Abstract
IL-23 regulation is a central event in the pathogenesis of the inflammatory bowel diseases. We demonstrate that IFN-γ has anti-inflammatory properties in the initiation phase of IL-23–mediated experimental colitis. IFN-γ attenuates LPS-mediated IL-23 expression in murine macrophages. Mechanistically, IFN-γ inhibits Il23a promoter activation through altering NF-κB binding and histone modification. Moreover, intestinal inflammation is inhibited by IFN-γ signaling through attenuation of Il23a gene expression. In germ-free wild-type mice colonized with enteric microbiota, inhibition of colonic Il23a temporally correlates with induction of IFN-γ. IFN-γR1/IL-10 double-deficient mice demonstrate markedly increased colonic inflammation and IL23a expression compared with those of IL-10−/− mice. Colonic CD11b+ cells are the primary source of IL-23 and a target for IFN-γ. This study describes an important anti-inflammatory role for IFN-γ through inhibition of IL-23. Converging genetic and functional findings suggest that IL-23 and IFN-γ are important pathogenic molecules in human inflammatory bowel disease.
The inflammatory bowel diseases (IBDs) result from inappropriately directed inflammatory responses to the enteric microbiota in a genetically susceptible host. Key participants in the innate immune response to the enteric microbiota are macrophages (1). Of the inflammatory genes induced in macrophages, IL-12 family members play a central role in mediating intestinal inflammation. IL-12 and IL-23 are heterodimeric cytokines composed of a common p40 subunit (Il12b) and a p35 and p19 (Il23a) subunit, respectively (2). Recently IL-23 has been strongly implicated in the pathogenesis of human IBD (3).
IL-23, unlike IL-12, promotes a distinct CD4+ T cell phenotype characterized by the production of the cytokine IL-17, denoted Th17 cells. IL-23 enhances Th17 function and survival by acting on differentiated Th17 cells that express the IL-23 receptor. Development of Th1, Th2, and Th17 cells are mutually exclusive because differentiation of one subset is inhibited by the presence of another (4). Indeed, IFN-γ, the signature Th1 cytokine induced by IL-12, inhibits Th17 development (4). IFN-γ strongly synergizes with bacterial products to activate and sustain production of IL-12 by dendritic cells and macrophages (5). Importantly, IFN-γ as a proinflammatory cytokine has been implicated in the pathogenesis of multiple chronic inflammatory conditions, including IBD (1). In this study, we demonstrate that IFN-γ is a negative regulator of IL-23 in murine macrophages and experimental colitis.
Materials and Methods
Mice
Wild-type (WT), IL-10−/−, and IFN-γ R1−/− mice on the C57BL/6 background were matched for age in all experiments. IFN-γR1/IL-10−/− mice were obtained by crossing IL-10−/− and IFN-γR1−/− mice. Heterozygous offspring were then bred to obtain homozygous IFN-γR1/IL-10−/− mice. Littermates were used as controls. 129S6/SvEv germ-free (GF) mice (WT and IL-10−/−) were Cesarian-derived and maintained at the Gnotobiotic Facility at the University of North Carolina. Mice were housed in accordance with guidelines from the American Association for Laboratory Animal Care and Research Protocols, and experiments were approved by the Institutional Animal Care and Use Committee of the University of North Carolina.
General reagents and methods
Murine IL-12 p40, IL-12 p70, IFN-γ (R&D Systems, Minneapolis, MN) and IL-23 (eBioscience, San Diego, CA) ELISA kits were used according to the manufacturers’ instructions. The Il23a 1.8 kb luciferase reporter plasmid was provided Y.H. Chen (University of Pennsylvania School of Medicine) (6). Quantitative real-time PCR was performed as described previously (7).
Bone marrow-derived macrophages
Bone marrow-derived macrophages (BMMs) were harvested and cultured as described (7).
Transient transfections
BMMs were transiently transfected using AMAXA Nucleofector Technology (AMAXA, Walkersville, MD) using the protocol for murine macrophages. Transfection efficiencies of >50% are routinely obtained (data not shown). Luciferase activity was determined as described (7).
Colonic tissue explant cultures
Colonic sections were processed as described previously (7). Supernatants were collected after 24 h for cytokine ELISAs.
Colonic macrophages
Lamina propria mononuclear cells were isolated from mouse colon by an enzymatic method and density gradient centrifugation, as described previously (8). Lamina propria mononuclear cells were further separated into CD11b+ cells using anti-CD11b microbeads (Miltenyi Biotec, Auburn, CA). Purity was >90% by flow cytometric analysis (data not shown).
Histology
Colitis scoring was performed as described (7). Histological scores were determined by a pathologist (T.R.) blinded to experimental protocols.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) was performed with ChIP-IT Express kit (Active Motif, Carlsbad, CA) according to the manufacturer’s protocol. DNA–protein complexes were immunoprecipitated with Abs (RelA, p50, rabbit polyclonal IgG, Santa Cruz Biotechnology, Santa Cruz, CA; acetylated core histone H4 [H4Act], Upstate Biotechnology, Lake Placid, NY). Real-time PCR primers for the Il23a promoter and the Nuc1 region of Il12b promoter were used to amplify immunoprecipitated and input DNA (diluted 10-fold).
Statistical analysis
Statistical significance from experiments in cells was determined using Wilcoxon signed-rank tests. Results were considered statistically significant if p < 0.05. Statistical significance for in vivo data was assessed by the Mann-Whitney U test (SPSS, Chicago, IL) with Bonferroni correction.
Results and Discussion
IFN-γ inhibits LPS-induced IL-23 in murine macrophages
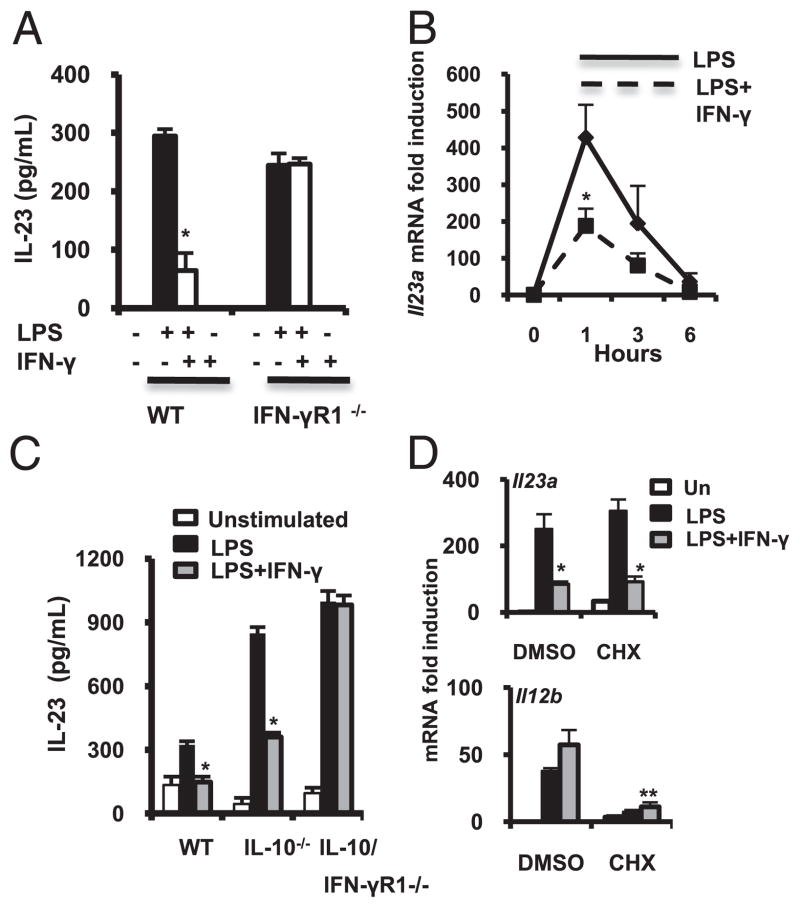
In BMMs from C57BL/6 mice, LPS-induced IL-23 protein secretion and IFN-γ significantly inhibited LPS-stimulated IL-23 (Fig. 1A). IFN-γ–mediated inhibition of IL-23 was in notable contrast to enhancement of LPS-induced IL-12 p40 and IL-12 p70 by IFN-γ (Supplemental Fig. 1A, 1B). Consequently, in LPS-activated BMMs from IFN-γR1−/− mice, a loss of IFN-γ–mediated IL-23 inhibition (Fig. 1A) and IL-12 (Supplemental Fig. 1A, 1B) induction were observed.
FIGURE 1.
IFN-γ negatively regulates LPS-mediated IL-23 expression in macrophages. A, BMMs from WT and IFN-γR1−/− mice were cultured in the presence of LPS (100 ng/ml, black bars) with or without IFN-γ (10 ng/ml, white bars). Supernatants were analyzed for IL-23 by ELISA. Results are expressed as mean ± SEM of three independent experiments. *p < 0.05 versus LPS-stimulated BMMs. B, WT BMMs were stimulated with LPS (100 ng/ml, solid line) and IFN-γ (10 ng/ml, dash line). Il23a mRNA expression was quantified by real-time RT-PCR. Results are expressed as fold induction normalized to β-actin. Error bars represent mean ± SEM of three independent experiments. *p < 0.05 versus LPS-stimulated BMMs. C, BMMs from WT, IL-10−/−, and IFN-γR1/IL-10/−/− mice were cultured with LPS (black bars, 100 ng/ml) plus IFN-γ (gray bars, 10 ng/ml). Supernatants were analyzed for IL-23 by ELISA. Results are expressed as mean ± SEM of three independent experiments. *p < 0.05 versus LPS-stimulated BMMs. D, WT BMMs were incubated for 30 min with DMSO or CHX (5 μg/ml) and then stimulated with LPS (100 ng/ml, black bars) ± IFN-γ (10 ng/ml, grey bars). Il23a (upper panel) and Il12b (bottom panel) mRNA was analyzed by real-time RT-PCR after 1 h. Results are expressed as fold induction normalized to β-actin. Error bars represent mean ± SEM of three independent experiments. *p < 0.05 versus LPS-stimulated BMMs.
Il23a (IL-23 p19) mRNA expression was rapidly induced by LPS by 1 h. IFN-γ inhibited LPS-activated Il23a (Fig. 1B) and augmented Il12b (IL-12 p40) expression (Supplemental Fig. 1C). Il23a expression levels returned to baseline by 6 h, whereas LPS plus IFN-γ–induced Il12b continued to rise (Supplemental Fig. 1C).
Whether effects of IFN-γ on IL-23 expression were dependent on the production of IL-10 was next determined. IL-10−/− macrophages demonstrate significantly enhanced LPS-induced IL-23 secretion compared with that of WT BMMs. IFN-γ–inhibited IL-23 expression in IL-10−/− BMMs, and inhibition was abrogated in IFN-γR1/IL-10−/− BMMs (Fig. 1C). IFN-γ–mediated synergistic induction of IL-12 p40 and IL-12 p70 (Supplemental Fig. 1D, 1E) was also abrogated in IL-10/IFN-γR1−/− BMMs.
Primary response genes have promoters that exist in an open chromatin structure and/or undergo rapid nucleosome remodeling. In contrast, secondary response genes have delayed induction kinetics, requiring ATP-dependent nucleosome remodeling and new protein synthesis prior to transcription initiation. In the presence of the protein synthesis inhibitor cyclohexamide (CHX), Il23a was rapidly induced by LPS in BMMs (Fig. 1D), characteristic of a primary response gene. CHX alone induced Il23a compared with vehicle-treated BMMs, as reported for other primary response genes (9). Unlike Il23a, Il12b induction is reduced in the absence of new protein synthesis, as described (9). In the presence of CHX, IFN-γ–mediated inhibition of LPS-induced Il23a is preserved. In contrast, Il12b induction by LPS and IFN-γ remains inhibited in the absence of new protein synthesis (Fig. 1D).
IFN-γ prevents RelA binding to the Il23a promoter
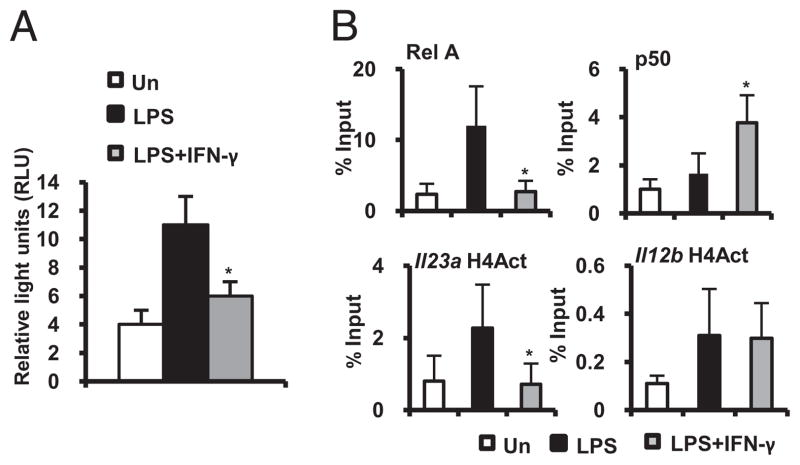
Two NF-κB sites have been reported to mediate LPS-induced Il23a promoter activity in murine macrophages (6). A 1.8 kb promoter luciferase reporter plasmid containing the NF-κB sites (κ1 −82 and κ2 −618 from the transcription initiation site) was transiently transfected into BMMs. LPS strongly induced and IFN-γ inhibited Il23a promoter activity (Fig. 2A). Inhibition of NF-κB transactivation by IFN-γ likely occurs at the Il23a promoter because IFN-γ fails to block phosphorylation of NF-κB p65 and degradation of IκBα (Supplemental Fig. 2).
FIGURE 2.
IFN-γ inhibits Il23a promoter activity. A, BMMs were transfected with an Il23a promoter luciferase reporter plasmid and cultured with LPS (100 ng/ml) ± IFN-γ (10 ng/ml) for 18 h. Reporter activity is represented as luciferase units normalized to heat shock protein promoter β-galactosidase activity. Data represent mean ± SEM of three independent experiments. *p < 0.05 versus LPS-stimulated Il23a promoter. B, Binding of RelA, p50, and H4Act to the distal NF-κB site on the endogenous WT Il23a promoter and H4Act at the nucleosome 1 position of the Il12b promoter was assessed by ChIP 1 h after incubation with LPS (100 ng/ml) ± IFN-γ (10 ng/ml). Real-time PCR was performed on anti-RelA, anti-p50, and anti-H4Act precipitated DNA samples, respectively. Results are presented as enrichment (percentage input) of RelA, p50, or H4Act DNA binding. Error bars represent mean ± SEM of three independent chromatin preparations from three independent experiments. *p < 0.05 versus LPS-stimulated BMMs
Next, BMMs were cultured with LPS plus IFN-γ, and occupancy of RelA on the distal (κ2) Il23a NF-κB binding site was analyzed by ChIP using PCR primers that span the Il23a promoter sequence from −549 to −680. RelA promoter occupancy was demonstrated 1 h after LPS stimulation. IFN-γ inhibited LPS-induced RelA recruitment to the Il23a promoter and enhanced the recruitment of NF-κB p50 (Fig. 2B, upper panels).
Histone acetylation is associated with transcriptionally active chromatin. The core histone H4 was acetylated 1 h after LPS stimulation at the distal Il23a NF-κB binding site. IFN-γ inhibited LPS-induced histone H4 acetylation (Fig. 2B, lower panels). In contrast, LPS plus IFN-γ stimulation was associated with histone H4 acetylation at an NF-κB site in the Il12b proximal promoter (Fig. 2B, lower panels). Therefore, IFN-γ may limit RelA access to the Il23a promoter by altering the dynamics of NF-κB subunit recruitment and by regulating covalent histone modifications.
These results provide new insights into transcriptional inhibition of Il23a. Il23a expression has markedly different kinetics of induction and is regulated through notably divergent mechanisms compared with those of another NF-κB–dependent gene, Il12b. Where IFN-γ potently synergizes with bacterial products for optimal induction of Il12b gene expression (10), IFN-γ inhibits LPS-mediated Il23a expression, surprisingly, through effects on NF-κB DNA binding and histone acetylation.
Enteric microbiota induce colonic IL-23 expression in experimental colitis
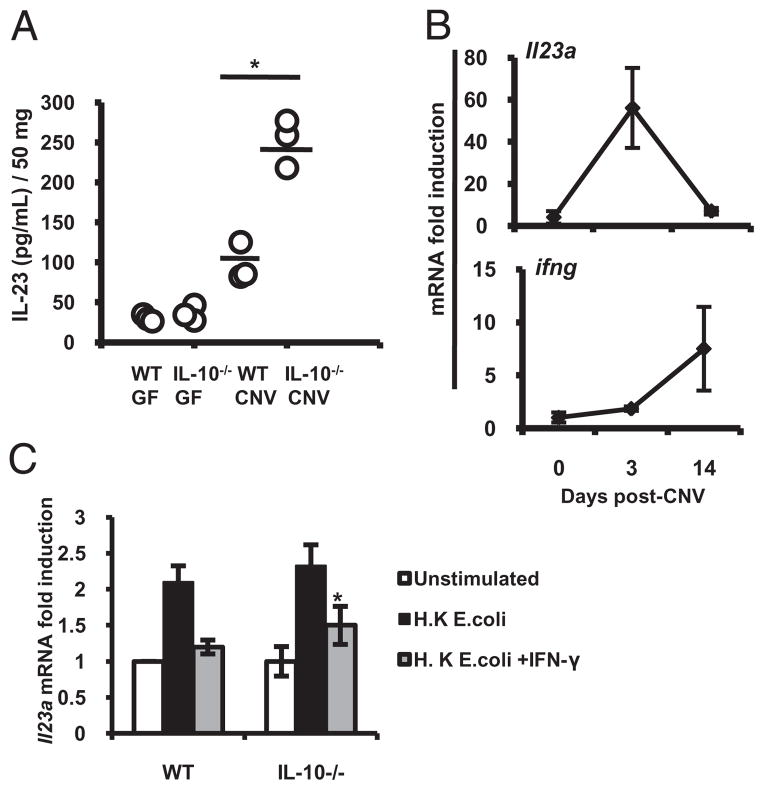
IL-10−/− mice develop chronic intestinal inflammation mediated by IL-23 (11) and dependent on the presence of the enteric microbiota (12). We investigated the role of the enteric microbiota in the regulation of mucosal IL-23 in WT and IL-10−/− mice raised germ-free (GF) and transitioned to a conventionalized (CNV) specific pathogen free microbiota at 8 wk of age. Two weeks after transition, colonic explants from CNV IL-10−/− mice secreted significantly more IL-23 (Fig. 3A) than GF WT, GF IL-10−/−, and CNV WT mice.
FIGURE 3.
Enteric microbiota induce colonic IL-23 expression in experimental colitis. WT and IL-10−/− mice raised in GF conditions were colonized with enteric microbiota from CNV mice at 8 wk of age. A, IL-23 secretion at day 14 postcolonization in colonic explant cultures was determined by ELISA. B, Colonic Il23a (upper panel) and ifng mRNA (bottom panel) expression was detected by real time RT-PCR in GF WT mice (day 0) and at days 3 and 14 postcolonization. Each time point includes three individual mouse colons and is representative of three independent experiments. C, CD11b+ LPMCs were isolated from WT and IL-10−/− mouse colons. CD11b+ LPMCs were activated with heat-killed E. coli ± IFN-γ (10 ng/ml). Il23a and β-actin mRNA expression was detected by real-time RT-PCR. Results are expressed as fold induction normalized to β-actin. Error bars represent mean ± SEM of three independent experiments. *p < 0.05 versus heat-killed E. coli-stimulated IL-10−/− CD11b+ LPMCs.
WT mice transitioned to a CNV microbiota revealed an increase in colonic Il23a mRNA after 3 d (Fig. 3B, top panel) that returned to baseline levels by day 14. Downregulation of colonic Il23a temporally correlated with increased colonic IFN-γ (ifng) mRNA (Fig. 3B, bottom panel). These results suggest that IFN-γ expression is a homeostatic checkpoint controlling the initiation of mucosal innate immune responses to the enteric microbiota. Consistent with these results, the enteric microbiota was recently shown to inhibit expression of IL-23 with subsequent effects on expansion and survival of Th17 cells in the colon (13).
In IL-10−/− mice, 2 wk after transition to a CNV microbiota, an increase in colonic Il23a and ifng mRNA was detected (Supplemental Fig. 3A). Increased colonic expression of Il23a correlated with the development of intestinal inflammation (Supplemental Fig. 3B). Therefore, in IL-10−/− mice, IFN-γ was insufficient to completely inhibit colonic Il23a expression.
IFN-γ inhibits Il23a expression in colonic CD11b+ lamina propria cells from IL-10−/− mice
Colonic CD11b+ lamina propria mononuclear cells (LPMCs) were the primary source of Il23a (Fig. 3C). IFN-γ inhibited heat-killed Escherichia coli-induced expression of Il23a in colonic CD11b+ WT and IL-10−/− LPMCs (Fig. 3C), whereas Il12b expression was not inhibited (Supplemental Fig. 3C). Thus, IFN-γ and IL-10 are negative regulators of IL-23 in colonic macrophages.
Increased mucosal expression of IL-23 correlates with severity of colonic inflammation in IFN-γR1/IL-10−/− mice
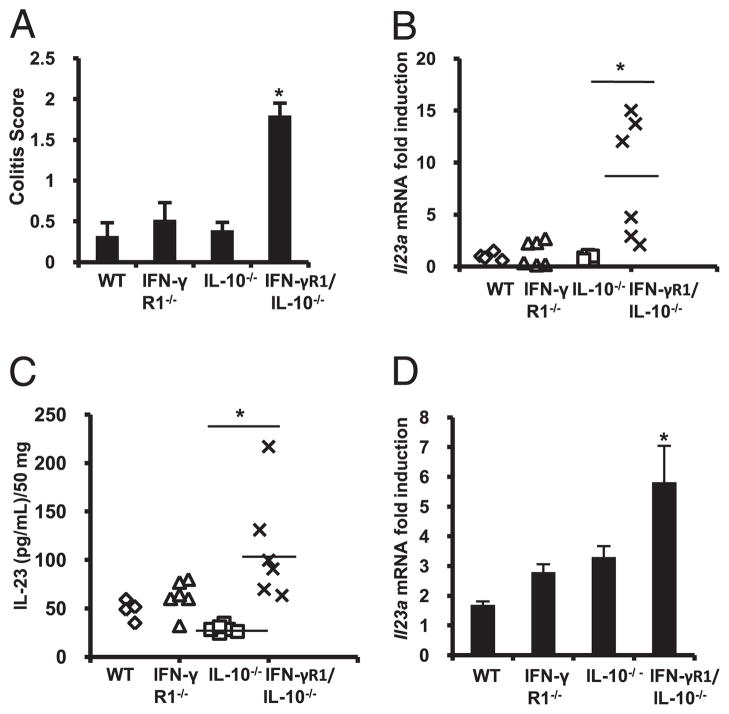
To understand functional consequences of IFN-γ deficiency in the development of colitis, colonic inflammation and IL-23 expression were determined in IL-10−/− and IFN-γR1/IL-10−/− mice. Eight-week-old IL-10−/− mice demonstrated minimal or no inflammatory changes. However, age-matched littermate IFN-γR1/IL-10/−/− mice developed significant colonic inflammation (Fig. 4A). Severity of colonic inflammation correlated with increased colonic Il23a expression (Fig. 4B) and IL-23 secretion (Fig. 4C) in colon explant cultures. There were no significant differences in colonic Il12b and Il12a expression between IL-10−/− and IFN-γR1/IL-10−/− mice (Supplemental Fig. 4A, 4B). Moreover, IFN-γ/IL-10−/− colonic CD11b+ LPMCs demonstrated increased heat-killed E. coli-activated Il23a induction compared with that of IL-10−/− CD11b+LPMCs (Fig. 4D). These results suggest that a primary defect in Il23a inhibition in colonic macrophages may mediate the development of severe IBD in IFN-γR1/IL-10−/− mice.
FIGURE 4.
Increased colonic inflammation and expression of IL-23 in IFNγR1/IL-10−/− mice. A, Colitis scores of WT, IFN-γR1−/−, IL-10−/−, and IFN-γR1/IL-10−/− mice at 8 wk of age. B, Colonic Il23a mRNA was examined by real-time RT-PCR. C, IL-23 protein in supernatants from colon explants cultures were analyzed using cytokine-specific ELISA and from WT, IFN-γR1−/−, IL-10−/−, and IFN-γR1/IL-10−/− mice. Results are expressed as mean ± SEM from four to six mice per group. *p < 0.05 versus IL-10−/− mice. D, CD11b+ LPMCs isolated from WT, IFN-γR1−/−, IL-10−/−, and IFN-γR1/IL-10−/− mice at 8 wk of age were activated with heat-killed E. coli, multiplicity of infection (10:1). Il23a mRNA expression was detected by realtime RT-PCR. Results are expressed as fold induction relative to unstimulated CD11b+ LPMCs normalized to β-actin and represent mean ± SEM of three independent experiments. *p < 0.05 versus heat-killed E. coli-stimulated IL-10−/− CD11b+ LPMCs.
These studies identify protective effects of IFN-γ in two models of experimental colitis. IL-10−/− mice on a C57BL/6 background raised in a CNV environment are relatively resistant to spontaneous colitis (14). In the absence of IFN-γ signaling, development of colitis is accelerated. To specifically test the role of the enteric microbiota in colitis initiation events, GF IL-10−/− mice colonized with microbiota were used. In this model, colitis is rapidly induced upon introduction of the enteric microbiota (15). A robust increase in colonic Il23a was observed in GF IL-10−/− mice upon colonization that correlated with colitis severity. Protective properties of IFN-γ have been described in other chronic inflammatory disease models. IFN-γ gene deletion or administration of anti–IFN-γ Abs leads to increased severity of experimental autoimmune encephalomyelitis and collagen-induced arthritis (16–18). A number of mechanisms have been proposed to explain protection afforded by IFN-γ in autoimmunity. For example, IFN-γ directly inhibits Th17 differentiation (19). Beyond T cell responses, little has been described about homeostatic effects of IFN-γ on innate immunity in chronic inflammation.
This study focused on events during the initiation of colitis. By utilizing GF mice transitioned to a CNV microbiota and through studies in IL-10−/−, IFN-γR1/IL-10−/−, and IRF-1/IL-10−/− mice, we have clarified mechanisms that may be operative at disease onset. It is possible that with long-standing inflammation, as in human IBD, other mechanisms become more relevant, and in fact IFN-γ may promote inflammation (20).
Regulation of Il23a is an important in vivo checkpoint to determine the subsequent T cell response. Hypothetically, as Th1 and Th17 responses are counterregulatory, IFN-γ may act upon the macrophage to attenuate Th17 responses through inhibition of IL-23. We also further implicate macrophage-derived IL-23 in the initiation of experimental colitis, highlighting the protective effects of IFN-γ signaling in IL-10−/− mice. Recently, genome-wide association studies in human IBD have elucidated contributions of single nucleotide polymorphisms located in relevant genomic loci, including the IL23R, IL12B (3), and IFNG (IFN-γ) genes (21). Thus, converging genetic and functional findings suggest that IL-23 and IFN-γ may be important therapeutic targets in human IBD.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants RO1 DK054452 (to S.E.P.), T32 DK007737 (to S.Z.S.), National Research Service Award F32 DK083186 (to S.Z.S.), P40 RR018603, and P30 DK034987 (Immunotechnologies, Gnotobiotic, and Histology Cores), and a Crohn’s and Colitis Foundation of America Research Fellowship Award (to K.M.).
Abbreviations used in this paper
- BMM
bone marrow-derived macrophage
- ChIP
chromatin immunoprecipitation
- CHX
cyclohexamide
- CNV
conventionalized
- GF
germ-free
- H4Act
acetylated core histone H4
- IBD
inflammatory bowel disease
- LPMC
lamina propria mononuclear cell
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, Robinson N, Buonocore S, Tlaskalova-Hogenova H, Cua DJ, Powrie F. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 4.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 5.Zhu C, Rao K, Xiong H, Gagnidze K, Li F, Horvath C, Plevy S. Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J Biol Chem. 2003;278:39372–39382. doi: 10.1074/jbc.M306441200. [DOI] [PubMed] [Google Scholar]
- 6.Carmody RJ, Ruan Q, Liou HC, Chen YH. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J Immunol. 2007;178:186–191. doi: 10.4049/jimmunol.178.1.186. [DOI] [PubMed] [Google Scholar]
- 7.Hegazi RA, Rao KN, Mayle A, Sepulveda AR, Otterbein LE, Plevy SE. Carbon monoxide ameliorates chronic murine colitis through a heme oxygenase 1-dependent pathway. J Exp Med. 2005;202:1703–1713. doi: 10.1084/jem.20051047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi T, Okamoto S, Hisamatsu T, Kamada N, Chinen H, Saito R, Kitazume MT, Nakazawa A, Sugita A, Koganei K, et al. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn’s disease. Gut. 2008;57:1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plevy SE, Gemberling JH, Hsu S, Dorner AJ, Smale ST. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Zaph C, Du Y, Saenz SA, Nair MG, Perrigoue JG, Taylor BC, Troy AE, Kobuley DE, Kastelein RA, Cua DJ, et al. Commensal-dependent expression of IL-25 regulates the IL-23–IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mähler M, Leiter EH. Genetic and environmental context determines the course of colitis developing in IL-10-deficient mice. Inflamm Bowel Dis. 2002;8:347–355. doi: 10.1097/00054725-200209000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Billiau A, Heremans H, Vandekerckhove F, Dijkmans R, Sobis H, Meulepas E, Carton H. Enhancement of experimental allergic encephalomyelitis in mice by antibodies against IFN-gamma. J Immunol. 1988;140:1506–1510. [PubMed] [Google Scholar]
- 17.Kelchtermans H, Struyf S, De Klerck B, Mitera T, Alen M, Geboes L, Van Balen M, Dillen C, Put W, Gysemans C, et al. Protective role of IFN-gamma in collagen-induced arthritis conferred by inhibition of mycobacteria-induced granulocyte chemotactic protein-2 production. J Leukoc Biol. 2007;81:1044–1053. doi: 10.1189/jlb.0806486. [DOI] [PubMed] [Google Scholar]
- 18.Vermeire K, Heremans H, Vandeputte M, Huang S, Billiau A, Matthys P. Accelerated collagen-induced arthritis in IFN-gamma receptor-deficient mice. J Immunol. 1997;158:5507–5513. [PubMed] [Google Scholar]
- 19.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer DM, Veldman GM, Banerjee S, Willis J, Levine AD. Distinct inflammatory mechanisms mediate early versus late colitis in mice. Gastroenterology. 2002;122:94–105. doi: 10.1053/gast.2002.30308. [DOI] [PubMed] [Google Scholar]
- 21.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, et al. Ulcerative colitis—risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.