littermates were treated for 4 h with IFN-γ and RNA was analyzed by the Northern method for the expression of ip-10, irf1, and gapdh mRNAs. (C) IRF1-null cells or the same cells in which IRF1 expression was stably restored with a retrovirus expressing an estrogen receptor-IRF1 fusion protein were treated with 1 μM tamoxifen, IFN-γ, or both, and ip-10 and gapdh mRNA expression were measured by the Northern method. (D) Wild-type MEFs or (E) IKK-β-null cells or a pool of the same cells in which IKK-β expression was stably restored were treated with either IFN-γ or IL-1β and IRF1 binding to the ip-10 promoter was analyzed by ChIP. The lane at the far right of (D) represents a control in which the lysate was immunoprecipitated with an isotype control antibody.
These results led us to investigate whether IRF1-null MEFs induce ip-10 in response to IFN-γ. As shown in Figure 3B, these cells expressed much less ip-10 in response to IFN-γ after 4 h compared to match wild-type MEFs. The fact that a little IP-10 protein is still induced in these IRF1-null cells suggests partial compensation, perhaps through different IRFs. To determine if IRF1 expression could restore ip-10 induction to wild-type levels, we created an expression construct encoding a variant of the estrogen receptor (ER) fused to IRF1. The rationale is that constitutively expressed IRF1 translocates to the nucleus, causing cellular senescence. On the other hand, the nuclear translocation of IRF1-ER is tightly regulated and, in the absence of estrogen (or an estrogen mimic), it remains in the cytoplasm (Kroger and others 2003). IRF1-null cells expressed very little ip-10 mRNA in response to IFN-γ, either alone or in combination with tamoxifen (Fig. 3C). Interestingly, when IRF1-null cells stably expressing the IRF1-ER protein were treated with tamoxifen alone (which causes IRF1 to translocate to the nucleus), ip-10 was still not expressed. In response to IFN-γ alone, those cells express similar amounts of ip-10 mRNA as do IRF1-null cells. Therefore, IRF1 expression and nuclear translocation alone are insufficient to drive ip-10 induction and an additional IFN-γ-dependent signal is required. When the restored cells were treated with both IFN-γ and tamoxifen, ip-10 mRNA was fully expressed. By ChIP analysis (Fig. 3D), IRF1 does indeed bind to the ip-10 promoter in response to treatment with IFN-γ for 2 h, but not in response to either IFN-γ or IL-1β for 0.5 h. IKK proteins activate IRF proteins in several different signaling pathways (Sharma and others 2003; McWhirter and others 2004; Hoshino and others 2006); therefore, we tested the requirement for IKK-β for IRF1 activation in response to IFN-γ. IKK-β-null MEFs and the same cells in which IKK-β expression was restored stably, in pools, to approximate physiological levels by using a retroviral expression vector (Shultz and others 2007) were treated for 2 h with IFN-γ and analyzed by ChIP (Fig. 3E). IRF1 in IKK-β-restored cells, but not IKK-β-null cells, bound to the ip-10 promoter in response to IFN-γ.
IL-1β and IFN-γ induce ip-10 synergistically, but not through classical NF-κB activation
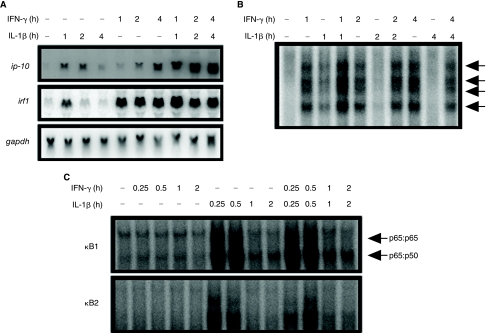
Many IKK-β-dependent ISGs, including ip-10, are induced synergistically by the combination of IFN-γ- and NF-κB-dependent signals. For induction of ip-10 by IFN-γ and TNF-α, synergy is due to increased transcription (Ohmori and Hamilton 1995). To investigate how the IFN-γ- and NF-κB-dependent signals synergize, we analyzed ip-10 induction by IL-1β alone or in combination with IFN-γ. In contrast to IFN-γ, IL-1β induced ip-10 mRNA most strongly after 1 or 2 h, followed by a decrease between 2 and 4 h (Fig. 4A). Together, IL-1β and IFN-γ were strongly synergistic and induced ip-10 with kinetics similar to those seen with IFN-γ alone, but with increased expression. Likewise, irf1 mRNA is induced transiently by IL-1β, while induction by IFN-γ shows sustained transcription, beginning at 1 h. In combination, IFN-γ and IL-1β induce increased amounts of irf1 mRNA at 1 h, but thereafter the amount of irf1 mRNA induced is equivalent to amounts induced by IFN-γ alone. Next, EMSA was used to analyze the pattern of IRF1 binding in response to IFN-γ, IL-1β, or the two in combination (Fig. 4B). As shown previously, IFN-γ alone induces sustained binding over a 4-h time course. IL-1β, on the other hand, induced binding at 1 h but not at later time points. The combination of IL-1β and IFN-γ together enhanced binding at 1 h compared to either stimulus alone but otherwise appeared similar to IFN-γ alone. To determine whether IL-1β-induced activation of NF-κB is affected by concomitant IFN-γ-dependent signaling, we performed EMSAs, using probes representing each of the two κB elements from the ip-10 promoter (Ohmori and Hamilton 1995), confirming that activated NF-κB was not found in nuclear lysates from cells treated with IFN-γ over a 4-h time course (Fig. 4C). Binding of p65 or p50 to either of the ip-10 κB elements in response to IL-1β alone was strong after 15 and 30 min but was gone by 1 h. In combination with IFN-γ, the kinetics of NF-κB binding to either element in response to IL-1β was unchanged in comparison to IL-1β alone. This result indicates strongly that the additional component required for IFN-γ-induced ip-10 transcription shown in Figure 3C, as well as the signal provided by IL-1β for synergy with IFN-γ, is not classical NF-κB.
FIG. 4.
Interferon (IFN)-γ and interleukin (IL)-1β induce ip-10 synergistically. (A) Wild-type MEFs were treated with murine IL-1β (10 mg/mL) or IFN-γ (1,000 IU/mL) and RNA was analyzed by the Northern method. (B) Electrophoretic mobility shift assay (EMSA) was used to measure binding to the IFN-stimulated regulatory elements (ISRE) of ip-10 in response to treatment with IL-1β, IFN-γ, or both. (C) EMSA was used to measure binding to the two κB elements of ip-10 in response to IL-1β, IFN-γ, or both. By supershift analysis, the lower band was found to be a heterodimer of p65 and p50 and the upper band a p65 homodimer (data not shown).
Discussion
IFN-γ binds to its receptor, followed by the sequential activation of Jaks 2 and 1, which in turn phosphorylate the cytoplasmic tail of the receptor at conserved tyrosine residues. One site in particular is essential for docking Stat1 (Qing and others 2005). Following binding, Stat1 is itself phosphorylated by the Jaks, causing it to homodimerize and translocate to the nucleus, where it binds to GAS elements in the promoters of ISGs. By this mechanism, IFN-γ induces sustained IRF1 expression, which participates in the transcription of a subset of ISGs, represented here by ip-10, by binding to their ISREs. It is clear from the work shown here that other IFN-γ signals are required in order for ip-10 to be induced. We have shown previously that irf1 expression does not require IKK-β. Therefore, at least one other IKK-β-dependent signal is required, and we show here that the binding of IRF1 to the ip-10 promoter requires IKK-β. We have also shown previously that p65 expression is required for ip-10 expression (Shultz and others 2007) and we now show that p65 binds to the ip-10 promoter in response to IFN-γ, and that this response also requires IKK-β.
IL-1β induces the expression of irf1 by activating classical NF-κB. Activation of the IKK complex leads to the phosphorylation of IκBα, followed by its proteasome-mediated destruction, liberating NF-κB. Activation of NF-κB by IL-1β is relatively short-lived and, therefore, so is the IL-1β-induced transcription of the irf1 gene. IRF1 synthesized in response to IL-1β can bind to the ip-10 ISRE but, since the IRF1 protein and mRNA have half-lives of only 30 min (Taniguchi and others 2001), that aspect of IL-1β signaling is transient, like the activation of NF-κB.
At least two possibilities might explain why IKK-β is required for the induction of ip-10 expression in response to IFN-γ. One is that IKK-β enhances the transactivating function of IRF1 through direct phosphorylating, affecting its ability to bind to DNA. Another is that IKK-β is required for the phosphorylation of p65, which then interacts with IRF1 directly or indirectly to affect its binding and, in turn, ip-10 transcription. EMSA experiments here reveal multiple IRF1-containing DNA-binding elements in response to IFN-γ, although none appear to be shifted using p65 antibody.
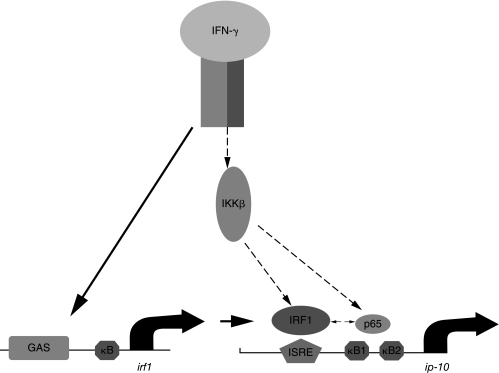
An IKK-β-dependent, p65-containing complex that might cooperate with IRF1 is the classical NF-κB p65:50 heterodimer. A comparison of the binding of IRF1 to the ip-10 ISRE in response to IFN-γ alone or in combination with IL-1β was helpful in testing this hypothesis. Whereas IFN-γ alone induced strong steady IRF1 binding, IL-1β and IFN-γ together induced stronger binding of IRF1 than seen with IFN-γ alone, but only for the first hour of treatment. The IL-1β-induced activation of NF-κB is also short-lived, as shown not only by EMSA analysis of the binding of p65 and p50 to κB elements but also by the kinetics of transcription of the target gene irf1. IFN-γ alone does not activate NF-κB at all. On the other hand, synergism between IFN-γ and IL-1β for ip-10 induction occurs for at least 4 h, long after NF-κB activity has extinguished. Together, these results suggest that a p65-containing component different from classical NF-κB increases IFN-γ-, or IFN-γ plus IL-1β-induced ip-10 expression. IRF1 and p65 bind to the ip-10 promoter in response to IFN-γ and may interact with one another or act separately. Both scenarios are represented in Figure 5. Although we would have liked to determine, by modifying the promoter, which elements might be required for such interactions (ie, the ISRE versus the κB elements), the ip-10 promoter, despite repeated attempts, was not amenable to this method of investigation.
FIG. 5.
A model of ip-10 induction by interferon (IFN)-γ. Solid lines represent known pathways; for example, IRF1 induction by IFN-γ (requiring Stat1) and subsequent IRF1 binding to the IFN-stimulated regulatory element (ISRE) of ip-10. Dashed lines represent pathways revealed by the present work. The pathway connecting the IFN-γ receptor to IKK-β is unknown, as are the pathways connecting IKK-β to IRF1 and p65. IRF1 and p65 may interact with one another when bound to the ip-10 promoter.
There is ample evidence that components of IFN- and NF-κB-dependent signaling cross-interact to promote or repress gene expression by the other pathway. Treatment of human fibrosarcoma cells with IFN-β promotes p65 phosphorylation without activating NF-κB (Rani and others 2001). PIAS1, an inhibitor of both IFN- and NF-κB-dependent signaling, is phosphorylated by IKKα in response to inflammatory cytokines, allowing it to block the binding of both NF-κB and Stat1 to promoters (Liu and others 2007). Two other proteins in the IKK family, IKKɛ and TBK1, phosphorylate IRF3 and IRF7 in response to TLR-dependent signaling (Honda and Taniguchi 2006) and IKKα phosphorylates IRF7 (Hoshino and others 2006). Finally, IRF3 has been shown to bind to and affect the transactivating properties of p65, and vice versa (Leung and others 2004). Logical next steps are to explore the possibility of a similar relationship between IRF1 and p65 and to delineate the mechanism by which IFN-γ activates both proteins, especially whether p65 and IRF1 are phosphorylated by IKK-β in response to IFN-γ.
Acknowledgments
This work was supported by NIH grant PO1 CA 062220 to G.R.S. D.B.S. was supported in part by NIH grant T32 GM07250 to the Case Medical Scientist Training Program. The authors are grateful for technical assistance by Jennifer Shrock.
References
- Bluyssen H. Muzaffar R. Vlieststra R. van der Made A. Leung S. Stark GR. Kerr IM. Trapman J. Levy DE. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc Natl Acad Sci USA. 1995;92(12):5645–5649. doi: 10.1073/pnas.92.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H-M. Paulson M. Holko M. Rice CM. Williams BRG. Marie I. Levy DE. Induction of interferon-stimulated gene expression ant antiviral responses require protein deacetylase activity. Proc Natl Acad Sci USA. 2004;101(26):9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K. Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Hoshino K. Sugiyama T. Matsumoto M. Tanaka T. Saito M. Hemmi H. Ohara O. Akira S. Kaisho T. IkB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- Hurgin V. Novick D. Werman A. Dinarello CA. Rubinstein M. Antiviral and immunoregulatory activities of IFN-gamma depend upon constitutively expressed IL-1alpha. Proc Natl Acad Sci USA. 2007;104(12):5044–5049. doi: 10.1073/pnas.0611608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger A. Dallugge A. Kirchoff S. Hauser H. IRF-1 reverts the transformed phenotype of oncogenically transformed cells in vitro and in vivo. Oncogene. 2003;22:1045–1056. doi: 10.1038/sj.onc.1206260. [DOI] [PubMed] [Google Scholar]
- Leung TH. Hoffmann A. Baltimore D. One nucleotide in a kB site can determine cofactor specificity for NFkB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Levy D. Larner A. Chaudhuri A. Babiss L. Darnell JE. Proc Natl Acad Sci USA. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. Yang Y. Chernishof V. Loo R. Jang H. Tahk S. Yang R. Mink S. Shultz D. Bellone C. Loo J. Shuai K. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129(5):903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- Matsumoto M. Tanaka N. Harada H. Kimura T. Yokochi T. Kitagawa M. Schindler C. Taniguchi T. Activation of Transcription factor ISGF3 by IFN-gamma. Biol Chem. 1999;380:699–603. doi: 10.1515/BC.1999.087. [DOI] [PubMed] [Google Scholar]
- McWhirter S. Fitzgerald K. Rosains J. Rowe D. Golenbock D. Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA. 2004;101(1):233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori Y. Hamilton T. Cooperative Interaction between ISRE and kB motifs controls IFN-gamma and LPS-stimulated transcription from the murine IP-10 promoter. J Biol Chem. 1993;268(9):6677–6688. [PubMed] [Google Scholar]
- Ohmori Y. Hamilton TA. The interferon-stimulated response element and kappa B site mediate synergisitic induction of murine IP-10 gene transcription by IFN-gamma and TNF-alpha. J Immunol. 1995;154(10):5235–5244. [PubMed] [Google Scholar]
- Qing Y. Costa-Pereira A. Watling D. Stark GR. Role of tyrosine 41 of INFGR1 in Socs-1-mediated attenuation of Stat1 activation. J Biol Chem. 2005;280:1849–1841. doi: 10.1074/jbc.M409863200. [DOI] [PubMed] [Google Scholar]
- Qing Y. Stark GR. Alternative Activation of Stat1 and Stat3 in response to Interferon-gamma. J Biol Chem. 2004;279(40):41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- Ramsauer K. Farlik M. Zupkovitz G. Seiser C. Kroger A. Hauser H. Decker T. Distinct models of action applied by transcription factors Stat1 and IRF1 to initiate transcription of the IFN-gamma inducible gpb2 gene. Proc Natl Acad Sci USA. 2007;104(8):2849–2854. doi: 10.1073/pnas.0610944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani MR. Asthagiri AR. Singh A. Sizemore N. Sathe SS. Li X. Donato JD. Stark GR. Ransohoff RM. A role for NF-kappa B in the induction of beta-R1 by interferon-beta. J Biol Chem. 2001;276(48):44365–44368. doi: 10.1074/jbc.C100417200. [DOI] [PubMed] [Google Scholar]
- Sakamoto S. Potla R. Larner A. Histone deacetylase activity is required to recruit RNA polymerase II to promoters of selected interferon-simulated early response genes. J Biol Chem. 2004;279:40362–40367. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- Schroder K. Hertzog PJ. Ravasi T. Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Sharma S. Tenoever B. Grandvaux N. Zhou G. Lin R. Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;16(5622):1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Shi S. Nathan C. Schappinger D. Drenkow D. Fuortes M. Block E. Ding A. Gingeras T. Schoolnik G. Akira S. Takeda K. Ehrt S. Myd88 primes macrophages for full scale activation by IFN-gamma yet mediates few responses to Mtb. J Exp Med. 2003;198(7):987–997. doi: 10.1084/jem.20030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz DB. Fuller JD. Yang Y. Sizemore N. Rani MRS. Stark GR. Activation of a subset of genes by IFN-gamma requires IKKbeta but not IFN-dependent activation of NFkB. J Interferon Res. 2007;27(10):875–884. doi: 10.1089/jir.2007.0031. [DOI] [PubMed] [Google Scholar]
- Sizemore N. Agarwal A. Das K. Lerner N. Sulak M. Rani S. Ransohoff R. Shultz D. Stark G. Inhibitor of kB kinase is required to activate a subset of interferon-gamma-stimulated genes. Proc Natl Acad Sci USA. 2004;101(21):7094–7098. doi: 10.1073/pnas.0401593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR. Kerr IM. Williams BRG. Silverman RH. Shreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Taniguchi T. Ogasawara K. Takaoka A. Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]