Abstract
We evaluated the performance of two plate readers (the Beckman Coulter [Fullerton, CA] DTX and the PerkinElmer [Wellesley, MA] EnVision™) and a plate imager (the General Electric [Fairfield, CT] IN Cell 1000 Analyzer™) in a primary fluorescent cellular screen of 10,000 Molecular Libraries Screening Center Network library compounds for up- and down-regulation of vascular cell adhesion molecule (VCAM)-1, which has been shown to be up-regulated in artherothrombotic vascular disease and is a general indicator of chronic inflammatory disease. Prior to screening, imaging of a twofold, six-step titration of fluorescent cells in a 384-well test plate showed greater consistency, sensitivity, and dynamic range of signal detection curves throughout the detection range, as compared to the plate readers. With the same 384-well test plate, the detection limits for fluorescent protein-labeled cells on the DTX and EnVision instruments were 2,250 and 560 fluorescent cells per well, respectively, as compared to 280 on the IN Cell 1000. During VCAM screening, sensitivity was critical for detection of antagonists, which reduced brightness of the primary immunofluorescence readout; inhibitor controls yielded Z′ values of 0.41 and 0.16 for the IN Cell 1000 and EnVision instruments, respectively. The best 1% of small molecule inhibitors from all platforms were visually confirmed using images from the IN Cell 1000. The EnVision and DTX plate readers mutually identified approximately 57% and 21%, respectively, of the VCAM-1 inhibitors visually confirmed in the IN Cell best 1% of inhibitors. Furthermore, the plate reader hits were largely exclusive, with only 6% agreement across all platforms (three hits out of 47). Taken together, the imager outperformed the plate readers at hit detection in this bimodal assay because of superior sensitivity and had the advantage of speeding hit confirmation during post-acquisition analysis.
Introduction
Cell-based screens are routinely performed by either quantitative analysis of images acquired by a high-throughput microscope platform or by direct quantification of whole-well signals using a plate reader. Operational ease, simple data output, and over a decade of use in HTS have established the plate reader as a workhorse technology. Plate readers are well suited for homogeneous assays (colorimetric, binding assays, fluorescent intensity, etc.), but are incapable of specifically measuring cellular subpopulations, intracellular patterns, and cellular heterogeneity across a sample well.
In contrast to the single whole-well measurements of plate readers, high-throughput microscopy (HTM) (also termed image cytometry, high content analysis, or high content screening [HCS]) is capable of compiling a well measurement from cell-by-cell metrics typically performed on tens to hundreds of cells. This enables pure pattern-change measurements, such as translocation from one cellular compartment to another, and object-by-object gating algorithms to select the target cellular subpopulation and eliminate artifacts (e.g., cellular and noncellular debris, dead cells, and fluorescent precipitates).1–4 Image cytometry enables measurements of small subcellular details and architecture that are beyond the reach of plate readers, and cytometric gating can also be used to detect and assay very rare cells (e.g., differentiation of a few percent of stem cells).5,6 Automated HTM was first characterized and validated for nuclear factor κB nuclear translocation induced by interleukin-1 and tumor necrosis factor-α (TNF-α) in 1998,1 and a later study added in-depth statistical analyses of this assay, including the dependence of 50% inhibitory concentration precision on cell-by-cell measurement variability.3 Both of these articles provide a more complete background of HCS using HTM than provided here. One challenge for rapid adoption of HCS has been algorithm development, in particular, incorporating the many recent software advances that enable the extraction of a broad array of numerical data from the images along with sophisticated downstream data analysis and reporting. Evolving software enables image data to be mined with increasing precision, but imaging can still be challenging in terms of time, computational complexity, and storage.
Many image-based assays, including those detecting changes in small subcellular organelles and pure translocations (that involve no change in overall cellular intensity), cannot be performed on plate readers, making it difficult to carry out head-to-head comparisons. Since both instrument platforms are often available in screening centers, we decided to compare the performances of the two platform types on test plates and a compound screen lacking subcellular content and rare event detection. Two plate reader platforms and one imaging platform were compared on bioassay test plates harboring cell populations labeled with fluorescent biosensors with emissions in blue, green, and red channels.
The screen was for regulation of vascular cell adhesion molecule (VCAM)-1, expressed on the membrane of endothelial cells. Anti-VCAM-1 administered in vivo alters the migration of T-lymphocytes.7 The assay used pooled human umbilical vein endothelial cells (HUVECs) primed with an optimized level of TNF-α to induce a baseline level of VCAM-1 expression on extracellular membranes. HUVECs exhibited both VCAM-1 expression and translocation modulations common to other plasma membrane localization assays (e.g., Prigozhina et al.4); however, the images were analyzed for whole-cell expression to enable fair head-to-head comparison with plate readers. The apparent signal strength and low requirement for subcellular detail did not suggest an a priori advantage for HTM over a simple plate reader. This created an opportunity to compare the performance of the two modalities directly on the same assay.
Materials and Methods
Test plate for cross platform analyses
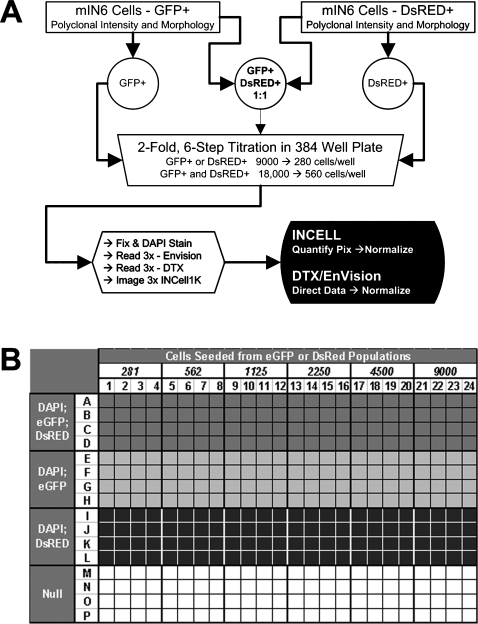
The test plate used to compare the performance of detection platforms was a black Greiner Bio-One (Monroe, NC) 384-well plate with a tissue culture-treated μClear® bottom seeded with cell lines engineered to express either enhanced green fluorescent protein (eGFP) or Discosoma sp. red fluorescent protein (DsRED) protein. The fluorescent cells were generated from the MIN6 mouse insulinoma cell line by stable transduction with lentiviral vectors directing expression of either eGFP from the human insulin (INS) promoter8 or DsRED from the minimal phosphoglycerate kinase (PGK) promoter.9,10 Figure 1 describes the plate layout and assay execution. The plate was arrayed as a six-step, twofold gradient of cells seeded with peak density of 9,000 cells per well. Cells were rinsed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS, and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize cell nuclei.
FIG. 1.
Description of cellular bioassay and platform detection test plate. (A) Process overview: evaluation of detection platforms. Cell lines with either INS promoter-eGFP or PGK promoter-DsRED expression constructs were derived from the MIN6 mouse insulinoma line and used in mock cellular bioassays. A 384-microtiter plate array was composed of samples containing either a single cell line or a mixture of the two. (B) Cells were arrayed in six-step, twofold dilutions (rows A–L) with 9,000 cells per well at the highest seed density. A reference image composed of images from null wells (rows M–P) was used to correct for fluctuations in image intensity resulting from illumination source and light path.
TNF-α/VCAM-1 high-throughput screen
The screen was submitted as part of the National Institutes of Health Molecular Libraries Screening Center Network (MLSCN) initiative and adapted based on guidelines provided by Dr. Thomas Mayer (Columbia College of Physicians and Surgeons, New York, NY) in the original submission (X01 MH076343) and was performed against the first release (10,000 compounds) of the MLSCN chemical library. The screen is described in detail on PubChem (http://pubchem.ncbi.nlm.nih.gov/) with assay identification numbers 454–457. Although originally designed to identify compounds that inhibit expression of VCAM-1 in TNF-α-sensitized HUVECs, the assay was modified during development at the San Diego Center for Chemical Genomics (http://sdccg.burnham.org/metadot/index.pl) to screen for both inhibitors and agonists of TNF-α-induced cell-surface VCAM-1 as visualized by specific immunostaining using an antibody generated against full-length VCAM-1 (sc-13506, Santa Cruz Biotechnology, Santa Cruz, CA) followed by a fluorophore-conjugated secondary. The assay was run at half-maximal level of TNF-α to detect both agonists and antagonists of the VCAM-1 response. Z′ values for the agonist and antagonist assays were calculated from this common baseline control, using the p38 antagonist (SB302580) to inhibit TNF-α-induced VCAM-1 expression for the antagonist assay and a maximal dose of TNF-α for the agonist assay. Cells were fixed with 4% paraformaldehyde and counterstained with DAPI. A Hamilton (Reno, NV) STAR workstation was used for all liquid handling tasks.
Instrument parameters
All microtiter plates were scanned on the Beckman Coulter (Fullerton, CA) DTX and the PerkinElmer (Wellesley, MA) EnVision™. Plate imaging was handled by the General Electric (Fairfield, CT) IN Cell 1000 Analyzer™. The IN Cell 1000 collected four images per well (approximately one-third the area of a 384-format well) with 12-bit depth and 4 × 4 pixel binning using a 10× objective, numerical aperture (NA) 0.45. Blue, green, and red channels were captured on both the test plate and the primary screen. The collected intensities were recorded using the following filters: DAPI 360/40 and 460/40, eGFP 475/20 and 535/50, DsRED 535/50 and 620/60, excitation/bandpass and emission/bandpass, respectively.
Quantification of pixel intensities was performed using the Developer Toolbox software package (General Electric) for all imaging applications. TIFF images collected by the IN Cell 1000 were batch-analyzed using a combination of custom and standard algorithms to gather pixel intensities from the three channels (red, green, and blue) of each image. The DAPI channel grayscale images of cell nuclei were segmented using the object segmentation module with kernel size = 3 and sensitivity setting at 50 using Developer Toolbox (see Fig. 2). All three image channels were first flat field-corrected from previously acquired blank reference images, and then an isotropic diffusion filter was passed over the images for five iterations to smooth image noise to create the “processed images” (Fig. 2, top panels), using standard algorithms in Developer Toolbox version 1.6. Images were then segmented using a threshold T = F × Mean_Image_Intensity, with 0.97 ≤ F ≤ 1.04 for eGFP and 0.89 ≤ F ≤ 1.04 for DsRED. The image segmentation parameters were set empirically depending on characteristics such as cell density (using Developer Toolbox software). Example image segmentation results for all three channels are shown in Fig. 2. Channel intensities were normalized for comparison on the same axis. The final readout for each well was the integrated intensity of the segmented pixels in the red, green, or blue channel, as appropriate. For the VCAM-1 assay, measurements of aberrant radii, standard deviation (SD), and circularity of objects were calculated using Developer Toolbox (see Fig. 5) to detect and eliminate abnormal wells (e.g., resulting from toxicity).
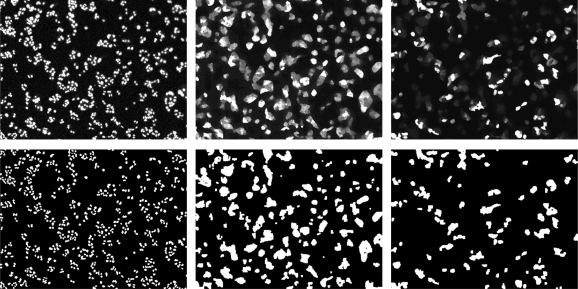
FIG. 2.
Image segmentation masks for fluorescence intensity quantification. The DAPI (left panels), eGFP (center panels), and DsRED (right panels) colors are shown separately with their processed images (top panels) and their segmentation image masks (bottom panels). Each three-color image is one of four fields of view collected per well, where one field is approximately 1/12 of the well area. The approximate cell seed density as shown was 2,250 cells per well (20 × 103/cm2).
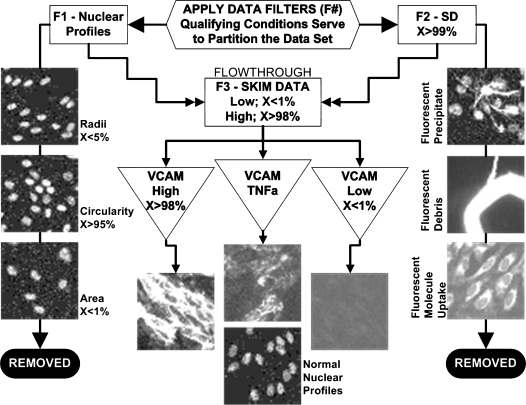
FIG. 5.
Application of data filters in the VCAM-1 screen. Filters are used to gate out wells containing artifacts. Nuclear filters (or gates) of mean radius below the 5th percentile, mean circularity above the 95th percentile, or mean area below the 1st percentile were used to detect and remove aberrant wells. X represents the percentile removed or retained. Compounds causing aberrant radii, circularity, and/or area, quantified using Developer Toolbox, were flagged as undesirable because of potential adverse effects on cell viability or growth. In addition, wells in which the SD of the object size in the top 1st percentile of SD values were flagged and removed from subsequent analysis. This metric was found empirically to be a reliable indicator of debris and other undesirable signals. Agonist and inhibitor hit thresholds were set to obtain the top 2nd and bottom 1st percentiles, respectively, of the wells that remained after these filters were applied.
Results
Dynamic range and sensitivity
We began by evaluating the dynamic ranges and sensitivities of the EnVision, DTX, and IN Cell 1000 using two polyclonal MIN6 mouse insulinoma cell lines engineered to express either eGFP from the human INS minimal promoter (INS-eGFP) or DsRED from the PGK minimal promoter (PGK-DsRED). The DNA-specific fluorochrome DAPI was used to enable image segmentation of the nuclei. All three fluorescent signals—DAPI (blue), eGFP (green), and DsRED (red)—were used to test the sensitivity of the instruments; brightness was modulated by plating a series of different cell numbers (0.28, 0.56, 1.13, 2.25, 4.50, and 9.00 × 103 cells per well). At MIN6 cell densities of 9,000 cells per well (∼8.2 × 104/cm2), the cells were 80% confluent. Promoter expression appeared muted at 18,000 cells per well because of excessive cell density (by visual inspection); thus, the 9,000 cells per well seed density approximates the maximum signal possible biologically and represents the upper end of the dynamic range for all fluorescent probes used. The intensity readouts (relative light units) are plotted as a function of cell density in Fig. 3 for each instrument. The error bars are SD values (n = 16 replicate wells for each condition). Student's t test was applied pairwise to the data plotted in Fig. 3 (as log10 vs. log2 bar graphs for linearity). The limit of detection was defined at P < 0.001. DAPI, which was by far the brightest of the three labels, produced the most similar detection curves across the three platforms. Accordingly, all platforms detected the lowest cell titer (280 cells per well) in the DAPI spectrum, demonstrating that DAPI was too bright to test the lower limits of any of the instruments. For both eGFP and DsRED, the DTX plate reader maintained P < 0.001 through a cell density of 2,250 cells per well. Note that P < 0.001 was then achieved again at 560 cells per well in the eGFP spectrum, making the sensitivity cutoff less clear. In DsRED, P < 0.001 was maintained through 2,250 cells per well for the DTX. The EnVision had a sensitivity limit of 560 cells per well in the eGFP and DsRED channels. Note that the DAPI plots tend to curve downward toward low cell densities, which may be a result of a greater proportion of cells lost at lower dilutions during plating or clustering at the well edges outside the image sampling area. Deviation from linearity in the other direction (tail curves upward toward lower cell densities) was observed for the DTX (DsRED) and EnVision (eGFP and DsRED). These flattening/upward trends were coincident with the worsening P values, lending further support to our conclusions about the sensitivity limits. For the IN Cell 1000, this test did not identify sensitivity limits for any of the three fluorescent labels.
FIG. 3.
Detection capability of the platforms in three channels. The detection limits of the (A) IN Cell 1000, (B) DTX, and (C) EnVision platforms are shown. Note that the DAPI signal was sufficiently bright that the dynamic range of each instrument could discern cell titers from the maximum to minimum plated. For eGFP and DsRED, the IN Cell 1000 exhibited a significant dynamic range advantage, whereas the DTX and EnVision could not discriminate the lowest cell titers (<2,250 and <560 cells per well, respectively), demonstrating a lower dynamic range than the IN Cell 1000. The DTX showed the greatest sample variation of the platforms tested. Asterisks indicate P values (paired t test) for resolving the twofold differences in signal intensities between the flanking cell seed densities as indicated: *P < 0.001, **P < 0.01, ***P < 0.1, ****P > 0.1.
Table 1 summarizes the observed dynamic response for the six-step cell titration of fluorescent cells as fold change in specific signal on each of the three platforms. Background was subtracted from each sample value by using the formula (Si − (Smin − 2SD)), where Si is the sample replicate average, Smin is the minimum sample average value, and SD is that of the minimum sample average. The response was recorded as the fold change in both cell number and signal range [equal to (Smax/Smin), where Smax and Smin are, respectively, the maximum and minimum signals surrounding intervening points that are distinguished by P < 0.001 in Fig. 3]. Table 1 summarizes the detection of cell number and fluorescent signal by each platform. The maximum cell titer range of the MIN6 test plates was 32-fold, as determined from the 6-step, twofold dilution. Note that the imager showed a clear advantage in quantifying dim signal and low cell titer in the green (eGFP) and (DsRED) channels, indicating better dynamic range and detection limit than the plate readers. Interestingly, both plate readers worked as well as the imager in resolving cell titer in the blue channel (DAPI), which exhibited an intense fluorescent signal. However, comparison of the signal dynamic range revealed that the imager nonetheless had a greater dynamic range than either plate reader in the blue channel.
Table 1.
Dynamic Range of Detected Signal as Fold Change (x)
| |
Platform |
|||||
|---|---|---|---|---|---|---|
| |
IN Cell 1000 |
DTX |
EnVision |
|||
| Dye | Signal | Cell titer | Signal | Cell titer | Signal | Cell titer |
| DAPI | 135.3x | 32x | 53.6x | 32x | 83.0x | 32x |
| eGFP | 23.3x | 32x | 2.2x | 4x | 3.9x | 16x |
| DsRED | 66.6x | 32x | 2.9x | 4x | 7.1x | 16x |
Dynamic range represents the quotient of instrument maximum and minimum values obtained from cell titers residing on a positive trendline and with P < 0.001.
Variations across sample and imaging/scan replicates
We next compared well-to-well variation across all three platforms. In addition to determining sample replicate variation, scan/imaging replicates were tested for signal variation by scanning or imaging the same plate three times. SDs were then calculated over the 16 sample replicates at each of six discrete cell titers (for 96 total samples). The coefficients of variation (CVs) were calculated [CV (%) = 100 × SD/mean; n = 16 for each of the six titers] and averaged (mean of n = 6 CVs) for Table 2. The EnVision showed the best performance for the DAPI channel for scan-to-scan replicates with a CV of 1%, whereas the DTX rated poorest for all tested channels. The IN Cell 1000 rated second for DAPI (2.2%) but scored best for eGFP (2.3%) and DsRED (3.3%) as compared to both plate readers. These observations support the data summarized in Table 1, further demonstrating that the lower sensitivity of the plate readers makes the measurements more susceptible to variation at low signal intensities. Scanning and imaging CVs were dependent on both the color/channel and signal strength, and the greatest CV across all platforms was for DsRED. As DAPI, eGFP, and DsRED signals decreased, the respective CVs increased most for the DTX (fourfold increase from DAPI to DsRED), whereas the IN Cell 1000 demonstrated the least increase in CVs from DAPI to eGFP and DsRED channels.
Table 2.
The Scan/Imaging CVs of Each Instrument
| |
Platform |
||
|---|---|---|---|
| Dye | IN Cell 1000 | DTX | EnVision |
| DAPI | 2.0% | 2.4% | 1.0% |
| eGFP | 2.3% | 6.4% | 2.7% |
| DsRED | 3.3% | 10.3% | 3.9% |
The test plate was read n = 3 times on each instrument to obtain the CV.
We then investigated the sources of variation in the IN Cell integrated intensity readout. The IN Cell 1000 acquired four images per well with a 10× objective to cover about one-third of the well area. The intensities were integrated across the segmented objects so only the areas containing cells (i.e., pixels with intensities above the threshold) contributed to the measurements. To provide a high level view of how CVs varied across platforms, the CVs in each channel were averaged across all cell seed densities. Figure 4A shows higher IN CELL 1000 integrated intensity (avg*area) CVs as compared with both plate readers. The majority of this IN CELL 1000 variation was contributed by the area measurements, which increased with cell titer (Fig. 4B). The lowest cell seed densities resulted in an approximate 40% variation (GFP/green) of the sampled area (∼100 ± 40 objects per well, or 25 ± 10 objects per image). Normalization to DAPI area in each well reduced variation across all three channels (Fig. 4C), whereas normalization to DAPI object count reduced variation in the red and blue, but not green, channels (Fig. 4D). We conclude that area variation due to cell seeding and clustering can have at least two components: (1) variations in object number, which could be reduced by normalization to object count in all but the lowest seed density for eGFP (green), and (2) variations in area, which was decreased in all but two data points (eGFP at 280 and 560 cells per well) by normalization to area. Thus, the primary source of variation in the IN Cell 1000 measurements was likely imperfect distribution during cell seeding and the tendency of the MIN6 cells to cluster. Plate readers are less susceptible to this source of error since they measure intensity from the entire well, rather than a portion of it (one-third in this example).
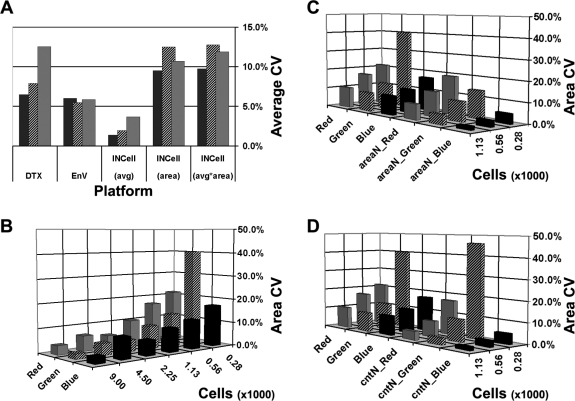
FIG. 4.
CVs of intensities for each instrument for DAPI (blue, ▪), eGFP (green  , and DsRED (red,
, and DsRED (red,  ) fluorescent labels are plotted. (A) The average readout CVs (over n = 6 seed densities) for each color on the DTX and EnVision (EnV) platforms are plotted with the areas (measured as the sum of the pixels in all segmented objects) and integrated intensities (average [avg]*area) per well (all four images) on the IN Cell 1000 (INCell) instrument. The integrated intensity from the INCell closely matches the DTX and EnV readouts. The overall sample CV was lowest on the EnV. The INCell had the highest CV of ∼12% for eGFP. The INCell avg area showed the smallest variation of all of the measurements, and the primary contribution of variation was the area component of the integrated intensity (avg × area). (B) The CVs of the total area of the objects (total number of pixels in objects in all four images per well) are plotted as a function of cell plating density for each fluorescent label. The area CVs increased with decreasing cell seed density. (C) CVs of the area measurements are plotted for the lower cell seed densities for each color channel, as raw data (red, green, blue) and as normalized by the nuclear area (number of pixels in segmented objects, DAPI) for each well (areaN) to correct for variations in the number and size of nuclei, which reduced CVs for all channels. (D) CVs of the area measurements are shown as raw data (red, green, blue) and as normalized to the number of segmented nuclear objects (DAPI) for each well (cntN). This normalization decreased the CVs for all but green at the lowest seed density (cntN_Green at 280 cells per well). Normalization by object count (D) had lower CVs than by area (C) for all red seeding densities and green at 1,130 cells per well. In contrast, higher CVs were observed in the green channel (280 and 560 cells per well), whereas the blue channel retained comparable CVs with both normalizations. Taken together, the results in (C) and (D) indicate that variation in both object number and area determination contributes to elevated area CVs at lower seed densities (see text).
) fluorescent labels are plotted. (A) The average readout CVs (over n = 6 seed densities) for each color on the DTX and EnVision (EnV) platforms are plotted with the areas (measured as the sum of the pixels in all segmented objects) and integrated intensities (average [avg]*area) per well (all four images) on the IN Cell 1000 (INCell) instrument. The integrated intensity from the INCell closely matches the DTX and EnV readouts. The overall sample CV was lowest on the EnV. The INCell had the highest CV of ∼12% for eGFP. The INCell avg area showed the smallest variation of all of the measurements, and the primary contribution of variation was the area component of the integrated intensity (avg × area). (B) The CVs of the total area of the objects (total number of pixels in objects in all four images per well) are plotted as a function of cell plating density for each fluorescent label. The area CVs increased with decreasing cell seed density. (C) CVs of the area measurements are plotted for the lower cell seed densities for each color channel, as raw data (red, green, blue) and as normalized by the nuclear area (number of pixels in segmented objects, DAPI) for each well (areaN) to correct for variations in the number and size of nuclei, which reduced CVs for all channels. (D) CVs of the area measurements are shown as raw data (red, green, blue) and as normalized to the number of segmented nuclear objects (DAPI) for each well (cntN). This normalization decreased the CVs for all but green at the lowest seed density (cntN_Green at 280 cells per well). Normalization by object count (D) had lower CVs than by area (C) for all red seeding densities and green at 1,130 cells per well. In contrast, higher CVs were observed in the green channel (280 and 560 cells per well), whereas the blue channel retained comparable CVs with both normalizations. Taken together, the results in (C) and (D) indicate that variation in both object number and area determination contributes to elevated area CVs at lower seed densities (see text).
In summary, the DTX and EnVision demonstrated greater variability at the lowest cell titers, where signal detection was not separable from noise. Furthermore, signal variation in plate reader measurements dramatically decreased when signal became separable from well noise (P < 0.001 was achieved at cell titers ≥2,250 for the DTX and ≥560 for the EnVision). In contrast, the IN Cell 1000 had a greater dynamic range at low signal thresholds, outstripping increased signal deviation (approximately 30% across three channels). However, the IN Cell 1000 measurements exhibited increased CVs at low cell densities that we attribute to inconsistent distribution of cells in the wells.
Primary screen tests confirm the test plate results
Next, we evaluated the platforms in an MLSCN screen of 10,000 small synthetic molecules for the ability to modulate TNF-α-dependent VCAM-1 expression on HUVECs. Experimental details of the agonist and antagonist assays as well as chemical structures of the hits are available on PubChem (http://pubchem.ncbi.nlm.nih.gov/), with separate entries for imaging and plate reader datasets. Images acquired on the IN Cell 1000 were visually inspected to confirm the top 2% (agonists) and bottom 1% (antagonists) of hits from each platform.
One advantage of the imaging platform is the ability to filter out wells containing sample artifacts that cannot be distinguished from hits by a plate reader. The process for filtering out wells with an artifact is diagrammed in Fig. 5. Wells were removed if they contained either (1) cells with misshapen nuclei (mean radius <5th percentile, mean circularity >95th percentile, and mean nuclear area <1st percentile; see F1 in Fig. 5) or (2) debris such as fibers, fluorescing compounds, or their breakdown products (SD of integrated intensity objects >95th percentile; see F2 in Fig. 5). The wells with integrated intensities in the 1st percentile (inhibitors of TNF-α-induced VCAM-1) or above the 98th percentile (agonists of TNF-α-induced VCAM-1) were then considered as possible hits (see “FLOWTHROUGH” in Fig. 5).
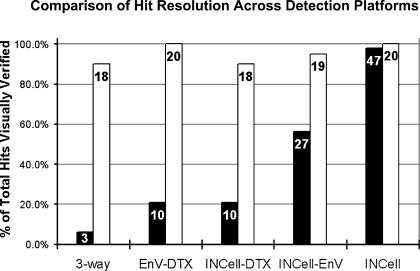
Together, all three instruments detected 48 inhibitors and 20 agonists, shown by instrument in Fig. 6. The IN Cell 1000 identified 47 of the 48 visually confirmed inhibitors. The single inhibitor not characterized as a hit by the IN Cell 1000 was automatically rejected in the data analysis because the nuclear radii were significantly reduced. Compounds that caused misshapen or small nuclei were considered undesirable as probes since such effects can be due to toxicity. The ability to use imaging criteria to automatically flag compounds for subsequent scrutiny underscores the advantage of HTM over plate reader platforms.
FIG. 6.
Performance summary of the IN Cell 1000 (INCell), DTX, and EnVision (EnV) instruments in the TNF-α/VCAM-1 screen. The number of hits identified for each platform is shown within the corresponding bars. Inhibitors (▪) or agonists (□) of TNF-α/VCAM-1 expression in HUVECs were identified using each platform from a primary screen of 10,000 compounds (see Materials and Methods). Of 48 inhibitors visually confirmed across all three platforms, the INCell image analysis identified 47. The single inhibitor not characterized as a hit by the INCell was rejected by the image and data analysis process as atypical in its nuclear morphology (see text). The EnV performed best between the two plate readers, identifying 57%, whereas the DTX identified 21% of the 48 inhibitors. Furthermore, discordance in the common inhibitor hits identified among all three platforms (6%) indicates impaired ability to discern inhibitors by the plate readers. In contrast, all platforms showed good concordance in detecting agonists.
Both plate reader platforms were less capable than the imaging platform at identifying hits that reduced VCAM-1 expression below baseline (Fig. 6). The EnVision and DTX recognized 57% and 21% of confirmed inhibitors, respectively. The Z′ for the inhibitor assay was 0.41 for the IN Cell 1000 versus 0.16 for the EnVision, correlating well with the results shown in Fig. 3A indicating that plate readers have lower dynamic range and sensitivity.
In contrast, identification of VCAM-1 expression agonists was nearly 100% congruent across the three platforms. This was not unexpected given that the Z′ values determined for the maximal induction of VCAM-1 during the assay development phase were comparable for the IN Cell 1000 and EnVision (0.57 and 0.65, respectively). This was also predicted from the MIN6 test plate results since the agonists enhanced VCAM-1 expression, thereby elevating fluorescent signal into a range where plate readers reliably detected signal. For this particular screen, the larger dynamic range of the imager was useful since it permitted detection of both agonists and antagonists in the same screen.
Discussion
Results obtained from the serial dilution of fluorescently labeled MIN6 cells demonstrated that the imaging platform has a significant advantage over both plate readers in terms of dynamic range and sensitivity. Specifically, the imaging platform was able to discern a 32-fold cell titer range in the three colors analyzed. Furthermore, slope and regression analyses showed that the imaging platform has a strong performance advantage in quantifying commonly used red and green fluorescent proteins. All three platforms resolved low-end signal comparably in the brightest channel (blue, DAPI). The intense DAPI signal results from high-affinity and high-occupancy binding of the probe to large concentrations of nucleic acid. Fluorescent protein intensity is typically weaker, primarily because of the lower numbers of fluorophores when used as reporters of gene expression or metabolic processes in living cells. The same is true for common fluorophore-conjugated antibodies as used in the bimodal TNF-α/VCAM-1 assay. For such reagents, the sensitivity and dynamic range of the imager were found to be essential to resolve low-end signal in our performance comparison in a primary screen to detect small molecule inducers or inhibitors of VCAM-1 by TNF-α. All three platforms detected a core number of TNF-α/VCAM-1 pathway agonists, consistent with their similar abilities to detect bright signals. However, the imaging platform had a higher Z′ (0.41) than did the plate readers (0.16 for the EnVision) and outperformed the plate readers in the detection of TNF-α/VCAM-1 inhibitors.
It is important to emphasize that data analysis was handled as equitably as possible given the different types of output from the imager and reader platforms. For plate readers, very little data filtering can be performed, with the exception of removing extreme outliers. This was done in the plate reader data from the TNF-α/VCAM-1 screen by removing wells that had extremely high or low levels of DAPI intensity. Although relatively insensitive, this nuclear channel intensity flag succeeded in removing some undesirable sample wells, such as those caused by debris and toxic compounds. Flagging and removal of extremely low cell density wells were critical for the TNF-α/VCAM-1 assay since VCAM-1 expression depends on cell–cell contact and was significantly reduced in wells with sparse cell seeding, toxicity, or growth inhibition. For the imaging dataset, IN Cell Developer Toolbox image analysis metrics were used to flag wells based on parameters of low total collected object areas, aberrant nuclear profiles, and aberrant SD among objects in a well. Specifically, inhibitors were rejected based on statistically low collected nuclear area, aberrant nuclear profile, or statistically low number of identified nuclei. Similarly, false agonists, such as molecular precipitates, inherently fluorescent molecules, and debris, were rejected based on high signal SD within the well. Thus, an additional benefit of the imager platform is its ability to use multiparametric data to classify samples, resulting in fewer false-positives.
We noted that the imager measurements showed greater well-to-well variance than did plate readers in sample wells with low cell titers. This was due primarily to variation in the total number of cells in the image area at low cell titers and, potentially, differences in defining the areas of interest (morphometry), rather than in quantification of pixel intensity per se. It was possible to reduce such variance by normalizing integrated density values relative to nuclear DAPI area and object counts (Fig. 4C and D). Cell-by-cell image cytometry, for which higher-resolution images are used to create better cell-by-cell segmentation, might further improve normalization by area and/or object counts and could provide additional intra-well filtering/gating, both of which might further improve the imaging results. In addition, use of a higher-resolution objective (e.g., 20×) would further increase the sensitivity of the imager because of the increased NA (fluorescent intensity is proportional to NA4 in a microscope).11 The possible advantages of higher-resolution images for any assay must always be balanced with the decrease in speed that may result if more images have to be acquired to measure the same number of cells.
In conclusion, this report has characterized the performance differences between two commonly used plate readers and a plate imager in the context of a fluorescent cellular bioassay. Although plate readers have strong advantages in speed and simplicity, automated microscopy platforms can provide superior performance for exacting resolution of fluorescent cellular bioassays. The EnVision and DTX platforms can scan a 384-well microplate in three colors in about 5 and 15 min, respectively and the data require no further processing besides normalization and analysis. In contrast, three-color imaging of a 384-well microplate by the IN Cell 1000 at one image per well requires approximately 25 min, and the downstream analysis of image data requires a rigorous process that is labor intensive to develop and validate. Furthermore, the complexity of image analysis algorithms increases for multiplexed assays, but this increased workload also brings substantial opportunities. Multiparametric analysis affords greater assay sensitivity and selectivity because of the ability to introduce filters/gates to refine hit calling. In some cases, this process can provide flexibility during assay development by increasing the range of parameters available to improve sensitivity and dynamic range or to correct and normalize data.
Lastly, image-based screening could lead to additional benefits. For instance, image datasets queried with one algorithm for one purpose could be revisited to quantify additional features. Thus, simple re-analysis of the existing dataset could provide selectivity or other useful information that might not have been foreseen prior to running the assay. Moreover, analysis using algorithms designed to detect features unrelated to the intent of the primary screen represents a new screen at marginal additional cost. While some assays are inappropriate (e.g., liquid phase/homogeneous assays) for an imaging platform, others are inappropriate for plate reader platforms (e.g., object morphometry; low-level signal detection as shown here). Ultimately, the choice of platform depends on assay demands and the investigator's resources and experience. In summary, image-based screening is a powerful technology that extends the potential of cell-based assays and will become more widespread as throughput and analysis techniques improve.12
Acknowledgments
We gratefully acknowledge the assistance of Dr. Thomas Mayer (Columbia University College of Physicians and Surgeons) who submitted the original assay (X01 MH076343) for development and screening at the Burnham Institute for Medical Research and Behrad Azimi, Nick Cosford, Susanne Heynen, Eduard Sergienko, Steve Vasile, and members of the BIMR chemical library screening center for many helpful discussions. This research was funded by grants U54HG003916, R21HL071913, R01DK068715, P30CA030199, R01EB006200, and R41DK076510 from the National Institutes of Health and grant RC1-00132 from the California Institute for Regenerative Medicine.
Abbreviations
- CV
coefficient of variation
- DAPI
4′,6-diamidino-2-phenylindole
- DsRED
Discosoma sp. red fluorescent protein
- eGFP
enhanced green fluorescent protein
- HCS
high content screening
- HTM
high-throughput microscopy
- HUVEC
human umbilical vein endothelial cell
- INS
insulin
- MLSCN
Molecular Libraries Screening Center Network
- NA
numerical aperture
- PBS
phosphate-buffered saline
- PGK
phosphoglycerate kinase
- SD
standard deviation
- TNF-α
tumor necrosis factor-α
- VCAM
vascular cell adhesion molecule
References
- 1.Ding GJ. Fischer PA. Boltz RC. Schmidt JA. Colaianne JJ. Gough A, et al. Characterization and quantitation of NF-kappaB nuclear translocation induced by interleukin-1 and tumor necrosis factor-alpha. Development and use of a high capacity fluorescence cytometric system. J Biol Chem. 1998;273:28897–28905. doi: 10.1074/jbc.273.44.28897. [DOI] [PubMed] [Google Scholar]
- 2.Li F. Zhou X. Ma J. Wong ST. An automated feedback system with the hybrid model of scoring and classification for solving over-segmentation problems in RNAi high content screening. J Microsc. 2007;226:121–132. doi: 10.1111/j.1365-2818.2007.01762.x. [DOI] [PubMed] [Google Scholar]
- 3.Morelock MM. Hunter EA. Moran TJ. Heynen S. Laris C. Thieleking M, et al. Statistics of assay validation in high throughput cell imaging of nuclear factor κB nuclear translocation. Assay Drug Dev Technol. 2005;3:483–499. doi: 10.1089/adt.2005.3.483. [DOI] [PubMed] [Google Scholar]
- 4.Prigozhina NL. Zhong L. Hunter EA. Mikic I. Callaway S. Roop DR, et al. Plasma membrane assays and three-compartment image cytometry for high content screening. Assay Drug Dev Technol. 2007;5:29–48. doi: 10.1089/adt.2006.024. [DOI] [PubMed] [Google Scholar]
- 5.Bushway PJ. Mercola M. High-throughput screening for modulators of stem cell differentiation. Methods Enzymol. 2006;414:300–316. doi: 10.1016/S0076-6879(06)14017-3. [DOI] [PubMed] [Google Scholar]
- 6.Marcelli M. Stenoien DL. Szafran AT. Simeoni S. Agoulnik IU. Weigel NL, et al. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J Cell Biochem. 2006;98:770–788. doi: 10.1002/jcb.20593. [DOI] [PubMed] [Google Scholar]
- 7.Gorczynski RM. Chung S. Hoang Y. Sullivan B. Chen Z. Altered patterns of migration of cytokine-producing T lymphocytes in skin-grafted naive or immune mice following in vivo administration of anti-VCAM-1 or -ICAM-1. Immunology. 1996;87:573–580. doi: 10.1046/j.1365-2567.1996.511581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odagiri H. Wang J. German MS. Function of the human insulin promoter in primary cultured islet cells. J Biol Chem. 1996;271:1909–1915. doi: 10.1074/jbc.271.4.1909. [DOI] [PubMed] [Google Scholar]
- 9.Deglon N. Tseng JL. Bensadoun JC. Zurn AD. Arsenijevic Y. Pereira de Almeida L, et al. Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson's disease. Hum Gene Ther. 2000;11:179–190. doi: 10.1089/10430340050016256. [DOI] [PubMed] [Google Scholar]
- 10.Hamaguchi I. Woods NB. Panagopoulos I. Andersson E. Mikkola H. Fahlman C, et al. Lentivirus vector gene expression during ES cell-derived hematopoietic development in vitro. J Virol. 2000;74:10778–10784. doi: 10.1128/jvi.74.22.10778-10784.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue S. Spring KR. Video Microscopy, The Fundamentals. Plenum; New York: 1997. [Google Scholar]
- 12.Fox S. Farr-Jones S. Sopchak L. Boggs A. Nicely HW. Khoury R, et al. High-throughput screening: update on practices and success. J Biomol Screen. 2006;11:864–869. doi: 10.1177/1087057106292473. [DOI] [PubMed] [Google Scholar]