Abstract
Purpose
To screen the myocilin (MYOC) and forkhead box protein C1 (FOXC1) genes for sequence variations in primary congenital glaucoma (PCG).
Methods
Seventy five PCG patients were screened for MYOC variations and 54 cases (negative or heterozygous for cytochrome P4501B1 mutations) for FOXC1 mutations by polymerase chain reaction (PCR) and DNA sequencing.
Results
Five single nucleotide polymorphisms (SNPs; −126T>C, −83G>A, p.R76K, IVS2+35G>A, and p.Y347Y) were identified in MYOC and two sequence variations (GGC375ins and GGC447ins) in FOXC1. No pathogenic variations were identified in MYOC and FOXC1 in our patients.
Conclusions
MYOC and FOXC1 mutations are not involved in pathogenesis of primary congenital glaucoma in our patients. Thus, it is important to screen other loci for involvement in congenital glaucoma in cases which are negative or heterozygous for CYP1B1 mutations to have a better insight in to disease pathogenesis.
Introduction
Glaucoma is an optic neuropathy, characterized by elevated intra-ocular pressure (IOP) which results in retinal ganglion cell (RGC) death and vision loss [1]. Primary congenital glaucoma (PCG; OMIM 231300) is a severe form of glaucoma which manifests in the neonatal/early infantile period with a classic triad of symptoms viz. epiphora (excessive tearing), photophobia (hypersensitivity to light), and blepharospasm [2]. The prevalence of congenital glaucoma varies across ethnic communities and geographical boundaries [3]. In the Indian state of Andhra Pradesh, its prevalence is estimated to be around 1:3,300 and accounts for 4.2% of all cases of childhood blindness [4].
Genetic heterogeneity is the hallmark of PCG and three chromosomal loci (I) 2p21 (GLC3A (GLC stands for glaucoma); OMIM 231300) [5], (II) 1p36 (GLC3B; OMIM 600975) [6], and (III) 14q24.3 (GLC3C) [7] have been mapped by linkage analysis, of which only the GLC3A locus harboring the human Cytochrome P450 gene (CYP1B1; OMIM 601771) has been characterized [8]. CYP1B1 exhibits a high degree of allelic heterogeneity and more than 79 different mutations associated with PCG have been identified [9]. The proportion of PCG cases with CYP1B1 mutations vary widely across different populations and are highest among the inbred Slovakian Gypsy [10] and Saudi Arabian populations [11] which exhibit allelic homogeneity.
Earlier we reported CYP1B1 mutations as a predominant cause for PCG phenotype [12,13]. We observed CYP1B1 mutations in 42.66% PCG cases (32/75; 13 cases were homozygous, 8 were compound heterozygous, and 11 were heterozygous for CYP1B1 mutations). However, 57.34% of these cases did not show the involvement of CYP1B1.
This led us to explore the role of other genes to understand their possible implications in the disease pathogenesis. The myocilin gene implicated in juvenile open angle glaucoma (JOAG) and in adult-onset primary open angle glaucoma (POAG) [14] was chosen as the potential candidate for screening these cases. The myocilin gene (MYOC) exhibits a wide spectrum of mutations and accounts for 2–5% cases of POAG [15] and in 5.5% of PCG [16]. MYOC is located on chromosome 1 at 1q25 and codes for myocilin/trabecular meshwork-induced glucocorticoid response (TIGR) protein. Most tissues of the eye express MYOC, including trabecular meshwork, sclera, ciliary body, and retina [17,18]. An earlier report showed that MYOC interacts with CYP1B1 through a digenic mechanism in causing JOAG [19].
Forkhead box protein C1 (FOXC1/FKHL7), another gene, was also selected for mutation analysis in PCG cases which were either negative or heterozygous for CYP1B1 mutations. A recent study from Southern India has reported involvement of FOXC1 mutations in PCG [20]. FOXC1 is a member of the winged helix/forkhead family of transcription factors. It is located on 6p25 and has single exon that codes for 553 amino acids long protein [21]. The FOXC1 protein is expressed in various ocular and non-ocular tissues [22,23] and is found in periocular mesenchyme cells that give rise to ocular drainage structures such as the iris, cornea, and trabecular meshwork [24]. Both the FOXC1 null (FOXC1−/−) and the heterozygous (FOXC1+/−) mice were found to have anterior segment abnormalities similar to those in humans with anterior segment dysgenesis (ASD) and congenital glaucoma [25]. In this study we have screened MYOC gene in 75 PCG cases and FOXC1 gene in 54 PCG cases (negative/heterozygous for CYP1B1 mutations) to understand the role of these two gene in the pathogenesis of this blinding disorder.
Methods
Congenital glaucoma cases presenting at the Dr. Rajendra Prasad Centre for Ophthalmic Sciences, All India Institute of Medical Sciences (AIIMS), New Delhi, India, were enrolled for this study. After ethical approval of the Institutional Review Board (IRB00006862; AIIMS) a total of 75 PCG cases were screened for MYOC sequence variations while FOXC1 was screened in 54 cases (either negative or heterozygous for CYP1B1 mutations; Table 1). The diagnosis involved clinical, ocular and systemic examination. Inclusion criteria of the patients were increased corneal diameter (>12.0 mmHg) and raised IOP (>21 mmHg) with presence/absence of Haab’s striae and optic disc changes (where examination was possible). Symptoms of epiphora and photophobia were the additional inclusion factors. The age of onset ranged from birth to 3 years.
Table 1. Clinical phenotype and CYP1B1 mutation status of PCG cases.
| Pt. ID | Age of onset of disease | Sex | Age at presentation/Sampling | Corneal Diameter (mm) OS/OD | Buphthalmos | IOP OS | IOP OD | Haabs’ striae | Last Cup Disc ratio OS/OD | Corneal edema | Mutations | Treatments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCG01 |
By birth |
M |
12 years |
15x15/15x15.5 |
OU |
40 |
30 |
OU |
Total cupping |
OU |
R390H (H) |
Medical and OU 3XTrab/Trab+MMC |
| PCG02 |
By birth |
F |
10 years |
13x13/13x13 |
OU |
36 |
40 |
OU |
0.5:1/0.6:1 |
OU |
R390H (H) |
Medical and OU 1XTrab/Trab+MMC |
| PCG03 |
By birth |
F |
7 months |
15x14/15x15 |
OU; OD>OS |
26 |
38 |
Absent |
Hazy MEDIA |
Absent |
R390H (H) |
Medical and OU 1XTrab/Trab+MMC; OS 1XTrab/Trab+MMC |
| PCG04 |
By birth |
M |
10 months |
12x11/12x12 |
Absent |
22 |
24 |
Absent |
0.4:1/0.4:1 |
Absent |
— |
Medical and OU Trab/Trab+MMC |
| PCG05 |
4 months |
M |
5 months |
14.5x15/15x15 |
OU; OD>OS |
28 |
28 |
OU |
0.7:1/0.7:1 |
OU |
R368H (H) |
Medical and OU 2XTrab/Trab+MMC |
| PCG06 |
By birth |
M |
4 months |
12x13/12x13 |
OU |
22 |
23 |
OU |
0.5:1/0.5:1 |
OU |
— |
Medical and OU Trab/Trab+MMC |
| PCG07 |
By birth |
F |
18 months |
12x13/11x11.5 |
OS |
38 |
14 |
Absent |
0.4:1/0.4:1 |
Absent |
— |
Medical and OU Trab/Trab+MMC |
| PCG08 |
1 month |
F |
NA |
OD |
23 |
25 |
Absent |
Hazy media |
One eye |
— |
Medical and OD Trab/Trab+MMC |
|
| PCG09 |
2 months |
13 months |
12x12/12x12 |
Absent |
22 |
22 |
Absent |
NA |
Absent |
— |
Medical and OU Trab/Trab+MMC |
|
| PCG010 |
1 month |
F |
1 month |
NA |
OD |
23 |
24 |
Absent |
NA |
One eye |
— |
Medical and OD Trab/Trab+MMC |
| PCG011 |
1 year |
M |
4 months |
14x14/14x15 |
OU; OD>OS |
26 |
22 |
Absent |
NA |
Absent |
E229K (h) |
Medical and OU 1XTrab/Trab+MMC |
| PCG012 |
By birth |
M |
58 months |
14x14.5/14x14.4 |
OU |
32 |
32 |
Absent |
NA |
Absent |
R368H (h) |
Medical and OU 1XTrab/Trab+MMC |
| PCG013 |
By birth |
F |
2 months |
14x14/14x14 |
OU |
31 |
30 |
Absent |
Hazy media |
Absent |
R390H (H) |
Medical and OU 2XTrab/Trab+MMC |
| PCG014 |
By birth |
F |
1 month |
NA |
OU |
25 |
24 |
Absent |
Hazy media |
Absent |
E229K (h) |
Medical and OU 2XTrab/Trab+MMC |
| PCG015 |
By birth |
M |
3 months |
14.5x14/13.5x13 |
OU; OS>ODR |
32 |
32 |
Absent |
0.6:1/0.6:1 |
Absent |
M132R (H) |
Medical and OU 2XTrab/Trab+MMC |
| PCG016 |
By birth |
M |
6 months |
11x11/12x12.5 |
OD |
18 |
26 |
Absent |
0.4:1/0.5:1 |
Absent |
— |
Medical and OD Trab/Trab+MMC |
| PCG017 |
By birth |
M |
9 months |
14x14/14x14.5 |
OU |
30 |
28 |
Absent |
NA |
Absent |
Ter@223 (H) |
Medical and OU 2XTrab/Trab+MMC |
| PCG018 |
By birth |
M |
3.4 years |
14.5x14/14x14 |
OU; OS>OD |
20 |
20 |
Absent |
NA |
Absent |
— |
Medical and OU Trab/Trab+MMC |
| PCG019 |
By birth |
F |
7 months |
12x12.5/12x12 |
OU |
22 |
22 |
Absent |
0.5:1 1 |
Absent |
Ter@223 (h) |
Medical and OU 2XTrab/Trab+MMC |
| PCG020 |
By birth |
M |
7 years |
12x13/13x13 |
OU; OD>OS |
18 |
37 |
Absent |
Hazy media |
Absent |
— |
Medical and OU1X Trab/Trab+MMC |
| PCG021 |
By birth |
M |
2 years |
15x16/11.5x12 |
OS |
32 |
15 |
OU |
Hazy media |
Absent |
Ter@223 (h) |
Medical and 2X Trab/Trab+MMC OS |
| PCG022 |
By birth |
F |
10 months |
15x15/16x16 |
OU; OD>OS |
28 |
28 |
OU |
0.7:1/0.5:1 |
Absent |
p.L24R (H) |
Medical and OS 2XTrab/Trab+MMC |
| PCG023 |
By birth |
F |
4 months |
14x15/14x15 |
OU |
34 |
36 |
OU |
Hazy media |
OU |
— |
Medical and OU 1X Trab/Trab+MMC |
| PCG024 |
By birth |
M |
1 months |
14x13/14x13 |
OU |
30 |
24 |
Absent |
0.7:1/0.7:1 |
OU |
— |
Medical and OU 1X Trab/Trab+MMC |
| PCG025 |
By birth |
F |
18 years |
NA |
Absent |
32 |
10 |
Absent |
NA |
Absent |
— |
Medical and OS Trab/Trab+MMC |
| PCG026 |
By birth |
M |
2 years |
14x15/14x15 |
OU |
22 |
22 |
Absent |
Hazy media |
OU |
— |
Medical and OU Trab/Trab+MMC |
| PCG027 |
By birth |
F |
2 years |
13x13.5/15x14.5 |
OU; |
22 |
22 |
Absent |
0.8:1/0.8:1 |
Absent |
— |
Medical and OU 1X Trab/Trab+MMC |
| PCG028 |
By birth |
M |
2.5 years |
13x13.5/13x13.5 |
OU |
20 |
20 |
Absent |
hazy media |
One eye |
— |
Medical and OU 1X Trab/Trab+MMC |
| PCG029 |
By birth |
F |
3 months |
15x15/14x14 |
OU |
28 |
27 |
OU |
Hazy media |
OU |
Ter@223 (h) |
Medical and 2X Trab/Trab+MMC |
| PCG030 |
By birth |
M |
1 month |
13x13.5/13.5x13 |
OU |
26 |
26 |
Absent |
0.7:1/0.7:1 |
One eye |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG031 |
By birth |
M |
4 months |
11x14/14x15 |
OU |
34 |
36 |
Absent |
Hazy media |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG032 |
By birth |
M |
10 months |
15x15/12x12 |
OS |
18 |
14 |
Absent |
0.3:1/0.3:1 |
Absent |
— |
Medical and 1X OS Trab/Trab+MMC |
| PCG033 |
By birth |
M |
1 year |
13x13/11x11 |
OS |
12 |
12 |
Absent |
0.2:1/NO glow |
One eye |
— |
Medical and 1X OS Trab/Trab+MMC |
| PCG034 |
By birth |
M |
2 months |
14x14/14x14 |
OU |
22 |
24 |
Absent |
NA |
Absent |
R390H (h); Ter@223 (h |
Medical and 1X OU Trab/Trab+MMC |
| PCG035 |
By birth |
M |
3 months |
13x13/13x12 |
OS |
18 |
16 |
Absent |
no glow |
Absent |
— |
Medical and 1X OS Trab/Trab+MMC |
| PCG036 |
By birth |
M |
1 month |
15x15/15x15.5 |
OU |
25 |
26 |
Absent |
0.8:1 1 |
Absent |
R390H (H); E229K (h) |
Medical and 1X OD Trab/Trab+MMC |
| PCG037 |
By birth |
M |
1 month |
14x14/11x11 |
OU;OS>OD |
18 |
22 |
Absent |
Hazy media |
One eye |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG038 |
By birth |
M |
3.5 years |
corneal: Dulcer/14x14 |
OD |
18 |
20 |
Absent |
hazy media |
OU |
— |
Medical and 1X OD Trab/Trab+MMC |
| PCG039 |
By birth |
M |
4.5 years |
13x13/12x13.5 |
OU |
36 |
34 |
Absent |
NT cupping |
OU |
R368H (h); Ter@223 (h) |
Medical and 1X OUTrab/Trab+MMC |
| PCG040 |
By birth |
F |
2 months |
11.5x12.5/12x13 |
OU; OD>OS |
23 |
23 |
Absent |
Hazy media |
OU |
Ter@223 (h) |
Medical and OD 1XTrab/Trab+MMC |
| PCG041 |
By birth |
M |
11 months |
12x10/12x12.5 |
OU; OD>OS |
40 |
26 |
Absent |
Hazy media |
OU |
R390H (H): Ter@223 (h) |
Medical and OU 1XTrab/Trab+MMC;
1XTrab+Trab+mmc OD, 1XPK |
| PCG042 |
By birth |
M |
5 days |
13x13/13x13 |
OU |
22 |
24 |
Absent |
Hazy media |
OU |
R368H (H) F190L (H) |
Medical and 1XTrab/Trab+MMC OU |
| PCG043 |
By birth |
F |
I2 months |
12x12/12x12 |
OU |
22 |
26 |
OU |
0.8:1 1 |
Absent |
R390H (h) G329D (h) |
Medical and 1X OU Trab/Trab+MMC |
| PCG044 |
By birth |
M |
4 years |
14x14/14x14 |
OU |
26 |
24 |
Absent |
0.8:1/0.9:1 |
Absent |
Ter@223(h) |
Medical and 1X OU Trab/Trab+MMC |
| PCG045 |
By birth |
M |
1 month |
12x12/12x12.5 |
OU |
20 |
22 |
Absent |
0.5:1/0.5:1 |
Absent |
E229K (h), R390H (h) |
Medical and 1X OU Trab/Trab+MMC |
| PCG046 |
By birth |
M |
6 months |
13x13/13x13.5 |
OU |
24 |
16 |
Absent |
0.5:1/0.3:1 |
OU |
— |
Medical and1X OU Trab/Trab+MMC |
| PCG047 |
By birth |
F |
5 months |
14.5x14/14x14 |
OU |
30 |
20 |
Absent |
0.7:1/0.9:1 |
One eye |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG048 |
By birth |
M |
11 months |
13x13.5/12x13 |
OU |
28 |
26 |
Absent |
NA |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG049 |
By birth |
M |
6 months |
18x20/14x16 |
OU |
24 |
22 |
Absent |
OS no glow/0.5:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG050 |
By birth |
F |
11x11.5/11x11.5 |
OU |
26 |
30 |
Absent |
0.3:1/0.3:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
|
| PCG051 |
By birth |
F |
36 months |
11x11.5/13x13 |
OU;OD>OS |
22 |
28 |
Absent |
0.8:1/0.9:1 |
OU |
— |
Medical and 1X OU Trab/Trab+MMC;
OU cataract surgery |
| PCG052 |
By Birth |
F |
2 months |
12.5x13/12.5x13 |
OU |
20 |
20 |
Absent |
Hazy media/0.5:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG053 |
By birth |
F |
4 months |
11.5x12/12x12 |
OU; OD>OS |
40 |
23 |
Absent |
Hazy media |
OU |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG054 |
By birth |
M |
9 months |
15x14.5/15x14.5 |
OU |
26 |
26 |
Absent |
Absent glow |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG055 |
By birth |
M |
8 months |
Phthisic eye/12x12 |
OD; OS Phthisic eye |
na |
37 |
Absent |
NA/0.9:1 |
OU |
p.I94X (H) |
Medical and 1X OD Trab/Trab+MMC |
| PCG056 |
By birth |
F |
12 months |
14.5x14.5/14x14 |
OU; OS>OD |
30 |
28 |
OU +ve |
Hazy media |
OU |
p.Q340H (H) + p.R390H (H) |
Medical and 1X OU Trab/Trab+MMC |
| PCG057 |
By birth |
M |
3 months |
13x13/13.5x13.5 |
OU; OD>OS |
28 |
30 |
Absent |
0.7:1/0.7:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG058 |
By birth |
M |
15 months |
14x14/12.5x12.5 |
OU; OS>OD |
20 |
16 |
Absent |
total cupping/ 0.5:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG059 |
By birth |
F |
10 months |
14x14.5/13.5x14 |
OU; OS>OD |
31 |
31 |
OS |
NA |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG060 |
7 months |
F |
41 months |
14x14/13x13.5 |
OU; OS>OD |
24 |
18 |
Absent |
0.4:1/0.6:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG061 |
By birth |
M |
4 months |
15.5x15/14x14 |
OU; OS>OD |
30 |
34 |
OU |
Not visible/0.9:1 |
OU |
p.H279D (h) |
Medical and 1X OU Trab/Trab+MMC |
| PCG062 |
By birth |
M |
8 months |
11x11.5/10x11.5 |
OU; OS>OD |
22 |
16 |
Absent |
0.6:1/0.6:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG063 |
3 months |
M |
12 months |
12x12/11x12.5 |
OU; OS>OD |
22 |
23 |
Absent |
0.4:1/0.4:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG064 |
By birth |
M |
1 month |
12x12/11.5x11.5 |
OU; OS>OD |
28 |
24 |
Absent |
not visible/0.7:1 |
Absent |
p.R390C (h) |
Medical and 1X OU Trab/Trab+MMC |
| PCG065 |
By birth |
M |
6 months |
13x13/12.5x13 |
OU; OS>OD |
25 |
26 |
Absent |
0.7:1/0.7:1 |
OU |
p.E229K (h) |
Medical and 1X OU Trab/Trab+MMC |
| PCG066 |
3 months |
M |
13 months |
12.5x12/12.5x12 |
OU |
22 |
24 |
Absent |
0.4:1/0.4:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG067 |
11 months |
M |
132 months |
12x13/13x14 |
OU; OD>OS |
21 |
24 |
Absent |
not visible |
OU |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG068 |
By birth |
M |
6 months |
13x13/13.5x14 |
OU; OD>OS |
26 |
28 |
Absent |
0.8:1/0.8:1 |
OU |
p.R368H (H) |
Medical and 1X OU Trab/Trab+MMC |
| PCG069 |
By birth |
M |
4 months |
13x13/12x13 |
OU; OS>OD |
24 |
26 |
Absent |
0.5:1/0.6:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG070 |
By birth |
M |
45 days |
15x15/15x14.5 |
OU/; OS>OD |
30 |
40 |
Not visible |
not visible |
OU |
p.R355X (H) |
Medical and 1X OU Trab/Trab+MMC |
| PCG071 |
13 months |
M |
18 months |
12x12/12x12 |
OU |
22 |
23 |
Absent |
0.4:1/0.4:1 |
Absent |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG072 |
By birth |
M |
45 days |
12x12/12x11.5 |
OU; OS>OD |
20 |
24 |
Absent |
no glow |
OU |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG073 |
By birth |
M |
2 months |
12.5x13/11x12 |
OU; OS>OD |
22 |
22 |
Absent |
Hazy media |
OU |
— |
Medical and 1X OU Trab/Trab+MMC |
| PCG074 |
By birth |
M |
8 months |
14x14.5/15x15 |
OU;OS>OD |
28 |
24 |
OU |
No glow |
OU |
p.R368 (H) |
Medical and 1X OU Trab/Trab+MMC |
| PCG075 | By birth | M | 1 year | 13x13.5/14x14 | 0U; OD>OS | 20 | 20 | Absent | No glow | OU | R390H (H) | Medical and 1X OU Trab/Trab+MMC |
Key: M- male; F- female; H- homozygous; h-heterozygous; X- times; Trab/Trab+MMC- combined trabeculotomy trabeculectomy and mitomycin C treatment; OD- right eye; OS- left eye; OU- both eyes; NA- not available.
All patients with history of blood transfusion, TORCH (Toxoplasmosis; Rubella; Cytomegalovirus; Herpes Simplex Virus) infection and drug intake by mother during pregnancy were excluded. Glaucoma cases other than PCG were excluded. Detailed family history of ocular or other hereditary disorders up to three generations were taken, and pedigree charts were constructed.
Control group
Seventy five ethnically matched normal individuals without any ocular/systemic disorders were enrolled as controls for MYOC and 50 for FOXC1 analysis. Peripheral blood samples were collected from patients and controls by venipuncture in ethylenediaminetetra-acetic acid (EDTA) vacutainers only after informed consent and stored in −80 °C until further use.
Mutation screening and sequence analysis
DNA was isolated from the peripheral blood using the Phenol chloroform method. All three exons with exon-intron boundaries were amplified from DNA using polymerase chain reaction (PCR) primers designed for MYOC (GenBank AB006688; available at Genbank) [26]. FOXC1 primers (Table 2) were designed using National Center for Biotechnology Information (NCBI) PRIMER3 program. PCR amplifications for FOXC1 primers were performed in a 40 μl volume containing 1.0 μl of 20 μM stock solution for each primer, 100 ng of genomic DNA, 1 unit of Taq polymerase (Banglore Genei P Ltd, Bengaluru, Karnataka, India), 0.1 mM of each dNTP and 4 μl of 10× PCR buffer (with 15 mM MgCl2), by means of 35 cycles of amplification, each consisting of 30 s denaturation at 94 °C, 50 s annealing at 56 °C −59 °C and 1 min extension at 72 °C and final extension at 72 °C for 5 min.
Table 2. FOXC1 primers used in this study.
| Serial number | Primer sequence | Product size |
|---|---|---|
| 1 |
1F- CCCGGACTCGGACTCGGC |
649 bp |
|
|
1R- TCTCCTCCTTGTCCTTCACC |
|
| 2 |
2F- GAGAACGGCAGCTTCCTG |
708 bp |
|
|
2R-TTGCAGGTTGCAGTGGTAGGT |
|
| 3 |
3F-GGCCAGAGCTCCCTCTACA |
636 bp |
| 3R-CTGCTTTGGGGTTCGATTTA |
All PCR products were analyzed on 1.8% agarose gel, stained with ethidium-bromide (EtBr 10 mg/ml). Agarose gel was analyzed using gel documentation system (Applied biosystems, Carlsbad, CA). Amplified PCR products were purified using gel/PCR DNA fragments extraction kit (Catalog number DF100; Geneaid Biotech Ltd., Sijhih City, Taiwan). Purified PCR product were sent for sequencing at MCLAB (Molecular Cloning Laboratories, South San Francisco, CA). DNA sequences were analyzed against the MYOC reference sequence and the FOXC1 reference sequence using ClustalW2 provided by the European Molecular Biology Laboratory (EMBL) European Bioinformatics Institute (EBI).
Computational assessment of missense mutations
Two homology based programs PolyPhen (Polymorphism Phenotyping) and SIFT (Sorting Intolerant From Tolerant) analysis tool were used to predict the functional impact of missense changes identified in this study [27,28]. The prediction is based on the position-specific independent counts (PSIC) score derived from multiple sequence alignments of observations. PolyPhen scores of >2.0 indicate the polymorphism is probably damaging to protein function. Scores of 1.5–2.0 are possibly damaging, and scores of <1.5 are likely benign. SIFT is a sequence homology-based tool that sorts intolerant from tolerant amino acid substitutions and predicts whether an amino acid substitution in a protein will have a phenotypic effect [29,30]. Positions with normalized probabilities less than 0.05 are predicted to be deleterious and, those greater than or equal to 0.05 are predicted to be tolerated. We have also used an improved splice site predictor tool [31] to predict whether a nucleotide change is likely to create a splice site.
Statistical analysis
The frequency of nucleotide variation between cases and controls was calculated by χ2/Fisher’s exact test and a p-value ≤0.05 was considered significant.
Results
A total of five nucleotide changes (2 in promoter region, 2 in coding region and 1 in intronic region) were observed in MYOC and two sequence variations (GGC375ins and GGC447ins) were observed in FOXC1 in this study. Details of all these changes are given below.
Myocilin (MYOC) gene
−126 Thymine to Cytosine (−126T>C)
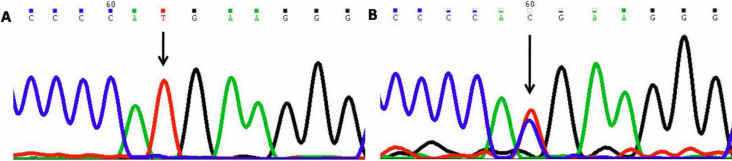
This nucleotide change resulted in thymine (T) being replaced by cytosine(C; Figure 1) at position g.169888563 or 126 base pairs upstream (−126 position). This change was homozygous in one (PCG043) and heterozygous in one case (PCG053) but was absent in controls.
Figure 1.
DNA sequence chromatogram of MYOC equivalent to bases −121 to −132. A: The reference sequence derived from control is shown. B: Sequence derived from congenital glaucoma patient PCG043 shows heterozygous −126T>C nucleotide change.
−83 Guanine to Adenine (−83G>A)
This nucleotide change resulted in guanine (G) being replaced by alanine (A; Figure 2) at position g.169888457 or 83 base pairs upstream (−83 position). This change was homozygous in five cases (PCG07, 015, 034, 040, and 062) and heterozygous (PCG06, 15, 34, and 40) in four cases. This change was also present in 2 controls.
Figure 2.
DNA sequence chromatogram of MYOC equivalent to bases −76 to −88. A: The reference sequence derived from control is shown. B: Sequence derived from congenital glaucoma patient PCG015 shows homozygous −83G>A nucleotide change.
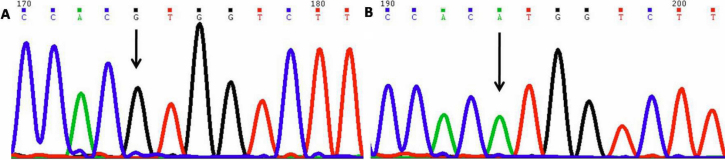
Arginine76Lysine (p.R76K)
This nucleotide change resulted in G being replaced by A (Figure 3) at position g.169888148; coding nucleotide number c.127. This resulted in a codon change from AGA to AAA and amino acid change from arginine to lysine (p.R76K) a non-synonymous mutation in MYOC. This change was homozygous in five patients (PCG007, 015, 034, 040, and 052) and heterozygous in twenty five patients (PCG005, 006, 011, 012, 0016–019, 021, 022, 027, 033, 036, 043, 044, 046, 049, 059, 062, and 065–069) and was also present in 12 controls.
Figure 3.
DNA sequence chromatogram of MYOC exon 1 equivalent to codon 75–78. A: The reference sequence derived from control is shown. B: Sequence derived from congenital glaucoma patient PCG005 shows homozygous c.127G>A, which predicts a codon change AGA>AAA and p.R76K change.
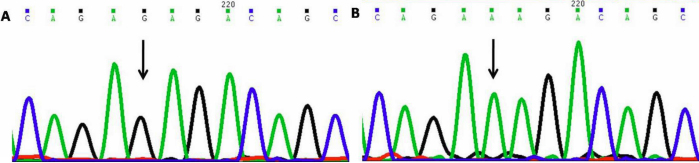
IVS2+35 Guanine to Adenine (IVS2+35G>A)
This nucleotide change resulted in G being replaced by A (Figure 4) at position g.169874325 or 35 base pairs downstream (IVS2+35 position) ; This change was homozygous in forty seven patients (PCG002, 004, 006–009, 011–013, 015, 017, 019–022, 026–029, 033–036, 039–041, 043–046, 049–060, 063–066, and 069) and heterozygous in thirteen patients (PCG001, 003, 005, 010, 016, 018, 023, 037, 042, 054, 061, 062, and 067) and was also present in 25 controls.
Figure 4.
DNA sequence chromatogram of MYOC equivalent to IVS2+31 to IVS2+42 A: The reference sequence derived from control is shown. B: Sequence derived from congenital glaucoma patient PCG004 shows homozygous IVS2+35G>A nucleotide change.
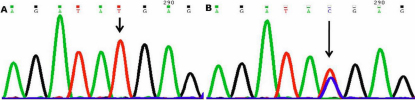
Tyrosine347Tyrosine (p.Y347Y)
This mutation resulted in T being replaced by C (Figure 5) at position g.169872162; coding nucleotide number c.1041. This resulted in a codon change from TAT to TAC with no amino acid change (synonymous mutation) at 347 (p.Y347Y) in MYOC protein. Five cases (PCG029, 036, 043, 053, and 056) were heterozygous for this change while the same was absent in controls. All MYOC sequencing results have been tabulated (Table 3).
Figure 5.
DNA sequence chromatogram of MYOC exon 3 equivalent to codon 346–348. A: The reference sequence derived from control is shown. B: Sequence derived from congenital glaucoma patient PCG029 shows heterozygous c.1041T>C, which predicts a codon change TAT>TAC and p.Y347Y change.
Table 3. MYOC gene variations identified in this study with p-value at 95% confidence interval by using Pearson χ2/Fisher’s exact test.
| Patient ID | ||||||||
|---|---|---|---|---|---|---|---|---|
|
Serial number |
Location |
Sequence change |
Codon change |
Mutation |
PCG (n=75) |
Controls (n=75) |
p-value |
Odds Ratio (at 95% CI) |
| 1 |
Promoter |
−126T>C |
- |
- |
2 |
0 |
0.496 |
- |
| 2 |
Promoter |
−83G>A |
- |
- |
9 |
2 |
0.055 |
4.97 (1.03–23.87) |
| 3 |
Exon 1 |
c.227 G>A |
AGA>AAA |
p.R76K |
30 |
12 |
0.001 |
3.50 (1.61–7.56) |
| 4 |
Intron 2 |
IVS2+35 G>A |
- |
- |
60 |
25 |
<0.001 |
8.00 (3.80–16.80) |
| 5 | Exon 3 | c.1041T>C | TAT>TAC | p.Y347Y | 5 | 0 | 0.058 | - |
Forkhead box protein C1 (FOXC1) gene
Insertion of GCG at g.1556820
Insertion of tri-nucleotide GCG at g.1556820 was observed in nine PCG cases (PCG006–008, 011, 023, 048, 051, 054, and 072) and four controls. This insertion was found in both alleles. This caused an insertion of an extra GGC triplet and an extra amino acid glycine at position 375 (GGC375ins). This leads to presence of seven glycine residues (generally six glycine are found) at amino acid position 375–381 instead of six glycine residues.
Insertion of CGG at g.1557040
Homozygous insertion of tri-nucleotide CGG at g.1557040 was observed in six PCG cases (PCG006, 007, 051, 059, 063, and 065) and five controls. This caused an insertion of an extra GGC triplet which inserts an extra amino acid glycine at amino acid position 447 (GGC447ins). This leads to presence of eleven glycine residues at amino acid position 447–457.
Three PCG cases (PCG006, 007, and 051) had both insertions (GGC375ins and GGC447ins). No other changes were identified in any PCG patient. All FOXC1 variations have been tabulated (Table 4).
Table 4. FOXC1 variations identified in this study with p-value at 95% confidence interval by using Pearson χ2/Fisher’s exact test.
| Patient ID | ||||||
|---|---|---|---|---|---|---|
|
Serial number |
Sequence change |
Mutation |
PCG (n=54) |
Controls (n=50) |
p-value |
Odds ratio (at 95% CI) |
| 1 |
Ins GCC |
GGC375ins |
5 |
4 |
0.240 |
2.3 (0.66–8.00) |
| 2 | Ins CGG | GGC447ins | 6 | 5 | 1 | 1.12 (0.32–3.94) |
SIFT and PolyPhen analysis
There was only one missense change in MYOC in our study. The PSIC score of the p.R76K mutation was <2 indicating that this change was benign to protein function. The SIFT score of p.R76K was 0.00 and was predicted to be deleterious for the protein function. Ideally, to call a change pathogenic both SIFT and PolyPhen results should be pathogenic. Since this change was present both in PCG and controls, it could be hypothesized that this change is non-pathogenic. Improved splice site prediction for IVS2+35G>A showed that this location (IVS2+35) is not present at splice site and may not create a splicing error in MYOC.
Discussion
PCG shows marked genetic heterogenity thus we have screened MYOC in 75 PCG cases to study digenic inheritance and FOXC1 in 54 cases which were either negative or heterozygous for the CYP1B1 mutation for their involvement. To the best of our knowledge this is the first study from north India which involves MYOC and FOXC1 screening in PCG. In our study three single nucleotide polymorphisms (SNPs; Table 3) were observed both in PCG cases as well as in controls while two were limited to cases only. The frequency of p.R76K, IVS2+35G>A, and p.Y347Y in MYOC was found to be statistically significant (p<0.05) in our study. However, no pathogenic MYOC mutation was detected. A recent study showed that a small proportion of PCG cases that do not harbor CYP1B1 mutations exhibit a heterozygous mutation in the MYOC gene [16]. Digenic inheritance of the mutant MYOC and CYP1B1 alleles has also been demonstrated in juvenile-onset POAG and CYP1B1 has been suggested to be a modifier of MYOC expression [19]. The heterozygous p.Gln48His MYOC mutation was first reported in two sporadic cases of JOAG and an adult-onset POAG case from eastern India. Later this change was also reported in a JOAG family and in a sporadic POAG case from India [32].
Although IVS2+35G>A was not at splice location but recent studies have revealed that intronic sequences may be associated with gene regulation. Intronic mutation, therefore, may be involved in the disease irrespective of whether the mutated base was at splice site [33]. Two promoter sequence variations (−126T>C and −83G>A) were observed in this study. These variations do not alter any known promoter or enhancer binding sites and are found in similar frequency in patients and controls [34]. There is no evidence from literature that the −83G>A and −126T>C variations are associated with either the steroid response or POAG [34]. Further, the majority (96%) of the steroid responders examined by Fingert et al. [34] harbored no variations that met the criteria for potential involvement in the steroid response phenotype, clearly indicating that variations in the proximal promoter are not a common cause of glaucoma. All these five changes of MYOC identified in this study are listed as a neutral polymorphisms and no pathogenic variation in MYOC was observed in this study.
Two changes (GGC375ins and GGC447ins) in FOXC1 were observed in cases as well as in controls. The frequency of these FOXC1 variations were found to be statistically non-significant (p>0.05) in our study. Both of these changes have already been described in the literature as neutral polymorphisms [20,35,36]. The presence of mutations in the FOXC1 gene in patients with PCG was initially described in an independent study conducted by Chakrabarti et al. [20] in 2009 from south India. In addition, FOXC1 mutations have been implicated in anterior segment dysgenesis (ASD) such as iridogoniodysgenesis, Axenfeld-Rieger syndrome (ARS) and Peter’s anomaly that progress to glaucoma in 50% to 75% of cases [37-40]. Some of these ASD cases are associated with congenital or early-onset glaucomas, whereas some have glaucoma secondary to anterior segment anomalies [39-41]. Pathogenic FOXC1 mutations were identified in PCG cases from southern India [20]. These findings indicate a potential role of the transcription factor FOXC1 in the development of ocular tissues including the drainage structures. Since neither CYP1B1 nor MYOC could explain the overall genetic contribution to PCG in earlier studies, we have also analyzed FOXC1. However, no pathogenic mutations were identified in MYOC and FOXC1 gene in our patients. It, therefore, gives us a premise to assume the non-involvement of both MYOC and FOXC1.
This difference in mutation spectrum of north Indian and south Indian population may be explained on the basis of different evolutionary history/ethnicity of both populations. India has a heterogeneous population with people from north and south India being ethnically different. The ethno-linguistic composition of the population of India, Pakistan, Bangladesh, Nepal, Bhutan, Maldives, and Sri Lanka mostly falls within two large groups; Dravidian and Indo-Aryan. These groups are further subdivided into numerous subgroups, castes, and tribes. Indo-Aryans form the predominant ethno-linguistic group in Pakistan, India (the central, eastern, western, and northern regions), Nepal, Sri Lanka, and the Maldives. Dravidians form the predominant ethno-linguistic group in southern India and the northern and eastern regions of Sri Lanka (The Indian Genome Variation database [IGVDb]) [42]. The north Indian population is predominantly Aryan population while the south Indian population is Dravidian with totally different morphological phenotype and genetic background. Recently it has been shown that ‘Ancestral North Indians’ (ANI), are genetically close to Middle Easterners, Central Asians, and Europeans, whereas the other, the ‘Ancestral South Indians’ (ASI), is distinct from ANI and East Asians as they are from each other [43]. Thus, though mutations have been identified in MYOC and FOXC1 in PCG cases from south India [16,20], these two genes are not involved in PCG in our patients.
Conclusion
This is the first study from north India showing non-involvement of MYOC and FOXC1 in the pathogenesis of primary congenital glaucoma. Thus, it is important to screen other loci for involvement in congenital glaucoma in cases which are either negative or heterozygous for CYP1B1 mutations to have a better insight in disease pathogenesis.
Acknowledgments
This work was financially supported by Department of Biotechnology, Govt. of India and a financial grant from AIIMS. The authors would like to thank the families of the patients for their cooperation. The author Mukesh Tanwar is a senior research fellow (SRF) of University Grants Commission (UGC), Govt. of India. The fellowship from UGC is gratefully acknowledged.
References
- 1.deLuise VP, Anderson DR. Primary infantile glaucoma (congenital glaucoma). Surv Ophthalmol. 1983;28:1–19. doi: 10.1016/0039-6257(83)90174-1. [DOI] [PubMed] [Google Scholar]
- 2.Sarfarazi M, Stoilov I. Molecular genetics of primary congenital glaucoma. Eye. 2000;14:422–8. doi: 10.1038/eye.2000.126. [DOI] [PubMed] [Google Scholar]
- 3.Gencik A, Gencikova A, Ferak V. Population genetical aspects of primary congenital glaucoma. I. Incidence, prevalence, gene frequency, and age of onset. Hum Genet. 1982;61:193–7. doi: 10.1007/BF00296440. [DOI] [PubMed] [Google Scholar]
- 4.Dandona L, Williams JD, Williams BC, Rao GN. Population-based assessment of childhood blindness in southern India. Arch Ophthalmol. 1998;116:545–6. [PubMed] [Google Scholar]
- 5.Sarfarazi M, Akarsu AN, Hossain A, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS. Assignment of a locus (GLC3A) for primary congenital glaucoma (buphthalmos) to 2p21and evidence for genetic heterogeneity. Genomics. 1995;30:171–7. doi: 10.1006/geno.1995.9888. [DOI] [PubMed] [Google Scholar]
- 6.Akarsu AN, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS, Sarfarazi M. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum Mol Genet. 1996;5:1199–203. doi: 10.1093/hmg/5.8.1199. [DOI] [PubMed] [Google Scholar]
- 7.Stoilov IR, Sarfarazi M. The third genetic locus (GLC3C) for primary congenital glaucoma (PCG) maps to chromosome 14q24.3. ARVO Annual Meeting; 2002 May 5–10; Fort Lauderdale (FL). [Google Scholar]
- 8.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6:641–7. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 9.The human gene mutation database. The Institute of Medical Genetics, Cardiff, Wales, UK (http://www.hgmd.cf.ac.uk/ac/gene.php?gene_CYP1B1 Accessed 11 August 2010.
- 10.Plasilova M, Stoilov I, Sarfarazi M, Kadasi L, Ferakova E, Ferak V. Identification of a single ancestral CYP1B1 mutation in Slovak Gypsies (Roms) affected with primary congenital glaucoma. J Med Genet. 1999;36:290–4. [PMC free article] [PubMed] [Google Scholar]
- 11.Bejjani BA, Stockton DW, Lewis RA, Tomey KF, Dueker DK, Jabak M, Astle WF, Lupski JR. Multiple CYP1B1 mutations and incomplete penetrance in an inbred population segregating primary congenital glaucoma suggest frequent de novo events and a dominant modifier locus. Hum Mol Genet. 2000;9:367–74. doi: 10.1093/hmg/9.3.367. [DOI] [PubMed] [Google Scholar]
- 12.Tanwar M, Dada T, Sihota R, Yadav U, Das TK, Dada R. Mutation spectrum of CYP1B1 in North Indian congenital glaucoma patients. Mol Vis. 2009;15:1200–9. [PMC free article] [PubMed] [Google Scholar]
- 13.Tanwar M, Dada T, Sihota R, Dada R. Identification of four novel CYP1B1 mutations (p.I94X, p.H279D, p.Q340H and p.K433K) in Congenital Glaucoma patients. Mol Vis. 2009;15:2926–37. [PMC free article] [PubMed] [Google Scholar]
- 14.Fingert JH, Heon E, Liebmann JM, Yamamoto T, Craig JE, Rait J, Kawase K, Hoh ST, Buys YM, Dickinson J, Hockey RR, Williams-Lyn D, Trope G, Kitazawa Y, Ritch R, Mackey DA, Alward WL, Sheffield VC, Stone EM. Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Hum Mol Genet. 1999;8:899–905. doi: 10.1093/hmg/8.5.899. [DOI] [PubMed] [Google Scholar]
- 15.Gong G, Kosoko-Lasaki O, Haynatzki GR, Wilson MR. Genetic dissection of myocilin glaucoma. Hum Mol Genet. 2004;13:R91–02. doi: 10.1093/hmg/ddh074. [DOI] [PubMed] [Google Scholar]
- 16.Kaur K, Reddy AB, Mukhopadhyay A, Mandal AK, Hasnain SE, Ray K, Thomas R, Balasubramanian D, Chakrabarti S. myocilin gene implicated in primary congenitl glaucoma. Clin Genet. 2005;67:335–40. doi: 10.1111/j.1399-0004.2005.00411.x. [DOI] [PubMed] [Google Scholar]
- 17.Kubota R, Noda S, Wang Y, Minoshima S, Asakawa S, Kudoh J, Mashima Y, Oguchi Y, Shimizu N. A novel myocin-like protein (myocilin) expressed in the connecting cilium of the photoreceptor: molecular cloning, tissue expression and chromosomal mapping. Genomics. 1997;41:360–9. doi: 10.1006/geno.1997.4682. [DOI] [PubMed] [Google Scholar]
- 18.Karali A, Russell P, Stefani FH, Tamm ER. Localization of myocilin/trabecular meshwork–inducible glucocorticoid response protein in the human eye. Invest Ophthalmol Vis Sci. 2000;41:729–40. [PubMed] [Google Scholar]
- 19.Vincent AL, Billingsley G, Buys Y, Levin AV, Priston M, Trope G, Williams-Lyn D, Héon E. Digenic inheritance of early-onset glaucoma: CYP1B1 gene a potential modifier gene. Am J Hum Genet. 2002;70:448–60. doi: 10.1086/338709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrabarti S, Kaur K, Rao KN, Mandal AK, Kaur I, Parikh RS, Thomas R. The transcription factor gene FOXC1 exhibits a limited role in primary congenital glaucoma. Invest Ophthalmol Vis Sci. 2009;50:75–83. doi: 10.1167/iovs.08-2253. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura DY, Swiderski RE, Alward WLM, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–7. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 22.Walter MA. PITs and FOXes in ocular genetics: The Cogan lecture. Invest Ophthalmol Vis Sci. 2003;44:1402–5. doi: 10.1167/iovs.02-0618. [DOI] [PubMed] [Google Scholar]
- 23.Gould DB, Smith RS, John SW. Anterior segment development relevant to glaucoma. Int J Dev Biol. 2004;48:1015–29. doi: 10.1387/ijdb.041865dg. [DOI] [PubMed] [Google Scholar]
- 24.Wang WH, McNatt LG, Shepard AR, Jacobson N, Nishimura DY, Stone EM, Sheffield VC, Clark AF. Optimal procedure for extracting RNA from human ocular tissues and expression profiling of the congenital glaucoma gene FOXC1 using quantitative RT-PCR. Mol Vis. 2001;7:89–94. [PubMed] [Google Scholar]
- 25.Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, Nishimura DY, Alward WL, Hogan BL, John SW. Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet. 2000;9:1021–32. doi: 10.1093/hmg/9.7.1021. [DOI] [PubMed] [Google Scholar]
- 26.Zhuo YH, Wang M, Wei YT, Huang YL, Ge J. Analysis of MYOC gene mutation in a Chinese glaucoma family with primary open-angle glaucoma and primary congenital glaucoma. Chin Med J (Engl) 2006;119:1210–4. [PubMed] [Google Scholar]
- 27.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunyaev S, Ramensky V, Koch I, Lathe W., III Kondrashiv, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 29.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 30.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved Splice Site Detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 32.Sripriya S, Uthra S, Sangeetha R, George RJ, Hemamalini A, Paul PG, Amali J, Vijaya L, Kumaramanickavel G. Low frequency of myocilin mutations in Indian primary open-angle glaucoma patients. Clin Genet. 2004;65:333–7. doi: 10.1111/j.1399-0004.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- 33.Klett CP, Bonner TI. Identification and characterization of the rat M1 muscarinic receptor promoter. J Neurochem. 1999;72:900–9. doi: 10.1046/j.1471-4159.1999.0720900.x. [DOI] [PubMed] [Google Scholar]
- 34.Fingert JH, Clark AF, Craig JE, Alward WL, Snibson GR, McLaughlin M, Tuttle L, Mackey DA, Sheffield VC, Stone EM. Evaluation of the myocilin (MYOC) glaucoma gene in monkey and human steroid-induced ocular hypertension. Invest Ophthalmol Vis Sci. 2001;42:145–52. [PubMed] [Google Scholar]
- 35.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerbäck S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63:1316–28. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cella W, de Vasconcellos JPC, de Melo MB, Kneipp B, Costa FF, Longui CA, Costa VP. Structural assessment of PITX2, FOXC1, CYP1B1, and GJA1 genes in patients with Axenfeld-Rieger syndrome with developmental glaucoma. Invest Ophthalmol Vis Sci. 2006;47:1803–9. doi: 10.1167/iovs.05-0979. [DOI] [PubMed] [Google Scholar]
- 37.Reese AB, Ellsworth RM. The anterior chamber cleavage syndrome. Arch Ophthalmol. 1996;75:307–18. doi: 10.1001/archopht.1966.00970050309003. [DOI] [PubMed] [Google Scholar]
- 38.Lines MA, Kozlowski K, Walter MA. Molecular genetics of Axenfeld-Rieger malformations. Hum Mol Genet. 2002;11:1177–84. doi: 10.1093/hmg/11.10.1177. [DOI] [PubMed] [Google Scholar]
- 39.Aldinger KA, Lehmann OJ, Hudgins L, Chizhikov VV, Bassuk AG, Ades LC, Krantz ID, Dobyns WB, Millen KJ. FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat Genet. 2009;41:1037–42. doi: 10.1038/ng.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strungaru MH, Dinu I, Walter MA. Genotype-phenotype correlations in Axenfeld-Rieger malformation and glaucoma patients with FOXC1 and PITX2 mutations. Invest Ophthalmol Vis Sci. 2007;48:228–37. doi: 10.1167/iovs.06-0472. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura DY, Swiderski RE, Alward WLM, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–7. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 42.The Indian Genome Variation Consortium The Indian Genome Variation database (IGVdb): a project overview. Hum Genet. 2005;118:1–11. doi: 10.1007/s00439-005-0009-9. [DOI] [PubMed] [Google Scholar]
- 43.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–94. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]