Abstract
Background & Aims
The pharmacokinetics and pharmacodynamics of pegylated-interferon-α-2a (PEG-IFN) have not been described in HCV/HIV co-infected patients. We sought to estimate the pharmacokinetics and pharmacodynamics of PEG-IFN and determine whether these parameters predict treatment outcome.
Methods
Twenty-six HCV/human immunodeficiency virus (HIV)-co-infected patients were treated with a 48-week regimen of PEG-IFN (180 µg/week) plus ribavirin (11 mg/kg/day). HCV-RNA and PEG-IFN concentrations were obtained from samples collected until week 12. A modeling framework that includes pharmacokinetic and pharmacodynamic parameters was developed.
Results
Five patients discontinued treatment. Seven patients achieved a sustained virological response (SVR). PEG-IFN concentrations at day 8 were similar to steady-state levels (p = 0.15) and overall pharmacokinetic parameters were similar in SVRs and non-SVRs. The maximum PEG-IFN effectiveness during the first PEG-IFN dose and the HCV-infected cell loss rate (δ), was significantly higher in SVRs compared to non-SVRs (median 95% vs 86% [p = 0.013], 0.27 vs 0.11 day−1 [p = 0.006], respectively). Patients infected with HCV genotype-1 had a significantly lower average first-week PEG-IFN effectiveness (median 70% vs. 88% [p = 0.043]), however, 4- to 12-week PEG-IFN effectiveness was not significantly different compared to those with genotype-3 (p = 0.114). Genotype-1 had a significantly lower δ compared to genotype-3 (median 0.14 vs. 0.23 day−1 [p = 0.021]). The PEG-IFN concentration that decreased HCV production by 50% (EC50) was lower in genotype-3 compared to genotype-1 (median 1.3 vs. 3.4 [p = 0.034]).
Conclusions
Both the HCV infected cell loss rate (δ) and the maximum effectiveness of the first dose of PEG-IFN-α-2a distinguished HIV co-infected patients and were highly predictive of SVR. Further studies are needed to validate these viral kinetic parameters as early on-treatment prognosticators of response in patients with HCV and HIV.
Keywords: Viral kinetics, Mathematical modeling
Introduction
Approximately 25% of persons infected with human immunodeficiency virus (HIV) are also infected with hepatitis C virus (HCV), and the prevalence of co-infection is >70% in certain at-risk populations, such as injection drug users and those who have received contaminated blood products [1,2]. HCV and HIV co-infection affects approximately 300,000 individuals in the United States [1,3]. In Brazil, there are about 100,000 co-infected individuals including an estimated 17.5% prevalence in general HIV-outpatient units and a prevalence of 85% in patients who contracted HCV through injection drug use [4,5]. HCV infection is more rapidly progressive in patient with HIV co-infection and liver disease is a major cause of morbidity and mortality in persons with HIV [3,6]. Moreover, sustained virological response (SVR) rates to pegylated-interferon (PEG-IFN) and ribavirin are lower in HIV/HCV co-infected [7–10] than in HCV mono-infected patients [11,12]. Among difficult-to-treat co-infected genotype-1 patients, SVR rates range from 8–38% [7–10,13]. Higher SVR rates of 43% to 73% were observed in genotype 2 and 3 HCV [3]. Thus, understanding the determinants of successful treatment outcomes and the early identification of patients who are likely to achieve SVR are important clinical challenges that are particularly relevant to HIV/HCV co-infected patients.
Kinetic modeling of HCV RNA response during treatment has proven to be useful in understanding HCV dynamics and predicting the mode of action of interferon-α [14] and ribavirin [15] (see review [16]). These models generally have not incorporated the effects of time-varying interferon-α concentrations. Indeed, pharmacokinetic data from weekly PEG-IFN-α-2a or PEG-IFN-α-2b based treatments - the only approved PEG-IFN formulations against HCV infection - showed that serum concentration peaked and then declined during the first dosing interval [17–20]. Such fluctuations in PEG-IFN levels could give rise to changes in drug effectiveness, making it important to incorporate time-varying interferon-α concentrations in current mathematical models. While Talal et al.[18] and more recently Rozenberg et al. [21] have modeled pharmacokinetic and pharmacodynamic parameters of PEG-IFN-α-2b in HCV/HIV co-infected patients, to the best of our knowledge, a comprehensive pharmacokinetic and pharmacodynamic analysis of PEG-IFN-α-2a in HCV/HIV co-infected patients has not been performed.
In this study, we provide a detailed analysis of PEG-IFN-α-2a and HCV-RNA kinetics in 21 HCV/HIV co-infected patients who were treated with PEG-IFN-α-2a and ribavirin for 48 weeks. We demonstrate how a recent model of HCV infection that includes hepatocytes proliferation [22], can be applied in order to evaluate HCV kinetic parameters as well as the pharmacodynamic parameters of PEG-IFN. In addition, we show in which cases one should use this model (e.g., when triphasic viral decline is observed [23,24]), and in which cases the standard biphasic model of HCV infection [14], that incorporates pharmacokinetic and pharmacodynamic parameters can be used. Furthermore, frequent sampling allowed us to relate PEG-IFN-α-2a pharmacokinetics to HCV RNA kinetics, via mathematical modeling, and identified viral kinetic and pharmacodynamic parameters that differed between SVR and non-SVR cases, and genotype 1 and genotype 3 HCV.
Patients and methods
The study was conducted at the University of São Paulo Hospital das Clínicas and was approved by the ethics in research committee. All subjects gave written informed consent.
Patients and Therapy
Twenty-six HIV-HCV co-infected patients were included in the study. All participants had well-controlled HIV infection with a CD4 count >300 cells/m3 and HIV-RNA level <10,000 cp/ml with or without highly active anti-retroviral therapy (HAART) for at least six months prior to study entry (Table 1). None were HBsAg positive or had evidence of another cause of chronic liver disease. All patients had a liver biopsy within 1-year of enrolling in the study, which was evaluated by a single pathologist to whom the subjects’ identity and history was hidden.
Table 1.
Baseline Characteristics.
| Patient No. |
Genotype | race | Age (yr) |
gender | Weight (kg) |
CD4 (cells/mm3) |
HIV RNA (cp/ml) |
HAART | Metavir Fibrosis Stage |
|---|---|---|---|---|---|---|---|---|---|
| 1* | 3 | W | 50 | M | 70 | 700 | 245 | AZT/ LAM | F1A2 |
| 2* | 3 | W | 34 | M | 66 | 533 | 405 | no | F1A 2 |
| 3* | 3 | W | 40 | M | 76 | 653 | 1850 | AZT/ LAM/ EFZ | F2A2 |
| 6* | 3 | B | 32 | M | 67 | 441 | < 50 | no | F1A1 |
| 7* | 3 | W | 34 | F | 52 | 455 | 1590 | AZT/ LAM/ EFZ | F0A1 |
| 4* | 1 | B | 56 | M | 60 | 611 | 325 | D4T/ LAM/ NFV | F4A1 |
| 5* | 1 | B | 37 | M | 86 | 400 | 9930 | no | F2A2 |
| 8 | 3 | W | 42 | M | 92 | 327 | < 50 | AZT/ LAM/ NFV | F4A1 |
| 9 | 3 | W | 35 | M | 71 | 664 | < 50 | AZT/ LAM/ EFZ | F2A2 |
| 13 | 3 | W | 39 | M | 89 | 436 | < 50 | AZT/ LAM/ NFV | F2A3 |
| 10 | 1 | B | 49 | F | 65 | 519 | 3920 | no | F0A1 |
| 11 | 1 | W | 39 | F | 76 | 587 | 170 | AZT/ LAM/ NFV | F0A1 |
| 12 | 1 | B | 43 | M | 60 | 612 | < 50 | AZT/ LAM/ NFV | F4A1 |
| 14 | 1 | W | 42 | M | 65 | 672 | < 50 | D4T/ LAM/ NVP | F2A2 |
| 15 | 1 | W | 35 | M | 69 | 690 | 1630 | no | F0A2 |
| 16 | 1 | B | 38 | M | 54 | 430 | < 50 | D4T / LAM/ LPVr | F1A2 |
| 17 | 1 | B | 47 | M | 58 | 929 | 792 | D4T/ LAM/ EFZ/ NFV | F1A2 |
| 18 | 1 | W | 39 | M | 79 | 579 | < 50 | AZT/ DDI/ NVP | F0A1 |
| 19 | 1 | W | 48 | M | 54 | 418 | 7350 | AZT/ LAM/ EFZ | F1A1 |
| 20 | 1 | B | 54 | M | 62 | 739 | < 50 | AZT/ LAM/ NVP | F0A1 |
| 21 | 1 | B | 36 | M | 65 | 956 | < 50 | AZT/ LAM/ NFV | F0A1 |
| p value | 0.056 @ | 1.0 | 0.37 | 1.0 | 0.8 | 0.5 | NS | 0.28 | 1.0 |
SVR. P values for the difference between SVRs and non-SVRs;
with Chi-square test p=0.026;
SD, standard deviation; W, white; B, black; M, male; HAART, highly active antiretroviral therapy; AZT, zidovudine; LAM, lamivudine; EFZ, efavirenz; D4T, stavudine; NFV, nelfinavir; LPVr, lopinavirritonavir; NVP, nevirapine; DDI, didanosine; Cirrhosis was defined based on Metavir fibrosis stage score, i.e., F4.
Patients received PEG-IFN-α-2a (180 µg/week) and weight based ribavirin (11 mg/kg/day) for 48 weeks. At the outset of treatment, patients were hospitalized for 48 hours in order to facilitate blood sample collection. To standardize the treatment regimen, each dose of PEG-IFN was given on Monday morning at 08:00. For the initial 4 weeks of treatment, patients took the PEG-IFN under direct observation. At each visit, ribavirin was dispensed in a quantity sufficient to last until the subsequent visit. Each patient received oral instructions and a written schedule for medication and sample collection. The end-of-treatment response and SVR were evaluated at 48 weeks and 72 weeks, respectively.
PEG-IFN-α-2a measurement
PEG-IFN-α-2a serum concentrations were measured at baseline, at 4, 8, 12, 18, 24, 30, 36 and 42 hours; on days 2, 3, 4, 7, 8, 15, 22, 29, 43, 57, and 84, using a quantitative sandwich interferon enzyme-linked immuno-sorbent assay (ELISA, Bender MedSystems Diagnostics GmbH, Vienna, Austria). Binding of (pegylated) interferon to a murine monoclonal antibody directed against interferon adsorbed onto micro wells was detected by an HRP-conjugated monoclonal anti-interferon antibody. Following 2 hours of incubation unbound complexes were removed by washing (three times) after which tetramethyl-benzidine was used to determine the amount of interferon in the sample. Absorbency was read using a spectro-photometer using 450 nm as the primary wave length. Standards were prepared from diluted series of PEG-IFN in normal human serum obtained from healthy volunteers. Patient sera and standards were tested in triplicate, on the same plate. Although optical densities obtained were related to a standard of pegylated interferon, the ELISA also may detect free recombinant interferon α-2a molecules and natural interferon-α. The detection limit of the assay is 35 pg/ml and less than 5% coefficient of variation.
HCV and HIV RNA Measurements and Sample Acquisition
Blood samples were collected as mentioned above. Serum was separated and aliquots were frozen at −80° C. Samples collected through day 84 (week 12 of treatment) were used for pharmacokinetic and pharmacodynamic analyses. All virologic assays were performed at TriCore Reference Laboratories (Albuquerque, NM). The COBAS TaqMan HCV test (Roche Molecular Systems Inc., Branchburg, NJ), with a detection range of 10 to 10,000,000 IU/ml was used to measure HCV RNA levels. Samples above the detection range were diluted and re-assayed. Hepatitis C genotyping was performed with the VERSANT genotype assay (LiPA; Bayer Corp., Tarrytown, NY). The COBAS Amplicor HIV-1 Monitor test (version 1.5; Roche Molecular Systems Inc.), with a detection range of 50 to 100,000 copies/ml, was used for HIV quantification (Table 1).
Mathematical Modeling
Pharmacokinetics
As detailed previously [18,25–27], the serum PEG-IFN concentration, C(t), after the first dose given at time t = 0, and before the second dose, is
| (Eq.1) |
where the PEG-IFN dose, D, reaches the injection site immediately, is absorbed into the blood with rate constant ka, is eliminated from the blood with rate constant ke, and is distributed through a volume Vd. Only, a fraction, F, of the drug is bioavailable. According to Eq. (1), C(t) is zero at t = 0, rises to a maximum concentration, Cmax, at t = tmax, and subsequently decreases to zero over an extended time period (t >tmax). It follows from Eq. (1) that
| (Eq.2) |
The area under the curve (AUC) was measured by summing the rectangle areas during the first dose (i.e., 7 days). Each area was measured by multiplying the average of two consecutive PEG-IFN concentrations by the time between the corresponding time points.
For longer time periods, i.e., for the administration of multiple doses, the C(t) solution[25] was used
| (Eq.3) |
where Id is the dosing interval and Nd = integer (t/Id)+1 is the number of doses until time t. The first dose administered at t = 0. We set Id = 7 as PEG-IFN was given every 7 days. Eq. 3 was also used to check if the estimated pharmacokinetic parameters derived from Eq. 1 can predict the PEG-IFN kinetics until week 12 of therapy.
Viral Dynamics
To analyze the observed HCV RNA decline patterns, the Neumann et al.[14] and the Dahari et al. [22] models were extended, respectively, by allowing the drug effectiveness, ε, to vary with PEG-IFN concentrations, C(t) (see Supplementary Material). To account for this variation in PEG-IFN effectiveness, we used the standard pharmacodynamic model [18, 26, 27]
| (Eq.4) |
where the constant EC50 is the PEG-IFN concentration at which the drug’s effectiveness in blocking viral production is half its maximum, n is a parameter called the Hill coefficient, which determines how steeply the effectiveness rises with increasing concentration, and the time delay τ takes into account that IFN must bind cellular receptors and initiate a signaling cascade before a drug affect is observed.
Data Fitting
Using nonlinear least-squares regression analysis (SPSS v. 17), we fitted the pharmacokinetic model (Eq. 1) to the PEG-IFN concentration for the first week to estimate ka7, ke7, and FD/Vd7 for each patient. In addition, we fitted the pharmacokinetic model for multiple doses, i.e., Eq. 3, to the PEG-IFN concentration for the first 12 weeks of treatment, using GraFit [28], to estimate the pharmacokinetic parameters (i.e., ka84, ke84, and FD/Vd84) of the PEG-IFN concentration C(t). Thus, we estimated the pharmacokinetic parameters for each patient based on data measured during the first week and during the first 12 weeks of PEG-IFN treatment.
As previously described by Neumann et al. [14], we first estimated the initial viral load, V0, and initial delay before viral decay, t0, and HCV clearance rate, c, by fitting a simple HCV kinetic model, i.e., Eq. 4 in Neumann et al.[14], to the initial decline phase of viral load before viral rebound (approximately 2 days post treatment initiation). These values, as well as the values of ka84, ke84 and FD/Vd84 estimated by the pharmacokinetic model, were fixed when the combined-pharmacodynamic models (see Eq. S1 and Eq. S2 in Supplementary Material) were fitted to the data. First, we fitted the combined-pharmacodynamic model that includes hepatocytes proliferation, i.e., Eq. S1, to each patient’s data and estimated the viral kinetic and pharmacodynamic parameters (see Table S1 in Supplementary Material). We did not fit Eq. S1 to the data of patients 3 and 15 in whom interferon-α concentration dropped from day 15 without evidence of non-adherence. We have recently showed that the estimate of productively HCV-infected cell loss, γ, is dependent on the fraction of HCV-infected hepatocytes among all susceptible hepatocytes, π, and the treatment effectiveness in slowing viral production, ε [29]. It was suggested that in cases of biphasic viral decline with ε~1 or π <40% the standard model of HCV infection [14] can be used to estimate δ. Thus, we re-fitted the data with a much simpler model (i.e., combined-biphasic-pharmacodynamic model, Eq. S2), to estimate δ, and EC50 whenever Eq. S1 predicted biphasic HCV decline (i.e, absence of a shoulder phase in viral kinetics [in silico] post first week of treatment), together with maximum first-dose PEG-IFN effectiveness, ε7max >95% or estimated fraction of HCV-infected hepatocytes, π <40% (Table S1). Additional information on the relationships among ε, π, and δ, can be found in Dahari et al. [29]. The delay τ was assumed to be equal to the initial delay t0. We fitted each patient’s data to the combined-pharmacodynamic models, i.e., Eq. S1 and S2, with the Hill coefficient, n, (range, 1 to 4) to determine the value of n that produced the best fit (i.e., the fit with the smallest residual sum of squares) for each patient. Data from patients 18, 19, and 21, in whom initial and/or final HCV RNA decline were not observed during 12 weeks of therapy, was not fitted to the combined-pharmacodynamic models. For the parameters estimated by Eq. S2, we calculated standard error, SE, (where ± ~2×SE corresponds to 95% confidence intervals) using the Marquardt algorithm [28,30].
Statistical Analysis
Medians and interquartile ranges (IQR), the difference between the third and first quartile, were calculated for all pharmacokinetic and pharmacodynamic parameters. Nonparametric methods were used to compare baseline, pharmacokinetic, and pharmacodynamic parameters among SVR, non-SVR, genotype 1 and 3 and race. To compare continuous variables and paired samples, we used a 2-tailed Mann-Whitney U test. To compare categorical variables, we used a 2-tailed Fisher exact test (SPSS v. 17). In all cases, a p value <0.05 was considered significant.
Results
Virological Response
Twenty-one patients completed the viral and pharmacokinetic portion of the study and provided frequent blood samples after the first dose and on a weekly basis thereafter (24 hr after each dose) until week 12 of therapy. We numbered the subjects according to their viral response: 1–7 had SVR, 8–18 relapsed (i.e., HCV RNA was undetectable at week 48 and relapsed during follow up), and 19–21 were nonresponders (i.e., HCV RNA was detectable at week 48). Five patients (subject numbers 22–26) were excluded from the analysis of PEG-IFN pharmacokinetics and pharmacodynamics because they dropped after completing 11 or 12 weeks of therapy. Their viral kinetics and baseline characteristics are shown in the supplementary material (Fig. S2 and Table S2).
Adherence to treatment was generally good based on patient self-reports. Seven of 26 patients (27%) achieved an SVR (Table 1). Only three patients (1, 2 and 7) were rapid virologic responders (RVR) with undetectable HCV RNA levels at week 4 (Fig. 2A), and they were all SVR and HCV genotype 3. Overall, RVR had a positive predictive value of 75% for SVR, a negative predictive value of 76%, a sensitivity of 43%, and a specificity of 93%. Ten patients (all genotype 3 subjects and 2 genotype 1) had complete early viral response (cEVR) with undetectable HCV RNA levels at week 12 (Fig. 2). Overall, cEVR had a positive predictive value of 60% for SVR, a negative predictive value of 91%, a sensitivity of 86%, and a specificity of 71%. At the end of treatment (week 48), HCV RNA was below detection (<10 IU/ml) in patients 1 through 18, whereas in patients 19–21 HCV RNA remained detectable (not shown). During follow up, patients 8–18 relapsed. Among baseline characteristics, HCV genotype 3 was associated with SVR (Table 1). HCV RNA level, HIV RNA level, CD4+ cell count, age, and weight were not significantly different between the SVR and non-SVR groups (Table 1).
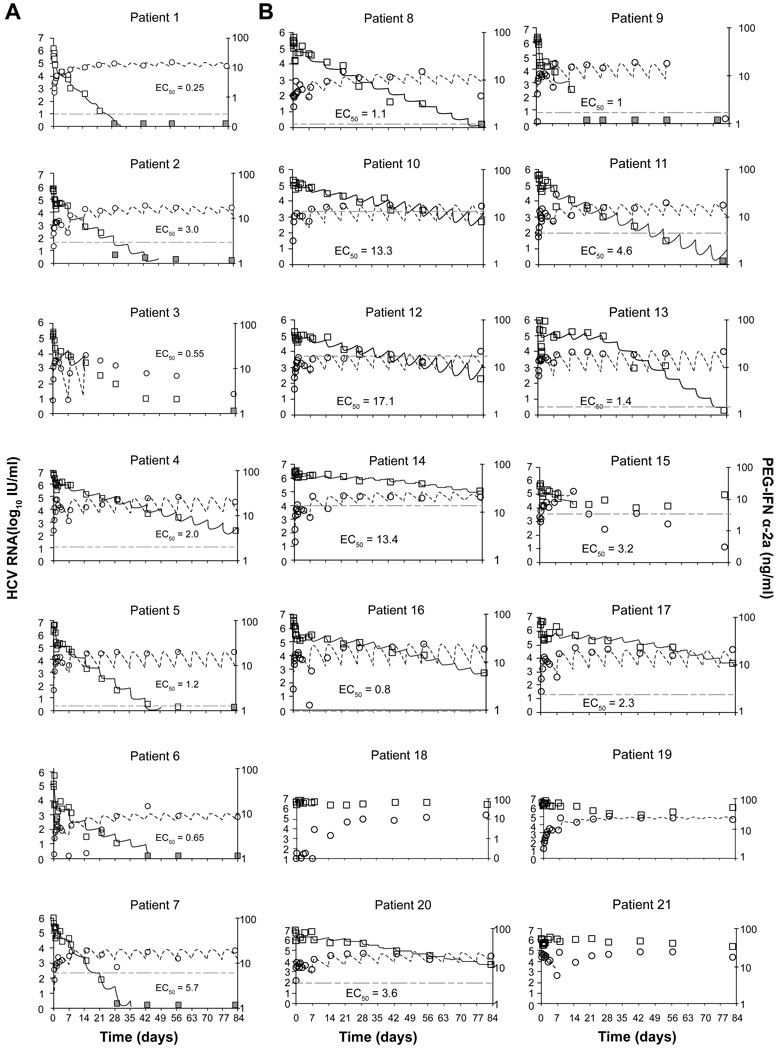
Fig. 2. PEG-IFN-α-2a serum concentrations and HCV-RNA levels during the first 12 weeks of treatment in (A) SVRs and (B) non-SVRs.
Graphs show drug concentration data (circles) and best-fit theoretical curve (Eq. 3, dashed line; right axis, ng/ml) and HCV RNA data (squares) and best-fit curve (solid line) (left axis, log10 IU/ml) from our combined triphasic or biphasic pharmacodynamic models (Eqs. S1 or S2, respectively; Table 3). Estimated EC50 is represented by horizontal dotted-dashed line. Gray squares indicate undetectable HCV RNA (<10 IU/ml). A 5-week version of these plots is shown in the Supplementary Material file.
Drug Concentration and Viral Profiles
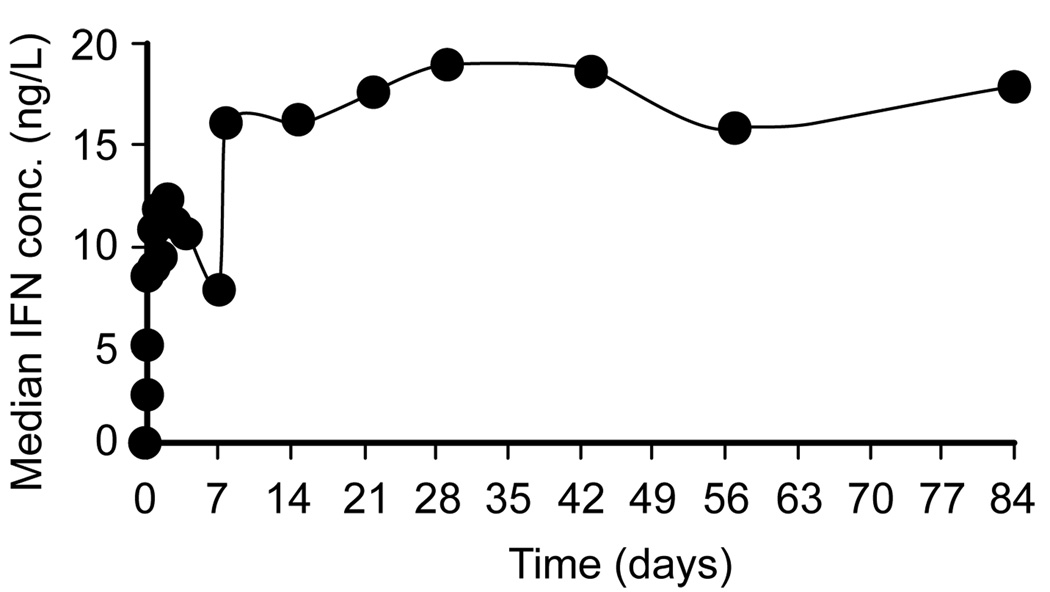
Five patients (8, 9, 12, 14, and 20) had baseline IFN concentrations above the detection limit (median 0.074 ng/ml) and all of them were non-SVR. Generally, PEG-IFN-α-2a concentration peaked and then declined during the first week of treatment (Fig. 1). On average, serum PEG-IFN levels peaked at 2.4 ± 1.8 days after the first dose at 12.9 ± 5.3 ng/ml, and decreased to 6.6 ± 3.6 ng/ml on day 7, immediately preceding the second dose. The pharmacokinetic profile during the first dose differed in four patients; PEG-IFN concentrations remained very low (<0.2 ng/ml) in patient 18 and rapidly increased during the first 1–2 days followed by a plateau, or increased until day 7 as in patients 7, 15 and 19 (not shown). The average PEG-IFN concentration during week 1 (AUC [0, 7 days]/7) was 8.9 ± 3.5 ng/ml and 8.8 ± 3.5 ng/ml in the SVR and non-SVR groups, respectively. Interestingly, at day 8 (24 hours after the second dose), the median (range) PEG-IFN concentration of 15.8 (27.0) ng/ml was similar to the median PEG-IFN concentration of 17.8 (26.0) ng/ml at week 12 (Fig. 1). Moreover, the median PEG-IFN concentrations at day 8 and week 12 were not associated with SVR.
Fig. 1. Median PEG-IFN-α-2a serum concentration during the first 12 weeks of treatment.
Generally, serum HCV RNA concentration changed inversely with PEG-IFN concentration during the first week of therapy. After a delay of 0.1–1.1 days, HCV RNA declined from baseline to a nadir of 0.96 ± 0.59 log10 at 1.7 ± 0.6 days. Thereafter, virus levels rebounded by 0.55 ± 0.43 log10 (from nadir) to peak at 3.2 ± 0.8 days and then decreased again by 0.33 ± 0.23 log10 at day 7. Following the second dose of PEG-IFN and until week 12, we identified 4 major viral kinetic patterns: (i) a second phase viral decline (patients 1–11 [Fig. 2], and 23–25 [Fig. S1]), (ii) a second phase of viral decline followed by a viral plateau or viral increase (patients 15 [Fig. 2B], 22 and 26 [Fig. S1]), (iii) a viral plateau or shoulder (for 1–3 weeks) followed by a final phase of viral decline (patients 12, 13, 14, 16, 17 and 20 [Fig. 2B]), and (iv) a viral plateau (patients 18 and 21 [Fig. 2B]) or late transient viral decline (Patient 19 [Fig. 2B]).
Pharmacokinetic Parameters
The availability of both PEG-IFN and HCV RNA concentrations, in each patient, allowed us to relate drug pharmacokinetics to viral kinetic profiles to gain a better understanding of viral-host-drug dynamics. We first estimated PEG-IFN absorption, ka7, and elimination, ke7, rate constants and FD/Vd7 (see Patients and Methods) by fitting Eq. 1 to the data of each patient during the first 7 days of therapy (Table 2). In addition, we fitted the multiple dose solution, i.e., Eq. 3, to the data of each patient during the first 84 days of treatment and re-estimated the pharmacokinetic parameters ka84, ke84, and FD/Vd84 (Table 2). Both fits (i.e., 7 days and 84 days) generated approximately the same parameter estimates with overlapping standard errors (Table 2). Only in patients 5, 7, 10, 13 and 17, the estimated parameters from fitting Eq. 1 to the 7-day data did not reproduce the PEG-IFN concentrations from day 7 to day 84 (not shown). Overall, none of the parameters were significantly different between the SVR and non-SVR groups (Table 2) or between genotype 1 and 3 (not shown). Interestingly, however, genotype 1 had a trend toward a higher Cmax7 (median (min-max) 12.5 (6.6–21.7) ng/ml) compared to genotype 3 (8.3 (5.8–15.4) ng/ml, [p = 0.06]. In addition, genotype 1 non-SVRs showed a trend (p = 0.09) toward a lower Cmax7 (median (min-max) 11.8 (6.6–21.7) ng/ml), compared to genotype 1 SVRs (16.7 (14.0–19.4) ng/ml).
Table 2.
Pharmacokinetic parameter estimates.
| Patient No. |
Genotype | FD/ Vd7 [SE] (ng/ ml) |
FD/ Vd84 [SE] (ng/ ml) |
ka7 [ SE] (1/ day) |
ka84 [SE] (1/day) |
ke7 [SE] (1/day) |
ke84 [SE] (1/day) |
tmax7a [SE] (day) |
tmax84a [SE] (day) |
C7a max [SE] (ng/ml) |
Cmax84a [SE] (ng/ml) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 12.8 [3.3] | 9.1 [0.8] | 0.46 [0.22] | 0.68 [0.12] | 0.17 [0.09] | 0.10 [0.01 ] | 3.4 | 3.3 | 7.2 | 6.6 |
| 2 | 3 | 10.1 [0.9] | 9.6 [0.5] | 1.61 [0.30] | 1.74 [0.27] | 0.11 [0.04] | 0.09 [0.01 ] | 1.8 | 1.8 | 8.3 | 8.1 |
| 3 | 3 | 15.0* | NF | 2.40 [0.64] | NF | 0.10 [0.04] | NF | 1.4 | NF | 13.1 | NF |
| 6 | 3 | 15.7 [7.7] | 5.3 [1.6] | 0.51 [0.23] | 1.39 [1.19] | 0.51 [0.17] | 0.08 [0.03 ] | 2.0 | 2.2 | 5.8 | 4.4 |
| 7 | 3 | LSE | 29.9 [4.4] | LSE | 0.34 [0.08] | LSE | 0.26 [ 0.04] | 3.4 | 12.6 | ||
| 4 | 1 | 36.8 [8.1] | 28.4 [8.3] | 0.73 [0.64] | 0.82 [0.37] | 0.32 [0.07] | 0.21 [0.06] | 2.0 | 2.2 | 19.4 | 17.7 |
| 5 | 1 | 15.5 [1.7] | 20.5 [2.5] | 2.42 [0.78] | 1.19 [0.27] | 0.07 [0.03] | 0.19 [0.03 ] | 1.5 | 1.8 | 14.0 | 14.4 |
| Median (IQR) |
15.0 (3.5) | 15.0 (20.6) | 1.27 (1.88) | 1.00 (0.88) | 0.11 (0.25) | 0.14 (0.13 ) | 2.0 (0.9) | 2.1 (1.5) | 11.3 (8.5) | 10.4 (9.2) | |
| 8 | 3 | 6.90 [2.7] | 7.0 [1.0] | 2.54 [0.98] | 2.01 [0.91] | 0.05 [0.17] | 0.09 [0.02 ] | 1.6 | 1.6 | 6.4 | 6.1 |
| 9 | 3 | 15.1 [1.9] | 15.4 [1.5] | 1.26 [0.31] | 1.26 [0.24] | 0.16 [0.04] | 0.16 [0.02 ] | 1.9 | 1.9 | 11.2 | 11.4 |
| 1 3 | 3 | 16.2 [0.8] | 24.4 [3.5] | 3.04 [0.85] | 1.14 [0.28] | 0.03 [0 03] | 0.23 [0.04 ] | 1.5 | 1.8 | 15.4 | 1 6.4 |
| 1 0 | 1 | 12.8 [0.6] | 15.4 [1.5] | 2.60 [0.57] | 1.64 [0.37] | 0.06 [0.03] | 0.16 [0.02 ] | 1.5 | 1.6 | 11.7 | 12.0 |
| 1 1 | 1 | 12.9 [3.1] | 14.0 [0.1] | 1.8 [0.81] | 1.09 [0.03] | 0.07 [0.06] | 0.13 [0.001 ] | 1.9 | 2.2 | 11.4 | 10.5 |
| 1 2 | 1 | 16.2 [4.1] | 21.9 [5.1] | 1.29 [0.58] | 0.79 [0.28 | 0.10 [0.07] | 0.21 [0.05 ] | 2.2 | 2.3 | 13.1 | 13.5 |
| 1 4 | 1 | 13.8 1.7] | 15.2 [1.9] | 1.86 [0.60] | 1.62 [0.51] | 0.09 [0.04] | 0.13 [0.02 ] | 1.7 | 1.7 | 11.8 | 12.2 |
| 1 5 | 1 | 6.91 [1.5] | NF | 1.35 [0.36] | NF | 10−5 [0.07] | NF | 3.7 | NF | 6. 6 | NF |
| 1 6 | 1 | 23.5 [5.8] | 24.4 [3.5] | 1.07 [0.47] | 1.14 [0.28] | 0.34 [0.16] | 0.23 [0.04 ] | 1.6 | 1.8 | 13.7 | 16.4 |
| 1 7 | 1 | 21.6 [2.0] | 19.4 [2.0] | 0.92 [0.14] | 1.08 [0.20] | 0.24 [0.03] | 0.19 [0.02 ] | 2.0 | 2.0 | 13.5 | 13.5 |
| 1 8 | 1 | ND | N D | ND | ND | ND | ND | ND | N D | ND | ND |
| 1 9 | 1 | 15.0* | 16.4 [2.3] | 0.37 [0.02] | 0.33 [0.07] | 0.08 [0.01] | 0.10 [0.02 ] | 5.3 | 5.2 | 9.8 | 9.8 |
| 2 0 | 1 | 11.8 [0.7] | 11.9 [0.1] | 4.21 [1.08] | 3.88 [1.21] | 0.05 [0.02] | 0.10 [0.01 ] | 1.1 | 1.0 | 11.2 | 10.9 |
| 2 1 | 1 | 23.2 [1.2] | NF | 9.90 [2.97] | NF | 0.16 [0.03] | NF | 0.4 | NF | 21.7 | NF |
| Median (IQR) |
15.0 (6.5) | 15.4 (7.9) | 1.80 (1.66) | 1.14 (0.56) | 0.08 (0.11) | 0.16 (0.11 ) | 2.0 (0.6) | 1.8 (0.6) | 12.1 (3. 1 ) | 11.9 (3.0) | |
| p value | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
fixed due to large standard error [SE]; NF, Eq. 3 cannot be fitted due to continuous decline (patients 3 and 15) or large fluctuations (patient 21) of interferon-alpha concentration; ND, IFN concentration remained below 0.2 ng/ml during the first 7 days; LSE, large standard error. IQR, interquartile range, the difference between the third and first quartile;
calculated by Eq. 2; NS, not significant.
Viral Kinetic and Pharmacodynamic Parameters
The modeling results (Table 3 and Fig. 2) indicated that the maximum PEG-IFN effectiveness during the first week of therapy, ε7max, and from week 4 to week 12 (where PEG-IFN reached its highest concentration), εmax, and δ were significantly higher in SVR than in non-SVR cases (Table 3). An ε7max value of 95% had a positive predictive value of 100% for SVR, a negative predictive value of 73%, a sensitivity of 43%, and a specificity of 100%. A δ value of 0.26 (1/day) had a positive predictive value of 100% for SVR, a negative predictive value of 79%, a sensitivity of 57%, and a specificity of 100%. Patients infected with HCV genotype 1 had significantly lower first-week PEG-IFN effectiveness, ε7aver compared to genotype 3 (median (IQR) 70% (46%) vs. 88% (12%) [p = 0.043]), however, εmax was not significantly different (p = 0.114). In addition, genotype 1 patients had significantly lower δ compared to genotype 3 (median (IQR) 0.11 (0.08) vs. 0.21 (0.14) day−1 [p = 0.021]). The PEG-IFN concentration that decreased HCV production by 50% (EC50) was lower in genotype 3 compared with genotype 1 (median (IQR) 1.3 (2.1) vs. 3.4 (11.5) [p = 0.034]). If the analysis is restricted to HCV genotype 1: δ, ε7max and ε7aver were found to be higher (p = 0.034, p = 0.087 and p = 0.068, respectively) in SVRs than in non-SVRs. These parameters did not differ when the analysis was restricted to HCV genotype 3. Lastly, Black and White race did not have an effect on viral kinetic and pharmacodynamic parameters and was not associated with outcome of therapy.
Table 3.
Viral kinetic and pharmacodynamic parameter estimates.
| Patient No. |
Genotype | V0 [ SE] Log10 (IU/ml) |
t0 [ SE] (h) |
C [ SE] (1/day) |
Ec50a [SE] |
δa [ SE] (1/day) |
n | ε7max (%) |
εmax (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 * | 3 | 5.9 [0.2] | 7.2 [2.9] | 6.2 [2.9] | 0.25 [0.04] | 0.30 [0.02 ] | 1.0 | 97 | 98 |
| 2* | 3 | 5.7 [0.1] | 12.0 [3.0] | 6.0& | 2.97 [0.20] | 0.17 [0.01] | 2.6 | 94 | 99 |
| 3 * | 3 | 5.3 [0.1] | 13.2 [1.3] | 5.5 [0.7] | 0.55[0.0 4] | 0.14b | 1.0 | 96 | SF |
| 6* | 3 | 5.0 [0.4] | 6.9 [3.4] | 6.0& | 0.65 [0.15] | 0.13 [0.05] | 2.0 | 99 | 99 |
| 7* | 3 | 5.6 [0.2] | 15.0 [3.6] | 6.0& | 5.74 [2.16] | 0.40 [0.04] | 1.0 | 67 | 72 |
| 4* | 1 | 6.8 [0.04] | 9.3 [3.6] | 2.5 [0.8] | 2.0 | 0.27 | 1.0 | 91 | 93 |
| 5 * | 1 | 6.6 [0.1] | 15.0 [1.8] | 8.5 [2.6] | 1.24 [0.25] | 0.28 [0.02] | 1.2 | 9 5 | 97 |
| Median (IQR) |
5.7 (1.3) | 12.0 (7.8) | 6.0 (0.7) | 1.6 (3.1) | 0.28 (0.17) | 1.1 (1.2) | 95 (13) | 98 (11) | |
| 8 | 3 | 5.7 [0.03] | 4.0 [1.2] | 4.8 [0.9] | 1.1 | 0.18 | 1.0 | 86 | 91 |
| 9 | 3 | 6.1 [0.1] | 13.8 [2.1] | 5.9 [1.2] | 1.63 [0. 18] | 0.24 [0.04] | 1.0 | 8 7 | 89 |
| 13 | 3 | 6.1 [0.1] | 10.9 [1.7] | 5.8 [0.6] | 1.4 | 0.26 | 1.0 | 9 2 | 94 |
| 10 | 1 | 5.2 [0.1] | 17.5 [1.8] | 6.0& | 13.25 [3.15] | 0.09 [0.01] | 1.0 | 4 7 | 58 |
| 11 | 1 | 5.7 [0.03] | 14.0 [1.9] | 3.8 [0.9] | 4.63 [0.54] | 0.12 [0.01] | 2.0 | 86 | 93 |
| 12 | 1 | 5.2 [0.1] | 7.9 [4.2] | 6.0& | 17.12 [4.86] | 0.10 [0.01] | 1.0 | 43 | 54 |
| 14 | 1 | 6.5 [0.1] | 2 7.8 [1.9] | 6.0& | 13.4 | 0.11 | 1.0 | 47 | 62 |
| 15 | 1 | 5.7 [0.1] | 1 0.1 [1.7] | 6.0 [1.6] | 3.18 [0. 39] | 0.07 [0.03] | 1.3 | 72 | SF |
| 16 | 1 | 6.4 [0.2] | 7.9 [5.1] | 2.8 [1.1] | 0.8 | 0.11 | 1.0 | 95 | 96 |
| 17 | 1 | 6.4 [0.2] | 1 8.0 [2.4] | 8.1 [1.1] | 2.3 | 0.11 | 1.0 | 86 | 89 |
| 18** | 1 | 6.5 | NF | NF | NF | NF | NF | NF | NF |
| 19**, $ | 1 | 6.4 | NF | NF | NF | NF | NF | NF | NF |
| 20$ | 1 | 6.8 [0.2] | 0 [4.4] | 6.0& | 3.6 | 0.15 | 1.0 | 76 | 84 |
| 21$ | 1 | 6.1 [0.04] | 7.7 [0.6] | 8.2 [2.4] | NF | NF | NF | NF | NF |
| Median (IQR) |
6.1 (1.0) | 10.9 (9.9) | 6.0 (0.7) | 3.0 (12.0) | 0.12 (0.09) | 1.0 (0.2) | 86 (41) | 89 (32) | |
| p value | NS | NS | 0.085 | 0.011 | NS | 0.011 | 0.022 |
SVRs;
Null responders during 12 weeks of treatment;
due to large standard error [SE] we fixed c = 6 day−1 as previously estimated[14];
detectable HCV RNA at the end of treatment (week 48);
parameters that include SE for δ or EC50 were estimated using Eq. S2 (see Methods and Supplementary Material), the remaining cases were estimated by a mathematical model that includes hepatocytes proliferation, see Eq. S1 and Table S1 in Supplementary Material;
due to large SE the decay rate was directly estimated by linear regression from day 7 to day 15; SD, standard deviation; NF, model cannot not be fitted to the data as explained in Methods; SF, since interferon-α declined from day 15 model fit was performed until that time.
Discussion
We showed that the PEG-IFN-α-2a concentrations tended to peak around 2 days after the first dose, then decreased only slightly by day 7, and reached near maximal concentrations 24 hours after administration of the second dose (Fig. 1). However, neither PEG-IFN concentration nor the pharmacokinetic parameters were found to be different between SVRs and non-SVRs. Our results were in agreement with previous PEG-IFN-α-2a pharmacokinetic study in HCV genotype 1 mono-infected patients [17] (Neumann A.U. unpublished data). Interestingly, the lack of predictable pharmacokinetic factors was previously found in HCV/HIV co-infected patients treated with PEG-IFN-α-2b [18,21]. The results suggest that neither PEG-IFN α-2a nor α-2b concentrations can be used to predict treatment outcome. Using a model that includes hepatocyte proliferation (Eq. S1) we were able to estimate the fraction of HCV-infected hepatocytes [29,31], π, and predict the nature of the shoulder phase (or triphasic viral decay pattern [22]) seen in several patients. Interestingly, while a shoulder phase was not observed in SVRs, it appeared in 6 of 14 non-SVR patients (12, 13, 14, 16, 17 (who were relapsers), and 20 (who was a non-responder); Fig. 2B). Of note, HCV RNA was undetectable at the end of treatment in these patients except in patient 20. Given that in patients 13, 16, and 17, PEG-IFN concentration was much higher than EC50 (Fig. 2B), it is possible that these patients could have benefited from longer therapy. Among the relapsers it is interesting to note that patients 8, 9, and 11 seemed to have a good early viral response, i.e., HCV RNA dropped in a biphasic manner, high death/loss rate of HCV-infected cells (δ >0.12 day−1 as previously suggested [14]), high ratio between PEG-IFN concentration and EC50, and undetectable viral load at week 12 (Fig. 2B). It was surprising that these patients did not achieve SVR. One possibility could be the lack of continuous high drug effectiveness between weeks 12 and 48, as may have been the case for patient 9 in whom the PEG-IFN concentration fell below EC50 at week 12 (Fig. 2B). It was not possible to discern whether non-adherence played a role in such cases. In other relapse patients, such as 10, 12, 14 and 15, the fact that PEG-IFN concentration fluctuated close to the EC50 may be an indication that these patients could have benefitted from a higher dose of PEG-IFN or a more potent therapy.
Both Talal et al.[18] and Rozenberg et al. [21] described PEG-IFN-α-2b pharmacodynamic and viral-kinetic parameters that might identify patients likely to achieve SVR as early as 3 doses of treatment. In the current study, we found that PEG-IFN-α-2a pharmacodynamic and viral-kinetic parameter estimations during the first 3 doses of treatment (not shown) were similar to the 12-dose estimations (Table 3) in patients exhibiting biphasic viral decline using the standard model (Eq. S2). In cases of triphasic viral decline, later data points are needed in order to estimate δ with the model that includes the proliferation of hepatocytes (Eq. S1).
A primary challenge in HCV management is the identification of factors that distinguish between SVRs and non-SVRs, and that might be used for the early prediction of a successful treatment outcome, especially in difficult-to-treat HIV/HCV-co-infected patients [7–10,13,18,32]. In particular, HCV genotype 1 and 3 pose a challenge due to lower SVR rates for genotype 1 and recent data suggesting an association between genotype 3 and more rapid progression of fibrosis [33]. Here, we used HCV dynamic models that incorporate PEG-IFN-α-2a pharmacodynamic and pharmacokinetic parameters to reproduce the changes in effectiveness of drug during weekly doses. We found that the maximum PEG-IFN effectiveness during the first PEG-IFN dose, ε7max, and the HCV-infected cell loss rate, δ, were significantly higher in SVRs compared with non-SVRs; an ε7max value of 95% or a δ value of 0.26 (1/day) had positive predictive values of 100% for SVR. Patients infected with HCV genotype 1 had significantly lower δ compared to genotype 3 and the drug concentration that decreased HCV production by 50% (EC50) was lower in genotype 3 compared with genotype 1. These findings provide insight into determinants of SVR in HIV/HCV-co-infected patients and identify potential early predictors of response that should be more fully evaluated in larger studies. Lastly, it is important to note that triphasic viral decline has been observed with combination therapy including new direct acting agents (e.g., Kieffer et al. [34] and Banbang et al. [35]), Therefore, our study provides a framework for future modeling studies and may help in developing more effective treatment strategies for hepatitis C.
In conclusion, the HCV infected cell loss rate (δ) and the maximum effectiveness of the first dose of PEG-IFN (ε7max) differed by genotype and were predictive of SVR in HIV co-infected patients. These key viral kinetic and pharmacodynamic parameters were estimated as early as two weeks of therapy using conventional modeling in patients with a biphasic response. In contrast, estimation of δ in patients with a triphasic response, which includes a flat phase between the first phase and the final phase of viral decline, requires use of a new model that includes hepatocyte proliferation and a more prolonged monitoring. Further studies are needed to validate these viral kinetic parameters as early on-treatment prognosticators of response in patients with HCV and HIV.
Supplementary Material
Acknowledgments
The authors thank Lisette Provacia, Norma Paula Cavalheiro and Carlos Eduardo Melo for excellent technical assistance, and the patients and their family members for the trust and participation.
Financial Support
H.D is supported by the University of Illinois Gastrointestinal and Liver Disease (UIC GILD) Association. Grant support (AAB and ESAA): Study carried out with the assistance of Roche Laboratories of Brazil, which provided financial support in the form of the pegylated interferon α-2a, kits for molecular assays and the shipping of samples.
List of Abbreviations
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- PEG-IFN
Pegylated interferon
- NPV
negative predictive value
- PPV
positive predictive value
- HAART
highly active antiretroviral therapy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
ESAA has received support for travel to international conferences, honoraria for scientific activities and research support from Roche Brazil.
References
- 1.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US Adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–837. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 2.Thomas DL. Hepatitis C and human immunodeficiency virus infection. Hepatology. 2002;36:S201–S209. doi: 10.1053/jhep.2002.36380. [DOI] [PubMed] [Google Scholar]
- 3.O'Leary JG, Chung RT. Management of hepatitis C virus coinfection in HIV-infected persons. AIDS Read. 2006;16:313–316. 318–320. [PubMed] [Google Scholar]
- 4.Ferreira P, Navarro R, Araujo E, Barone A, Hepatite C. In: Ed M, editor. Volume 1. 2009. pp. 280–308. [Google Scholar]
- 5.Ferreira P, Navarro R, Araujo E, Barone A. Boletim Epidemiológico Aids. Brazil: Health Ministry; 2009. [Google Scholar]
- 6.Bica I, McGovern B, Dhar R, Stone D, McGowan K, Scheib R, et al. Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;32:492–497. doi: 10.1086/318501. Epub 2001 Jan 2023. [DOI] [PubMed] [Google Scholar]
- 7.Chung RT, Andersen J, Volberding P, Robbins GK, Liu T, Sherman KE, et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-co-infected persons. N Engl J Med. 2004;351:451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, Lissen E, Gonzalez-Garcia J, Lazzarin A, et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–450. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 9.Carrat F, Bani-Sadr F, Pol S, Rosenthal E, Lunel-Fabiani F, Benzekri A, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. Jama. 2004;292:2839–2848. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 10.Ballesteros AL, Franco S, Fuster D, Planas R, Martinez MA, Acosta L, et al. Early HCV dynamics on Peg-interferon and ribavirin in HIV/HCV co-infection: indications for the investigation of new treatment approaches. Aids. 2004;18:59–66. doi: 10.1097/00002030-200401020-00007. [DOI] [PubMed] [Google Scholar]
- 11.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 12.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 13.Laguno M, Murillas J, Blanco JL, Martinez E, Miquel R, Sanchez-Tapias JM, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for treatment of HIV/HCV co-infected patients. Aids. 2004;18:F27–F36. doi: 10.1097/00002030-200409030-00003. [DOI] [PubMed] [Google Scholar]
- 14.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 15.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 16.Dahari H, Shudo E, Ribeiro RM, Perelson AS. Mathematical modeling of HCV infection and treatment. Methods Mol Biol. 2009;510:439–453. doi: 10.1007/978-1-59745-394-3_33. [DOI] [PubMed] [Google Scholar]
- 17.Levy-Drummer RS, Haagmans B, Soulier A, Germanidis G, Lurie Y, Hezode C, et al. Pharmacodynamic modeling of HCV kinetics during PEG-interferon-alfa-2A (40kD) and ribavirin treatment of chronic hepatitis C genotype 1 patients in the DITTO-HCV study [Abstract] Hepatology. 2004;40 Suppl 390A. [Google Scholar]
- 18.Talal AH, Ribeiro RM, Powers KA, Grace M, Cullen C, Hussain M, et al. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV co-infected sustained virological responders from nonresponders. Hepatology. 2006;43:943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 19.Glue P, Fang JW, Rouzier-Panis R, Raffanel C, Sabo R, Gupta SK, et al. Pegylated interferon-alpha2b: pharmacokinetics, pharmacodynamics, safety, and preliminary efficacy data. Hepatitis C Intervention Therapy Group. Clin Pharmacol Ther. 2000;68:556–567. doi: 10.1067/mcp.2000.110973. [DOI] [PubMed] [Google Scholar]
- 20.Matthews SJ, McCoy C. Peginterferon alfa-2a: a review of approved and investigational uses. Clin Ther. 2004;26:991–1025. doi: 10.1016/s0149-2918(04)90173-7. [DOI] [PubMed] [Google Scholar]
- 21.Rozenberg L, Haagmans BL, Neumann AU, Chen G, McLaughlin M, Levy-Drummer RS, et al. Therapeutic response to peg-IFN-alpha-2b and ribavirin in HIV/HCV co-infected African-American and Caucasian patients as a function of HCV viral kinetics and interferon pharmacodynamics. Aids. 2009;23:2439–2450. doi: 10.1097/QAD.0b013e32832ff1c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;46:16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 23.Dahari H, Shudo E, Ribeiro RM, Perelson AS. Mathematical modeling of HCV RNA kinetics. In: Tang H, editor. Hepatitis C Protocols. 2nd ed. NJ: Humana Press; 2009. [Google Scholar]
- 24.Herrmann E, Lee JH, Marinos G, Modi M, Zeuzem S. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003;37:1351–1358. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 25.Dixit NM, Perelson AS. Complex patterns of viral load decay under antiretroviral therapy: influence of pharmacokinetics and intracellular delay. J Theor Biol. 2004;226:95–109. doi: 10.1016/j.jtbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Welling P. Pharmacokinetics: Processes and Mathematics. Washington, DC: American Chemical Society; 1986. [Google Scholar]
- 27.Gabrielson J. Pharmacokinetic and Pharmacodynamic Data Analysis: Concepts and Applications. Stockholm: Swedish Pharmaceutical Press; 2000. [Google Scholar]
- 28.Leatherbarrow RJ. GraFit Version 6. Horley, U.K: Erithacus Software Ltd; 2007. [Google Scholar]
- 29.Dahari H, Shudo E, Cotler SJ, Layden TJ, Perelson AS. Modelling hepatitis C virus kinetics: the relationship between the infected cell loss rate and the final slope of viral decay. Antiviral Therapy. 2009;14(3):459–464. [PMC free article] [PubMed] [Google Scholar]
- 30.Marquardt DW. J. Soc. Ind. Appl. Math. 1963;11:431–441. [Google Scholar]
- 31.Dahari H, Layden-Almer JE, Kallwitz E, Ribeiro RM, Cotler SJ, Layden TJ, et al. A mathematical model of hepatitis C virus dynamics in patients with high baseline viral loads or advanced liver disease. Gastroenterology. 2009;136(4):1402–1409. doi: 10.1053/j.gastro.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahari H, Markatou M, Zeremski M, Haller I, Ribeiro RM, Licholai T, et al. Early ribavirin pharmacokinetics, HCV RNA and alanine aminotransferase kinetics in HIV/HCV co-infected patients during treatment with pegylated interferon and ribavirin. J Hepatol. 2007;47:23–30. doi: 10.1016/j.jhep.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bochud PY, Cai T, Overbeck K, Bochud M, Dufour JF, Mullhaupt B, et al. Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J Hepatol. 2009;51:655–666. doi: 10.1016/j.jhep.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46:631–639. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 35.Adiwijaya BS, Hare B, Caron PR, Randle JC, Neumann AU, Reesink HW, et al. Rapid decrease of wild-type hepatitis C virus on telaprevir treatment. Antivir Ther. 2009;14:591–595. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.