Abstract
Flow cytometry has emerged as a powerful tool for quantitative, single-cell analysis of both surface markers and intracellular antigens, including phosphoproteins and kinase signaling cascades, with the flexibility to process hundreds of samples in multiwell plate format. Quantitative flow cytometric analysis is being applied in many areas of biology, from the study of immunology in animal models or human patients to high-content drug screening of pharmacologically active compounds. However, these experiments generate thousands of data points per sample, each with multiple measured parameters, leading to data management and analysis challenges. We developed WebFlow (http://webflow.stanford.edu), a web server-based software package to manage, analyze, and visualize data from flow cytometry experiments. WebFlow is accessible via standard web browsers and does not require users to install software on their personal computers. The software enables plate-based annotation of large data sets, which provides the basis for exploratory data analysis tools and rapid visualization of multiple different parameters. These tools include custom user-defined statistics to normalize data to other wells or other channels, as well as interactive, user-selectable heat maps for viewing the underlying single-cell data. The web-based approach of WebFlow allows for sharing of data with collaborators or the general public. WebFlow provides a novel platform for quantitative analysis of flow cytometric data from high-throughput drug screening or disease profiling experiments.
Introduction
From its inception, flow cytometry has provided a means of assaying each of millions of individual cells within a sample. By measuring multiple fluorescence parameters, flow cytometric analysis yields an n-dimensional distribution of points that cannot be effectively represented in a single statistic. Recent developments in flow cytometry machinery, antibodies, and fluorophores have increased the number of parameters available for analysis while simultaneously simplifying the experimental process, allowing more researchers to perform complex multidimensional experiments.1–4 In addition, flow cytometers can now be used to measure intracellular signaling cascades and phosphorylation events and are employed extensively in high-throughput drug screening.5–10 Moreover, primary cell populations, such as human clinical samples or murine splenocytes, are routinely analyzed with flow cytometry in studies of basic immunology and human diseases.11–16
In many instances, these new applications of the technology rely on quantitative flow cytometric analysis of surface or intracellular markers, rather than traditional qualitative analyses, e.g., a “positive” or “negative” score for a cell lineage marker.17,18 Indeed, analyses of signaling cascades, drug screening, and clinical sample monitoring increasingly require quantitative analysis tools to distinguish controls from treated or diseased samples.
The need for quantitative analysis, coupled with a large number of samples per experiment, presents a considerable challenge for current data analysis tools and is a major bottleneck in application of flow cytometry to high-throughput systems.7 In our experience with large data sets, experimental setup and data acquisition are rapid compared to the process of managing, annotating, analyzing, and displaying the results of the experiment. Many of the existing software packages have separated the steps of flow cytometry file analysis, sample annotation, and statistics generation, making them more suited to experiments with fewer samples. In our laboratory, we typically must use several different programs to yield “figure-ready” flow cytometry data representations, including flow cytometry programs, spreadsheet software, and heat map generating programs.
Thus, there is need for an approach to flow cytometry analysis designed specifically for analysis of large data sets. In particular, the software must be able to read and analyze flow cytometry files, calculate statistics from those files, and generate data visualizations that enable the researcher to identify experimental trends and analyze results. Optimally, the underlying flow cytometry data files (FCS files) would be accessible at any stage of the analysis, allowing the user to perform adjustments such as editing the gating for one sample, with the software regenerating a heat map without requiring the user to bring the newly analyzed data through multiple programs in order to interpret it. Such software would enable handling of experiments with hundreds of samples, reducing current limitations at the data analysis stage.
To address these needs, we have developed a web server-based flow cytometry analysis software package called WebFlow (http://webflow.stanford.edu). WebFlow is able to handle large data sets, specifically those from the multiwell plate experiments commonly used in drug screening and high-throughput flow cytometry analysis. During analysis, the user assigns the flow cytometry data files to their cognate positions on a representational “plate” (e.g., 96-well) and then directs the software to calculate normalization statistics that are either provided inherently in the program or specified by user-defined formulae. Displaying the statistical output as a heat map allows users to scan the data to validate that samples were stained uniformly, find hits in a drug screen, and/or identify trends in disease profiles or cell types. Importantly, the results of a plate-based experiment can be analyzed rapidly, with the entire process from data upload to generation of a heat map taking only 15–30 min for a 96-well plate. In addition to displaying statistics, all heat maps are clickable, allowing the user to see the underlying flow cytometry data that were used to calculate the statistic; this paradigm, similar to the exploratory data analysis of microarrays,19 provides simultaneous access to “big picture” trends and the detailed biological data. Thus, by allowing for direct switching between the underlying flow cytometry data and statistical readouts, WebFlow attempts to reduce errors and facilitate analysis of quantitative multiparameter flow cytometry data.
Another critical feature of the WebFlow approach is that it is designed to operate over the internet in a distributed data environment; users access a central server from their own web browsers, and the data can reside on that server or at distant secure sites. By employing a web-based interface, users are not required to have an advanced analysis machine at their desktop because all computationally intensive data analysis is done on a server optimized for this purpose. In addition, such a model provides computer- and researcher-independent access to the data and analysis; because all of the data and analyses are centralized, they can be viewed or edited by anyone with the proper permissions from any computer in the world.
The overall analysis paradigm we have employed in WebFlow serves as a template for how high-throughput analyses can be accomplished with flow cytometry. This approach will enable the application of flow cytometry to systems biology and other proteomics initiatives by reducing the bottlenecks of data management, analysis, sharing, and presentation, such that focus can be returned to experimental design and data acquisition.
Materials and Methods
Software Design
WebFlow was written as a Java™ (Sun Microsystems, Inc., Santa Clara, CA) web application, employing JavaServer Pages as well as independent server-side Java (servlets) and client-side Java (applets). WebFlow is designed to be compatible with Java version 1.5. Our production platform was the Tomcat JavaServer Page server (version 5.5) connected to an Apache web server (version 2) via JK (version 1.2.15) (all from Apache Software Foundation, Forest Hill, MD); the server ran on Windows XP SP2 (Microsoft Corp., Redmond, WA) (3 GHz Pentium 4; 2 GB RAM). Test clients were Internet Explorer version 7.x (Microsoft Corp.) or Firefox (Mozilla Corp., Mountain View, CA) browsers running on Windows XP or Mac OS version 10.4 (Apple Computer, Cupertino, CA), with the Java Runtime Environment (JRE v1.41, Sun Microsystems) installed.
All source code, as well as the complete compiled application, is made available to the academic community (under the GNU GPL license) and can be accessed on our laboratory website (http://webflow.stanford.edu). As a basic overview of the internals, data are stored in the original FCS files, while all analysis is stored in XML files as serialized Java objects. Flow cytometry plots are generated by a servlet that sends images to an applet on the client's browser; this applet provides the interactivity of changing views and designing gates. Experiment annotation is done within an applet running on the client's browser. All other analysis is done using JavaServer Pages to allow for web-based interactivity on the client side with a Java backend for data processing. Mathematical expression parsing for the custom statistics was done with the JEP package (Singular Systems, Edmonton, AB, Canada; http://www.singularsys.com/jep).
Sample Experiments
Inhibitor dose-response
U937 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. Approximately 500,000 cells were aliquoted to each well of a V-bottom 96-well plate. Janus kinase (Jak) inhibitor I (pan-Jak inhibitor from Calbiochem [La Jolla, CA]) was added in a 10-point dose–response curve from 0.25 to 5,000 nM final concentration (0.5% dimethyl sulfoxide [final concentration] added to all wells) across rows C and F. Cells were incubated for 30 min, followed by addition of interferon-γ (IFN-γ) and granulocyte-macrophage colony-stimulating factor (10 ng/ml each) for 15 min. Cells were fixed for 10 min with 1.6% formaldehyde (Electron Microscopy Sciences, Hatfield, PA), pelleted, and resuspended in ice-cold methanol. After 30 min, cells were washed twice with staining medium (phosphate-buffered saline, 0.5% bovine serum albumin, and 0.02% sodium azide) and then stained with phosphospecific monoclonal antibodies against signal transducer and activator of transcription (Stat) 1 (pY701, clone 4a) labeled with Alexa 488 and Stat5 (pY694, clone 47) labeled with Alexa 647 (both antibodies from BD Biosciences [San Jose, CA]). After 1 h, cells were washed and acquired on a BD LSRII flow cytometer (BD Biosciences) with HTS plate module and running Diva software. The cytometer was equipped with 405 nm, 488 nm, and 633 nm lasers. Data were exported as FCS version 3.0 files and uploaded directly into WebFlow for analysis.
Surface marker analysis
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats using Ficoll-Paque density gradient centrifugation. Cells were washed with staining medium, added to a V-bottom 96-well plate, and stained with antibodies against CD3 phycoerythrin (PE) (clone UCHT-1), CD8 PE-Cy7 (clone RPA-T8), and CD4 APC (clone RPA-T4) (antibodies from BD Biosciences). CD8 antibody was not added to column 8 of the plate. After washing, cells were acquired and analyzed as above.
Results
Data Management
Users are provided accounts on the server that correspond to a directory for their data. After login at the WebFlow site (example version at http://webflow.stanford.edu), the user is prompted to upload a new experiment or choose an existing experiment. For each experiment, the user can perform analysis (see below), duplicate the experiment to perform multiple different sets of analyses, and set permissions for other users to view or edit the analysis.
Overall Experiment Workflow
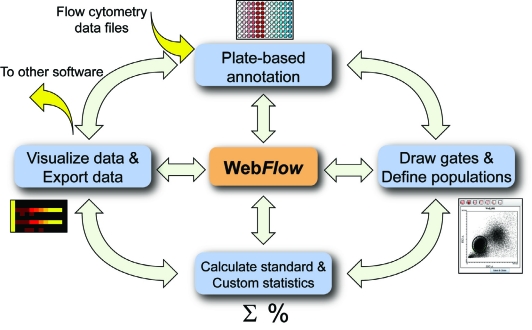
Once an experiment has been uploaded, WebFlow provides a list of analysis options, ordered corresponding to the suggested program flow (Fig. 1). The experiment must first be annotated, which involves placing the data files in “plates” and then adding optional keywords to describe the files. Most users of WebFlow perform their experiments in multiwell plates, reflecting the large-scale nature of an experiment; however, even experiments performed in tubes can be analyzed in WebFlow, with rows and columns serving to organize sample types, experimental conditions, and other variables from any experimental format (e.g., patients in rows and antibody staining sets in columns). After annotation, the user can then set up compensation if necessary—data will then be compensated appropriately using this matrix throughout the rest of the analysis. Next, the user views plots of the data files and draws gates to define the different cell types present in the sample. Once cell types are defined, WebFlow calculates standard statistics (mean, median, coefficient of variation [CV], percentage, and count) for each population on all fluorescence parameters.
FIG. 1.
Data analysis in WebFlow. WebFlow is designed with a stepwise interface that begins with data annotation and ends with exploratory data analysis via heat maps or data export as annotated lists. Importantly, WebFlow allows users to move back and forth between the analysis steps so that the underlying raw flow cytometry data are only “one click away” at any given point.
In addition to the standard statistics, the user has the option to define custom statistics for tasks such as normalization across a plate, calculation of percentage change relative to controls, or analysis of compound selectivity. At this point, all of the required information for the computational analysis is complete, and the user can explore different views of the results. WebFlow provides a number of visualizations, discussed further below, including viewing heat map versions of the plates, exporting the data as a text list for use in other programs, and printing plots representing the populations.
Throughout the Results section, we will use the example of analyzing two 96-well plate experiments to highlight the features of WebFlow. The first experiment highlights the utility of WebFlow in drug screening environments, showing the effects of a small molecule Jak inhibitor on Stat1 phosphorylation induced by IFN-γ in the U937 myeloid cell line. In this experiment, custom statistics are employed to calculate fold change in phosphorylation as well as percentage inhibition of signaling caused by the small molecule, allowing determination of the IC50 (concentration of compound that inhibits signaling by 50%). The second experiment highlights the use of WebFlow in exploratory data analysis and quality control of staining, by analysis of CD3, CD4, and CD8 surface staining of human peripheral blood. Here, we identified samples that were not stained properly and therefore should be excluded from further analysis.
Experiment Annotator
In many popular flow cytometry software analysis packages, samples are treated individually; however, in the context of a high-throughput, plate-based experiment, there is information in the plate layout itself that is then lost. In WebFlow, we designed the software to inherently regard samples as part of a plate; then, any plate-based information (e.g., serial dilutions across columns) is directly tagged to the samples. Central to the functioning of this feature is the experiment annotator, which is designed to be a convenient approach to describe experiments that were laid out in multi-well plates, e.g., 96- or 384-well plates. Samples that did not originate from plates must still be organized in a plate-shaped grid for analysis, but the size of the grid is user-defined and can therefore be used to analyze data sets of any size and organization.
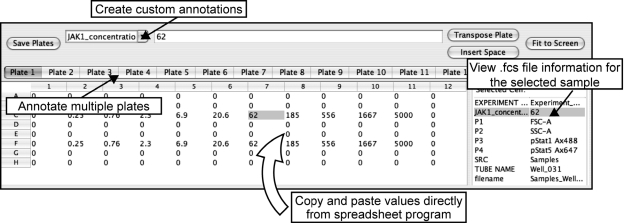
Features of the annotator applet are shown in Fig. 2, where the user is annotating the Jak inhibitor dose–response experiment performed in U937 cells. By implementing the copy and paste/fill features of spreadsheet programs, the annotator allows users to tag samples based on templates they may have created in Microsoft Excel or a similar spreadsheet program. In particular, the annotator allows the user to add as many annotations as desired, in any organization on the plate. For example, it is possible to annotate a dose–response in columns or, alternatively, to place different stimuli in the rows and different time points in columns (Fig. 2). In addition, the software allows a “copy and paste” of a given input pattern from one part of a plate to another, to fill the rest of the plate from a copied pattern, or to paste text from Microsoft Excel into a plate. Finally, WebFlow also reads and utilizes standard annotations placed in FCS files by flow cytometer software. All of the annotations assigned to files in the annotator can be used at the later stages of analysis, from heat map generation to data export; thus, the multiwell paradigm of the experiment is carried throughout the analysis. This ability to organize data by the annotations added in WebFlow removes the need to reorganize and annotate the data in subsequent analysis steps within WebFlow or in other programs for proper visualization.
FIG. 2.
Annotation of high-throughput flow cytometry experiments. Screenshot of WebFlow annotation page. WebFlow utilizes a plate layout for annotation and later analysis steps for editing and viewing of large, plate-based experiments (the size of the plate can be modified to accommodate non–plate-based experiments as well). The user creates annotations for each sample directly in WebFlow (entering Jak1 inhibitor concentration in this example), inputting values or descriptors either directly or by pasting from spreadsheet programs. In addition, the program reads annotations from FCS files that were added during acquisition (shown on the right side of the screen), allowing the user to annotate the experiment before, during, or after acquisition. Custom annotations added in this step are carried through later data analysis and export steps.
Compensation
Multiparameter flow cytometry experiments require compensation of fluorescent signals to eliminate “spillover” from one parameter into neighboring parameters.20,21 Compensation is often considered to be a challenging and confusing aspect of multiparameter flow cytometry applications. Therefore, in WebFlow, we attempted to make the compensation process as transparent as possible. Users are first directed to a page that requires them to specify which files contain the positive and negative compensation populations; then, by clicking the “Gate” button, the user is presented with a dot plot awaiting the user's gate of the relevant population. The user can also input universal negative populations if they were used. After all of the relevant populations are specified, the compensation matrix is calculated, and the user is presented with visual displays of the compensation results. The compensation matrix can be edited if the user wishes (after appropriate consideration of the pitfalls of manual high-dimensional compensation).
Plot Viewer and Gating
Viewing plots and drawing gates in WebFlow are similar to other popular flow cytometry analysis software packages. A single plot viewing applet is used throughout WebFlow to display dot plots, density plots, contour plots, and histograms (Fig. 3A). This viewer allows the user to specify the axes, plot mode, and gates for any particular sample. In addition, the plot viewer allows the user to draw gates (ellipse, rectangle, polygon, quadrant, and region), which can be accessed by clicking the proper button on the toolbar at the top of the plot. In WebFlow, gates are automatically applied to all data files (these are termed “global gates”). However, within any plot, the user has the option to change a gate for that particular sample, by selecting the “Show/Hide Gates” menu item and then changing the gate's values for that sample. As an added measure of security against errors, global gates are locked from editing by default; either they can be unlocked individually via the “Show/Hide Gates” menu item, or all gates can be unlocked by clicking the “Unlock All” toolbar button.
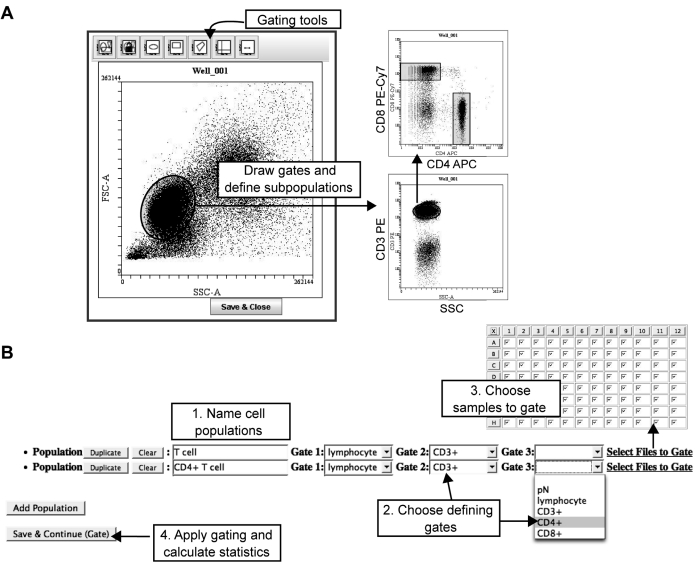
FIG. 3.
Definition of cell populations in WebFlow. Screenshots of WebFlow gating and populations definition pages. (A) Gating cell subsets. The gating applet of WebFlow provides multiple gating tools including ellipses, rectangles, polygon, quadrant, and histogram gates. The user can draw gates, gate the sample, and then define further subgates. Here, the user gated lymphocytes based on forward and side light scatter (FSC and SSC), then gated on CD3+ T cells. The CD4+ and CD8+ T cell subsets were gated based on CD4 and CD8 levels. (B) Population definition. Once gates have been drawn, the user names cell populations (Step 1) and chooses the gates that define that population (Step 2). The user then selects which samples to apply the population analysis to (Step 3), allowing different analyses to be applied to different plates. Once all populations have been defined, the final step instructs WebFlow to calculate standard statistics (mean, median, CV, percentage, number) for each parameter on each defined cell population (Step 4). In this example, CD4+ T cells are defined by the lymphocyte, CD3+, and CD4+ gates. All samples in the plate were stained the same way, so the entire plate is selected for analysis.
Defining Populations
In order for WebFlow to process experiments with large numbers of similar samples, the computational part of gating (what we call “defining populations”) occurs only once in the analysis, as opposed to occurring dynamically while the gate is drawn as it does in many other software packages. In this way, the statistics for each population are cached (i.e., stored in memory), and so viewing statistical data is extremely rapid; if the user changes particular gates, WebFlow will then recalculate the statistics for the affected populations. Because of this, the user interface for defining populations works somewhat differently in WebFlow than in common flow analysis software. Figure 3 displays the population definition process for the T lymphocyte staining experiment, where cells were stained with anti-CD3, -CD4, and -CD8 antibodies. The user first draws and names gates that define the cell populations; in this case, a lymphocyte size gate was drawn, and then CD3+ cells were gated followed by selection of CD4+ or CD8+ cells (Fig. 3A). In the next step, the user specifies a population name and selects the gates that define that population (Fig. 3B). For instance, CD4+ T cells are defined by the “lymphocyte,” “CD3+,” and “CD4+” gates. Once a population is defined, the user selects which files to perform the gating on. After all of the populations are defined, the “save” button is pressed, and standard statistics for each parameter are calculated for each population. A specific step to define populations is necessary in order to have a single gating calculation step; this allows for caching of the gating results without having to re-gate all of the populations each time the user wishes to view a different set of parameters, a critical feature of rapid high-throughput analysis.
Heat Maps
By employing a plate-based analysis throughout, WebFlow enables users to view their results in a plate-shaped layout. This eliminates a bottleneck in flow cytometric analysis that previously required exporting data into another program and subsequent annotation in order to visualize data in heat map format. After the data have been gated, the results are available for viewing in a number of different visualization modalities. The heat map format displays the samples in a plate-shaped grid, with each entry color-coded based on the numerical value of the sample's statistic in the cell (Fig. 4, top panel). Using predefined statistics including mean, median, CV, percentage, and cell number, users can get an overview of their experiment.
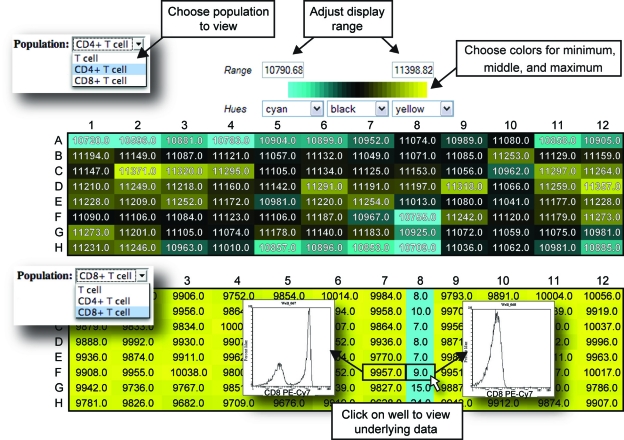
FIG. 4.
Exploratory data analysis of flow cytometry data for verification of staining integrity. Screenshot of WebFlow heat map page displays results from staining PBMCs with CD3, CD4, and CD8 antibodies, with the CD8 antibody omitted in column 8. Heat maps allow visualization of calculated statistics for each population across all of the samples in a plate. For each heat map, the user specifies the colors and minimum and maximum values that correspond to those colors. (Top panel) “Number in Gate” for CD4+ T cells. (Bottom panel) The same statistic for CD8+ T cells. Clicking on a sample in a heat map shows a plot of the underlying data (shown here for samples in F7 and F8). This allows for rapid confirmation of results displayed in the heat map, here clearly showing the lack of CD8+ cells in the T cell gate in sample F8.
This overview allows for visual verification of results to indicate problem areas on plates, as well as rapid determination of a variety of potential errors that might occur during the experimental process. For example, it is possible to check that all samples were properly stained with antibodies by viewing the “number in gate” parameter. In the example T-cell staining experiment, for instance, it was readily observed that column 8 appeared to be lacking CD8+ T cells (Fig. 4, bottom panel). By viewing the data in this way, we determined that the experimenter omitted the anti-CD8 antibody from that column during the staining.
An important feature of the interface design is the ability to interact with heat maps: clicking on a heat map cell opens a histogram of the data showing the relevant parameter across the x-axis (Fig. 4, bottom panel), which the user can then change to any desired one-or two-dimensional plot view. This allows for rapid transition from overview results back to the underlying flow data, thus providing visual confirmation and verification that the gates fit the data correctly or that the sample is not in some other manner aberrant. Additionally, as we have found in our own laboratory, the ease of access to primary data coupled with the ease of generating analyses encourages researchers to explore additional parameters of the data, discovering new patterns in the data or verifying the uniformity of samples in their experiments. With other software packages, performing what should be a routine verification of data quality requires time-consuming one-by-one analysis of each sample to ensure accurate and consistent staining.
Custom Statistics
By retaining the plate location information, WebFlow allows users to define custom statistics for analyzing their data that can use data from any plate position, parameter, or population (e.g., normalization across a row). As with the predefined statistics, these custom statistics can be viewed conveniently in a heat map (or exported for analysis in other programs). Custom statistics are created by entering a mathematical expression that can reference any of the other statistics of any other files in a plate. In particular, the equation processor will interpret sample positions in a plate, different populations, different channels, and different statistics.
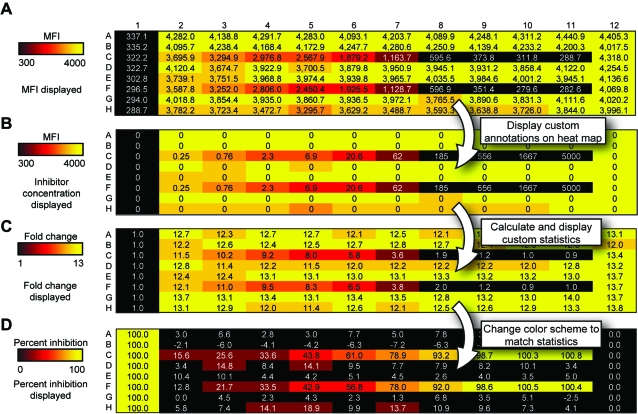
These custom statistics allow for a number of powerful analyses with relative ease. Importantly, the heat map visualization can be changed fluidly to visualize standard and custom statistics or even annotations. For instance, when visualizing the data from the drug screening experiment (in which an inhibitor was titrated across rows C and F), the user can view the median fluorescence intensity of phospho-Stat1 staining in each sample (Fig. 5A) and then switch to viewing the concentration of inhibitor (Fig. 5B). Colors were chosen such that negative controls (low values) are black, middle values are red, and positive controls (maximum values) are yellow. Another informative parameter in this type of experiment is the fold change in phosphorylation that is induced by addition of the IFN-γ stimulus. For this purpose, a custom statistic was defined that normalized the median fluorescence intensity of samples in columns 2–12 to the unstimulated sample in column 1 for each row (Fig. 5C). Finally, to analyze inhibitor activity, the user can define percentage inhibition (see Table 1 for equation) and display that statistic in the same plate-based heat map, allowing for identification of hits in screening experiments. Here, the user was able to visually determine the IC50 of the Jak inhibitor, approximately 10 nM (Fig. 5D). Note also that the color scheme was changed in Fig. 5D to reflect the different ranges of the data (from 0% to 100%). More details of how to specify equations are available in the program itself and in Appendix.
FIG. 5.
Heat map analysis of flow cytometric data for identification of pathway inhibitors. Screenshots of WebFlow heat maps generated from the sample U937 stimulation and inhibition experiment. U937 cells were treated with a dose–response of Jak inhibitor I (a pan-Jak kinase inhibitor) in rows C and F prior to stimulation with IFN-γ to induce Stat1 phosphorylation. Phosphorylation of Stat1 was measured by staining with an antibody against pStat1 (Y701). Demonstrated is the ability to change which statistic is represented by the heat map colors, and which is represented by the numbers on the heat map. (A) Heat map colored by median fluorescence intensity (MFI) in phospho-Stat1 channel, and numbers display pStat1 MFI values. (B) Heat map colored by median staining in pStat1, while numbers display Jak inhibitor I concentration. (C) Heat map colored by fold change in pStat1 relative to column 1, and numbers display those values. (D) Heat map colored by percentage inhibition of induced pStat1 levels relative to negative control in column 1 and positive control in column 12, and numbers display those values. In each case, the range of the heat map coloring was adjusted to reflect the range of the data for the calculated statistic.
Table 1.
Custom Calculation Equations
| Derived Value | Equation Used |
|---|---|
| Fold change | ‘File’.‘Population’.‘median’.‘Channel’/‘(getRow(),0)’.‘Population’.‘median’.‘Channel’ |
| Percentage inhibition | (1-(‘File’.‘Population’.‘median’.‘Channel’-‘(getRow(),0)’.‘Population’.‘median’.‘Channel’)/(‘(getRow(),11)’.‘Population’.‘median’.‘Channel’-‘(getRow(),0)’.‘Population’.‘median’.‘Channel’))*100 |
| CD4:CD8 ratio | ‘File’.‘CD4 + T cell’.‘number’/‘File’.‘CD8 + T cell’.‘number’ |
Other Data Output
Although WebFlow can perform custom calculations, other specialized software packages might be best for particular advanced analyses, such as clustering or multidimensional displays. Therefore, WebFlow can export data as a list in text format (Fig. 6), including all desired annotations and custom statistics, so that the user can then perform further analysis in specialized programs such as Microsoft Excel or Spotfire DecisionSite (TIBCO Software, Somerville, MA) without having to annotate data in a less convenient interface.
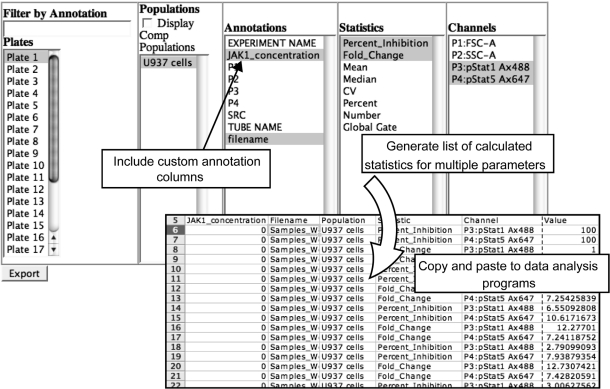
FIG. 6.
Export of custom annotations and statistics for high-content analysis. Screenshot of the WebFlow data export interface: the user chooses relevant plates and populations to export, as well as the desired annotations and statistics to include. Results of the export opened by Microsoft Excel are shown in the lower panel. Each row in the exported table includes one numerical value (with several annotations/descriptors), enabling pivoting in other software.
Indeed, movement of data from flow cytometry analysis programs into other data analysis programs previously involved large amounts of time and effort, since annotation of critical experiment variables (e.g., time, drug concentration, stimulations added, treatments performed, and patient identity) was not done in plate layouts that allow intuitive organization. As mentioned above, the primary goal of WebFlow is convenient annotation of large flow cytometry experiments followed by exploratory data analysis. The program therefore enables quick overviews of the data in heat map format and facile exporting of fully annotated data to more sophisticated analysis packages.
Discussion
The increased application of flow cytometry to high-throughput platforms requires software that is capable of analyzing such experiments conveniently. Most current flow cytometry analysis packages focus on analysis of individual samples, outside of the context of their original (e.g., plate-based) experimental organization. By employing a plate-based annotation scheme from the beginning,7,10 WebFlow enables analysis of experiments for data integrity, normalization across the plate, and export of annotated sample statistics to more sophisticated analysis software. In addition, the ability to rapidly switch among data views employing different statistics encourages researchers to verify their data quality and to explore potentially new patterns and associations in the data. Finally, the ease of directly accessing the primary flow cytometry data that underlie a given statistic encourages the experimenter to verify that the analysis correctly represents the data, i.e., that gates are correctly placed and that there are not subpopulations within an assumed homogeneous gate.
As a demonstration of WebFlow's utility, we performed two test 96-well flow cytometry experiments. In the first, a drug screening experiment, Stat1 phosphorylation was induced by treatment of U937 cells with IFN-γ. An inhibitor of Jak kinases was titrated across two rows of the plate. Prior experience in our laboratory is that a complete analysis, resulting in an IC50 value for the Jak inhibitor, would have taken several hours with currently available software (and would have required three programs: Flowjo, Microsoft Excel, and Spotfire). Using WebFlow, we were able to perform the analysis (annotation step shown in Fig. 2 and results shown in Fig. 5) with a start-to-finish time of 15 min. Thus, coordination of data analysis into a streamlined system allowed for a far more rapid, and less tedious, process. This reduces total man-hours spent on unnecessary and repetitive tasks, reducing the potential for errors while increasing the flexibility of the analysis options.
To highlight WebFlow's ability to detect errors via exploratory data analysis, we conducted a second 96-well plate experiment involving human PBMCs that were stained for the surface markers CD3, CD4, and CD8. In this experiment, we intentionally omitted one of the antibodies (CD8) during the staining of some samples (column 8 in the plate). By employing heat maps to verify that our staining was uniform (Fig. 4), WebFlow allowed us to quickly identify that there was a problem with column 8. Indeed, we easily noticed that there were no CD8+ cells in that column, which we confirmed by visualizing the underlying data to determine that there was staining in the CD4 channel but no staining in the CD8 channel. By allowing users to do this and similar checks quickly, this visualization tool will encourage researchers to routinely ensure that there were no systematic errors in the data. Currently, the time it takes to perform these overviews prevents many researchers from closely checking data integrity, and thus they often gate based on one sample and apply that gating scheme to the rest of the samples without verification. We believe that such approaches as we employ in this software package can heighten awareness of systematic errors in plate-based experiments and thus help researchers to perform more accurate experiments.
In summary, we present a web-based set of concepts and approaches for flow cytometry analysis, deployed currently as a software suite termed WebFlow (http://webflow.stanford.edu). The philosophical approach is to provide a system that handles data for high-throughput cytometry environments, enhancing speed of analysis and detection and reducing errors. By encouraging up-front organization, annotation, and analysis of samples in a plate-based format, which is the current standard for high-throughput experimentation, the system can reduce data corruption caused by mislabeling or other experimental errors. In addition, this same paradigm increases the speed of analysis and visualization of data for large experiments because it allows for the straightforward analysis steps of annotate, gate, view heat maps, and view sample. Finally, the option to export annotated statistics from large and complex experiments for further analysis in other software packages enables facile high-level analysis.
As flow cytometry moves into the realm of high-throughput drug screening and large-scale disease profiling, the number of researchers requiring advanced data analysis capabilities will greatly increase. The design paradigm of WebFlow is an initial step in this direction, providing a simple interface and minimal set of features designed to allow exploratory data analysis and data export. On a larger scale, our laboratory has taken these initial design philosophies several steps further in the development of the Cytobank project (authors' manuscript in preparation), which incorporates web-based flow analysis with experiment management features, such as sharing experiments with collaborators, searching annotated experiments, connecting flow data with external data (e.g., patient information in another database), and the publication of interactive illustrations. The concept of web-based data analysis, originally applied by others to the microarray field (e.g., Aach et al.,22 Marc et al.,23 and Sherlock et al.24), will be key to wider dissemination of flow cytometric analysis, especially in an era of distributed computing and collaborative interactions across institutions.
Acknowledgments
We would like to thank Ken Schulz and Matt Clutter for reviewing this manuscript, as well as members of the Nolan laboratory for critical testing of the software during development. This research was supported by National Heart Lung and Blood Institute contract N01-HV-28183 and National Institutes of Health grant AI35304.
Abbreviations
- CV
coefficient of variation
- IC50
concentration of compound that inhibits signaling by 50%
- IFN-γ
interferon-γ
- Jak
Janus kinase
- PBMC
peripheral blood mononuclear cell
- PE
phycoerythrin
- Stat
signal transducer and activator of transcription.
Appendix
Custom Statistics Equations
In an equation, the user can employ numbers, mathematical expressions, and references to statistics defined for any data files. In particular, the equation can contain the following key words:
usual mathematical operators and common functions (sin, exp, etc.)
getRow(), which returns the row of the current sample
getColumn(), which returns the column of the current sample
-
a statistic for the current sample or for another sample, with the syntax ‘file’.‘population’.‘statistic’[.‘channel ’].
In particular, file can either be “File,” in which case the current sample is used, a reference to a plate location as (row,column), or a reference to a particular sample name.
population can either be “Population,” in which case the current population is used, or it can refer to a particular population.
statistic can either be one of the predefined statistics (“Mean,” “Median,” “CV,” “Percent,” or “Number”) or one of the user-defined statistics.
Finally, channel can either be “Channel,” in which case the current channel is used, or it can specify a particular channel by name or number. Note that not all statistics require/allow a channel specification.
Calculations will fail gracefully for pathological cases, e.g., files without the requisite population, or circular definitions.
Disclosure Statement
No competing financial interests exist.
References
- 1.Baumgarth N. Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 2.Bigos M. Baumgarth N. Jager GC. Herman OC. Nozaki T. Stovel RT, et al. Nine color eleven parameter immunophenotyping using three laser flow cytometry. Cytometry. 1999;36:36–45. doi: 10.1002/(sici)1097-0320(19990501)36:1<36::aid-cyto5>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay PK. Price DA. Harper TF. Betts MR. Yu J. Gostick E, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 4.Perfetto SP. Chattopadhyay PK. Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 5.Krutzik PO. Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 6.Smith RA. Giorgio TD. Cell-based screening: a high throughput flow cytometry platform for identification of cell-specific targeting molecules. Comb Chem High Throughput Screen. 2004;7:141–151. doi: 10.2174/138620704773120829. [DOI] [PubMed] [Google Scholar]
- 7.Young SM. Bologa C. Prossnitz ER. Oprea TI. Sklar LA. Edwards BS. High-throughput screening with HyperCyt flow cytometry to detect small molecule formylpeptide receptor ligands. J Biomol Screen. 2005;10:374–382. doi: 10.1177/1087057105274532. [DOI] [PubMed] [Google Scholar]
- 8.Krutzik PO. Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 9.Krutzik PO. Crane JM. Clutter MR. Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 10.Ivnitski-Steele I. Larson RS. Lovato DM. Khawaja HM. Winter SS. Oprea TI, et al. High-throughput flow cytometry to detect selective inhibitors of ABCB1, ABCC1, and ABCG2 transporters. Assay Drug Dev Technol. 2008;6:263–276. doi: 10.1089/adt.2007.107. [DOI] [PubMed] [Google Scholar]
- 11.Dunphy CH. Applications of flow cytometry and immunohistochemistry to diagnostic hematopathology. Arch Pathol Lab Med. 2004;128:1004–1022. doi: 10.5858/2004-128-1004-AOFCAI. [DOI] [PubMed] [Google Scholar]
- 12.Irish JM. Hovland R. Krutzik PO. Perez OD. Bruserud O. Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 13.Krutzik PO. Hale MB. Nolan GP. Characterization of the murine immunological signaling network with phosphospecific flow cytometry. J Immunol. 2005;175:2366–2373. doi: 10.4049/jimmunol.175.4.2366. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Fuentes MP. Salama A. In vitro assays for immune monitoring in transplantation. Methods Mol Biol. 2006;333:269–290. doi: 10.1385/1-59745-049-9:269. [DOI] [PubMed] [Google Scholar]
- 15.Kaleem Z. Flow cytometric analysis of lymphomas: current status and usefulness. Arch Pathol Lab Med. 2006;130:1850–1858. doi: 10.5858/2006-130-1850-FCAOLC. [DOI] [PubMed] [Google Scholar]
- 16.Jacobberger JW. Sramkoski RM. Frisa PS. Ye PP. Gottlieb MA. Hedley DW, et al. Immunoreactivity of Stat5 phosphorylated on tyrosine as a cell-based measure of Bcr/Abl kinase activity. Cytometry A. 2003;54:75–88. doi: 10.1002/cyto.a.10063. [DOI] [PubMed] [Google Scholar]
- 17.Wang L. Gaigalas AK. Marti G. Abbasi F. Hoffman RA. Toward quantitative fluorescence measurements with multicolor flow cytometry. Cytometry A. 2008;73:279–288. doi: 10.1002/cyto.a.20507. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y. Campos SK. Lopez GP. Ozbun MA. Sklar LA. Buranda T. The development of quantum dot calibration beads and quantitative multicolor bioassays in flow cytometry and microscopy. Anal Biochem. 2007;364:180–192. doi: 10.1016/j.ab.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung YF. Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet. 2003;19:649–659. doi: 10.1016/j.tig.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Bagwell CB. Adams EG. Fluorescence spectral overlap compensation for any number of flow cytometry parameters. Ann N Y Acad Sci. 1993;677:167–184. doi: 10.1111/j.1749-6632.1993.tb38775.x. [DOI] [PubMed] [Google Scholar]
- 21.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 22.Aach J. Rindone W. Church GM. Systematic management and analysis of yeast gene expression data. Genome Res. 2000;10:431–445. doi: 10.1101/gr.10.4.431. [DOI] [PubMed] [Google Scholar]
- 23.Marc P. Devaux F. Jacq C. yMGV: a database for visualization and data mining of published genome-wide yeast expression data. Nucleic Acids Res. 2001;29:E63–E63. doi: 10.1093/nar/29.13.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherlock G. Hernandez-Boussard T. Kasarskis A. Binkley G. Matese JC. Dwight SS, et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]