Abstract
In mammals, proliferation is rapid in many tissues during early postnatal life, causing rapid somatic growth. This robust proliferation is then suppressed as the animal approaches adult size, bringing many tissues to a quiescent state where proliferation occurs only as needed to replace dying cells. Recent evidence suggests that the mechanism responsible for this decline in proliferation involves a multi-organ genetic program. We hypothesized that this genetic program continues to progress into later adult life, eventually suppressing proliferation to levels below those needed for tissue renewal, thus contributing to aging. We therefore used expression microarray to compare the temporal changes in gene expression that occur in adult mouse organs during aging to those occurring as juvenile proliferation slows. We found that many of the changes in gene expression that occur during the aging process originate during the period of juvenile growth deceleration. Bioinformatic analyses of the genes that show persistent decline in expression throughout postnatal life indicated that cell-cycle-related genes are strongly over-represented. Thus, the findings support the hypothesis that the genetic program that slows juvenile growth to limit body size persists into adulthood and thus may eventually hamper tissue maintenance and repair, contributing to the aging process.
Keywords: aging, postnatal growth, microarray, genetic program, proliferation
Introduction
Aging is generally regarded as a gradual decline in physiological function throughout the organism, decreasing the ability to respond to stress and homeostatic imbalance, and increasing susceptibility to disease. Despite its obvious relevance to human health, the biological basis of aging is not well understood. At the organ level, aging is thought to be associated with a progressive decline in cell proliferation (Burton 2009), one example being the decreasing capability of the aged liver to regenerate after partial hepatectomy (Stocker and Heine 1971; Timchenko 2009). This loss of renewal and regenerative potential may be in part due to decreased proliferative capacity of adult stem cells (Yui et al. 1998; Krtolica 2005; Sharpless and DePinho 2007) and to the accumulation of senescent cells with age (Jeyapalan et al. 2007).
Declining proliferative capacity in many tissues does not begin in mid or late adult life, as the aging process develops, but instead initiates much earlier (Chang et al. 2008). In embryonic and early postnatal life, many tissues show high replication rates, leading to rapid somatic growth of these organs. If this rate of proliferation were to remain constant, somatic growth would be exponential. Therefore, cellular mechanisms are required to slow this robust proliferation, such that, by adulthood, most tissues have entered a quiescent state where proliferation occurs only as needed to replace dying cells (Pellettieri and Sanchez 2007).
Recent evidence suggests that the mechanism responsible for this decline in juvenile cell proliferation involves a genetic program that is common to multiple organs (Lui et al. 2008; Finkielstain et al. 2009; Lui et al. 2010) and includes the downregulation of multiple growth-promoting genes with age. This genetic program appears to be driven by growth itself, since growth-inhibiting conditions delay the program (Finkielstain et al. 2009; Lui et al. 2010). Thus proliferation appears to cause downregulation growth-promoting genes, which in turn limits proliferation.
The developmental theory of aging proposes that aging could be caused in part by continuing actions of developmental processes, which eventually produce adverse effects in postreproductive life (de Magalhaes and Church 2005). In particular, there is evidence suggesting that cell proliferation beginning in early life may lead to damage or changes which in turn lead to a gradual loss renewal capacity, contributing to aging (de Magalhaes and Faragher 2008). Here, we considered the specific possibility that the mechanisms that progressively restrain juvenile growth may continue to progress into adult life, thus contributing to the decline in proliferative capacity associated with aging. Progression of the underlying proliferation-limiting genetic program might lead first to cellular quiescence in young adulthood, such that some cells can still proliferate when stimulated for tissue renewal, but then, during aging, continued progression of the program might further limit the ability of cells to proliferate to a level below that is needed for tissue renewal. This hypothesis, which links juvenile growth and aging, might provide an explanation for the observation that small mammals generally undergo both aging and suppression of juvenile growth on a far shorter time scale than do large mammals. Similarly, the hypothesis might help explain why conditions that inhibit proliferation such as caloric restriction, growth hormone deficiency, or insulin-like growth factor-I deficiency, might delay the genetic program and thus conserve proliferative capacity and slow aging (Mote et al. 1991; Brown-Borg et al. 1996; Coschigano et al. 2000; Flurkey et al. 2001; Smith et al. 2004; Spindler and Dhahbi 2007). Finally this hypothesis is consistent with the antagonistic pleiotropy theory, which proposes that aging results from genes that have beneficial effects in early life, when natural selection is strong, but harmful effects at a later age when selection pressure is weaker (Williams G.C. 1957).
As an initial test of this hypothesis, we used expression microarray to analyze changes in gene expression that occur in liver, kidney and lung of mice during aging. We then compared these changes to those occurring in early postnatal life, as somatic growth decelerates. We found that many of the changes in gene expression that occur during aging originate in early postnatal life, during the juvenile period of growth deceleration. Furthermore, bioinformatic analysis suggested that a subset of genes that show consistent changes in expression in multiple juvenile organs and also in aging organs regulate cell proliferation. Thus, the findings support the hypothesis that the genetic program that slows growth in juvenile life in order to limit adult body size persists into adulthood, and may eventually hamper maintenance and repair of multiple organs.
Materials and Methods
Animal procedures and tissue processing
C57BL/6 male mice at 1, 3, and 9 months of age were obtained from Charles River Laboratory (Charles River Laboratory, Wilmington, MA) and provided with regular chow (mouse chow 5R31, Purina LabDiet) and water ad libitum. All animals were maintained and used in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council 2003). Mice were killed by carbon dioxide inhalation at 3, 9 and 15 months of age. Liver, kidney, and lung were excised, homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA) and stored at −80°C for later use.
RNA Extraction and Purification
Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions followed by RNeasy Mini Kit purification (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. RNA concentration was determined by spectrophotometry at 260nm. All RNA samples had a 260/280 nm ratio between 1.9 and 2.1. RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and only high quality RNA (28S/18S ratio > 1.8) was used for further analysis.
Microarray Analyses
RNA was processed and analyzed by the NIDDK Core Facility at the National Institutes of Health using Affymetrix Mouse Genome 430 2.0 Array GeneChips (45,000 transcripts, Affymetrix, Santa Clara, CA). Three chips were used for each organ at each time point. Each microarray chip was used to analyze a pooled sample of RNA (2 μg) derived from three animals. Therefore, a total of 9 animals were used. This pooling served to decrease biological variability. Microarray signals were analyzed using the Affymetrix RMA algorithm. ANOVA was performed and FDR reports were generated using Partek Pro software (Partek, St. Charles, MO). The FDR was calculated collectively using data from all three organs. Pathway analyses were done using Ingenuity Pathways Analysis Software 7.1 (Ingenuity Systems Inc, Redwood City, CA). Correlation analyses (Pearson’s Pairwise Comparison and Spearman’s rank correlation) and heat maps were generated using JMP 8 software (SAS Institute Inc., Cary, NC).
Quantitative Real-time RT-PCR
Real-time PCR was used to assess specific mRNA levels in murine organs. Liver, kidney, and lung were dissected from C57BL/6 mice at 3, 9, and 15 months of age (n = 8–10 animals per time point). Total RNA (100–200 ng) was reverse transcribed using Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. The resulting cDNA solution was diluted 10-fold and stored at −20°C for later use. Quantitative real-time PCR was performed for 18S, Ccl5, Cxcl9, Peg3, and Ube2c using the following assays employing specific FAM or VIC-labeled (18S rRNA) TaqMan probes (Applied Biosystems, Foster City, CA): Ccl5, Mm01302427_m1; Cxcl9, Mm00434946_m1; Peg3, Mm01337379_m1; Ube2c, Mm00835439_g1; and 18S rRNA, 4319413E. Reactions were performed in triplicate on cDNA derived from individual animals, using the ABI Prism 7900 sequence detection system instrument (Applied Biosystems) according to the manufacturer’s instructions. The relative quantity of each mRNA was calculated using the formula: relative expression = 2−ΔCT ×106, where CT represents the threshold cycle and ΔCT = (CT of gene of interest) – (CT of 18S rRNA). Values were multiplied by 106 for convenience of comparison. Data are presented as mean ± SEM. The effect of age on relative expression was assessed by ANOVA.
Results
In this study, we analyzed the gene expression profile of aging C57BL/6 mice in liver, kidney and lung, representing organs with widely differing physiological functions and embryonic derivations. Time points were chosen to represent early (3-month-old), middle (9-month-old) and late (15-month-old) adulthood. The survival rate at 15 months of age was still 100% without apparent illness. We avoided time points close to the maximum life expectancy of these mice to reduce the possibility of picking up changes in gene expression associated with specific pathologic conditions rather than normal physiological aging.
Gene profiling of aging organs
Changes in gene expression during aging were analyzed using Affymetrix expression microarray that assesses 39,000 transcripts representing 34,000 genes in the mouse genome. We first identified genes that showed significant changes with age using a cutoff of P < 0.0077, which gave a false discovery rate of < 10%. An additional cutoff of fold change ≥1.5 was used, so that the genes included are more likely to have important physiological effects. In each organ, hundreds of genes changed significantly during aging, with more genes being upregulated with age than downregulated (Table 1). Using Ingenuity Pathway Analysis (IPA), we identified the biological pathways and molecular functions that were over-represented by these age-related genes. This analysis implicated more than 30 high-level biological functions that were being altered significantly with aging of each organ, many of which are disease related (Fig. 1). The levels of expression in aging organs and the fold-changes with age were generally consistent with similar prior microarray studies (Supplemental Table 1) (Misra et al. 2007; Schumacher et al. 2008).
Table 1.
Number of genes that showed significant change in expression (P < 0.0077, FDR < 10%, ≥1.5-fold) with age (3 to 15 month) in each organ
| Organ | Upregulated | Downreg ulated | Total |
|---|---|---|---|
| Liver | 381 (61%) | 247 (39%) | 628 |
| Kidney | 184 (71%) | 74 (29%) | 258 |
| Lung | 337 (64%) | 190 (34%) | 527 |
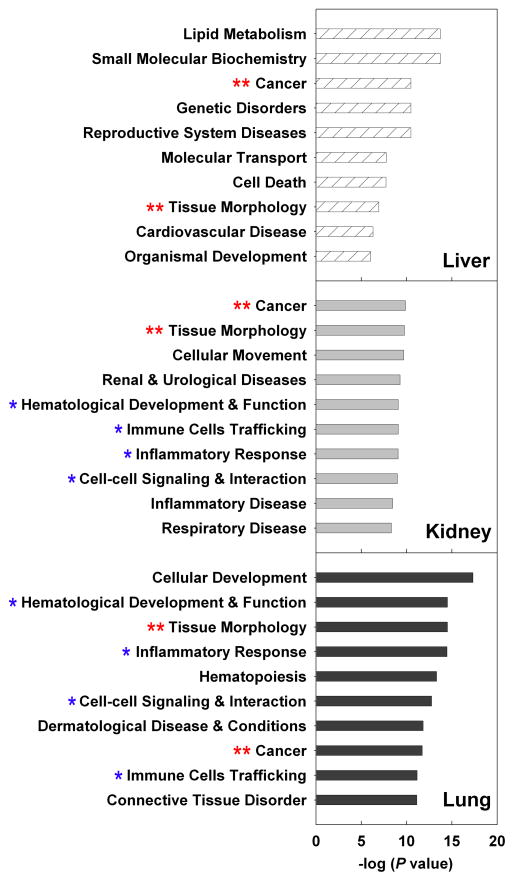
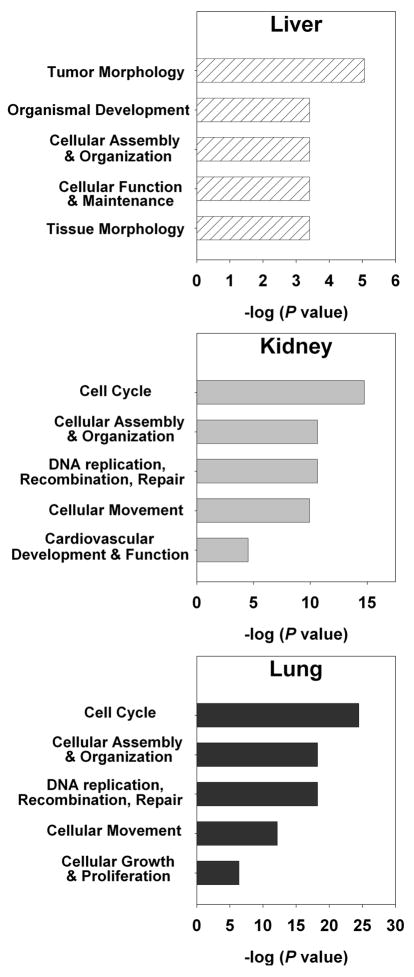
Fig 1.
Pathway analyses of genes significantly up- or downregulated during aging in liver (hatched bars), kidney (solid grey bars), or lung (solid black bars) using Ingenuity Pathway Analysis (IPA) 7.1. The 10 most overrepresented molecular, cellular, or physiological functions are shown. **, common biological function shared by all 3 organs; *, common biological function shared by kidney and lung.
In general, each organ showed a distinct pattern of age-related biological functions. For example, lipid metabolism was strongly over-represented only in the liver, while renal and urological disease was identified only in the kidney. However, we observed some similarities, particularly between kidney and lung. Amongst the ten most overrepresented functions in each organ, cancer and tissue morphology are shared between all three organs, while kidney and lung have additionally four functions in common (Fig. 1). Interestingly, we also observed in all three organs an increasing expression of multiple genes that are highly expressed in lymphocytes. One likely explanation for this observation is that infiltration of lymphocytes increases with age in many tissues (Phillips 1981; Hayashi et al. 1989; Baylis and Corman 1998; Singh et al. 2008).
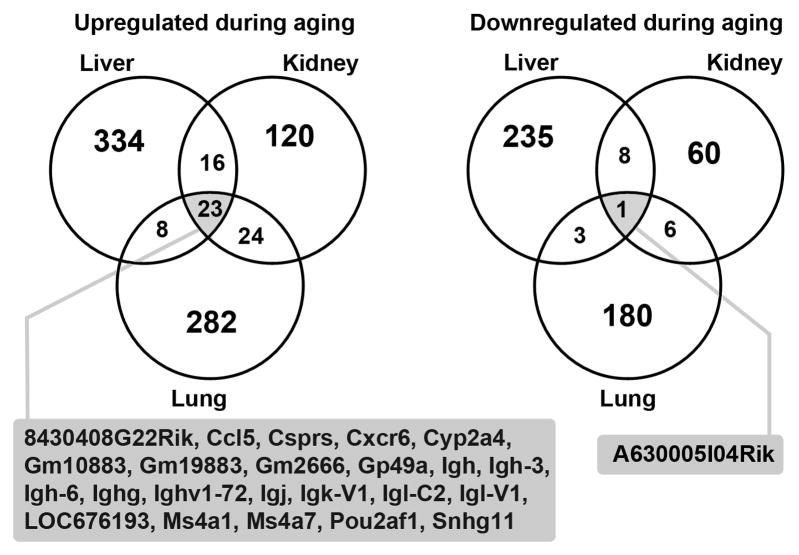
To further investigate whether there is a common genetic profile of aging among different organs, we performed correlation analysis, using two complimentary approaches. For example, to compare liver and kidney, we first focused on all genes that showed significant changes in expression in the liver and then asked whether similar changes were occurring in the kidney, using correlation analysis. Second, we focused on the genes that showed significant changes in expression in the kidney and asked whether similar changes were occurring in the liver. To minimize similarities attributable to infiltration of lymphocytes, we excluded all genes annotated by IPA to have immune-related functions from this analysis. As shown in table 2, there were positive correlations between each pair of organs assessed by both Pearson’s pairwise correlation (r value ranged from 0.17 to 0.58) and Spearman’s rank correlation (ρ value ranged from 0.14 to 0.43). However, as illustrated by a Venn diagram, (Fig. 2) only a small number of genes were upregulated (P < 0.0077, fold change ≥ 1.5) from 3 to 15 month in all three organs, and only one gene was substantially downregulated. Thus, the findings indicate that changes in gene expression that occur during aging are predominantly organ specific but also include a common component, shared between more than one organ.
Table 2.
Correlation between age-related changes in different organs
| Gene set analyzed: non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥1.5-fold) from 3 to 15 mo in liver (n=531) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Liver, 3 to 15 mo | vs | Kidney, 3 to 15 mo | r = 0.28 | ρ = 0.28 |
| P < 0.0001 | P < 0.0001 | |||
| Lung, 3 to 15 mo | r = 0.17 | ρ = 0.14 | ||
| P < 0.0001 | P = 0.0014 | |||
| Gene set analyzed: non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥ 1.5-fold) from 3 to 15 mo in kidney (n=178) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Kidney, 3 to 15 mo | vs | Liver, 3 to 15 mo | r = 0.47 | ρ = 0.26 |
| P < 0.0001 | P = 0.0004 | |||
| Lung, 3 to 15 mo | r = 0.58 | ρ = 0.43 | ||
| P < 0.0001 | P < 0.0001 | |||
| Gene set analyzed: non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥1.5-fold) from 3 to 15 mo in lung (n=381) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Lung, 3 to 15 mo | vs | Liver, 3 to 15 mo | r = 0.33 | ρ = 0.26 |
| P < 0.0001 | P < 0.0001 | |||
| Kidney, 3 to 15 mo | r = 0.50 | ρ = 0.39 | ||
| P < 0.0001 | P < 0.0001 | |||
Fig 2.
Venn diagrams showing the number of genes significantly upregulated and downregulated during aging (3 vs 15 mo) in mouse liver, kidney, and lung. Significant changes were defined by P < 0.0077, FDR < 10%, and ≥ 1.5-fold. Genes that showed significant up- or downregulation in all 3 organs are listed.
Similarities between aging and growth
The central hypothesis we considered here is that the mechanisms that progressively slow juvenile growth also contribute to the continued decline in proliferative capacity in aging organs. Taking advantage of our previous microarray studies done in 1-, 4-, and 8-wk old mice (heart, kidney and lung) (Finkielstain et al. 2009), we compared the genetic profile of the two biological processes, juvenile growth deceleration and aging.
We started by asking whether genes that showed a change in expression during aging also showed a change in the same direction during postnatal growth deceleration. Therefore, we performed correlation analysis between the aging dataset and the juvenile dataset, using the same gene sets that were used to compare overlap among different organs during aging, that is, non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥1.5-fold) from 3 to 15 month in each organ. There was a positive correlation between changes of gene expression during aging and juvenile growth (Table 3) in lung and kidney (3–15 mo versus 1–8 wk; lung, Pearson’s r = 0.45, Spearman’s ρ = 0.47; kidney, r = 0.24, ρ = 0.27; all P < 0.001). This similarity between growth and aging was not restricted to same-organ comparisons, as cross-organ comparisons also gave significant correlations (Table 3, for example 3–15 mo lung versus 1–4 wk heart, r = 0.32, ρ = 0.35, both P < 0.0001). Unlike the kidney and lung, the aging liver only showed weak but statistically significant correlation with juvenile organs, with r and ρ value close to 0.1.
Table 3.
Correlation between aging and postnatal growth (aging genes)
| Gene set analyzed: non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥1.5 -fold) from 3 to 15 mo in liver (n=531) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Liver, 3 to 15 mo | vs | Heart, 1 to 4wk | r = 0.10 | ρ = 0.10 |
| P = 0.02 | P = 0.02 | |||
| Kidney, 1 to 8wk | r = 0.13 | ρ = 0.13 | ||
| P = 0.003 | P = 0.003 | |||
| Lung, 1 to 8wk | r = 0.10 | ρ = 0.08 | ||
| P = 0.03 | P = 0.06 (not significant) | |||
| Gene set analyzed: non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥1.5-fold) from 3 to 15 mo in kidney (n=178) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Kidney, 3 to 15 mo | vs | Heart, 1 to 4wk | r = 0.21 | ρ = 0.24 |
| P = 0.006 | P = 0.001 | |||
| Kidney, 1 to 8wk | r = 0.24 | ρ = 0.27 | ||
| P = 0.001 | P = 0.0003 | |||
| Lung, 1 to 8wk | r = 0.28 | ρ = 0.26 | ||
| P = 0.0002 | P = 0.0005 | |||
| Gene set analyzed: non-immune-related genes that showed significant change (P < 0.0077, FDR < 10%, ≥ 1.5-fold) from 3 to 15 mo in lung (n=381) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Lung, 3 to 15 mo | vs | Heart, 1 to 4wk | r = 0.32 | ρ = 0.35 |
| P = <0.0001 | P = <0.0001 | |||
| Kidney, 1 to 8wk | r = 0.30 | ρ = 0.30 | ||
| P = <0.0001 | P = <0.0001 | |||
| Lung, 1 to 8wk | r = 0.45 | ρ = 0.47 | ||
| P = <0.0001 | P = <0.0001 | |||
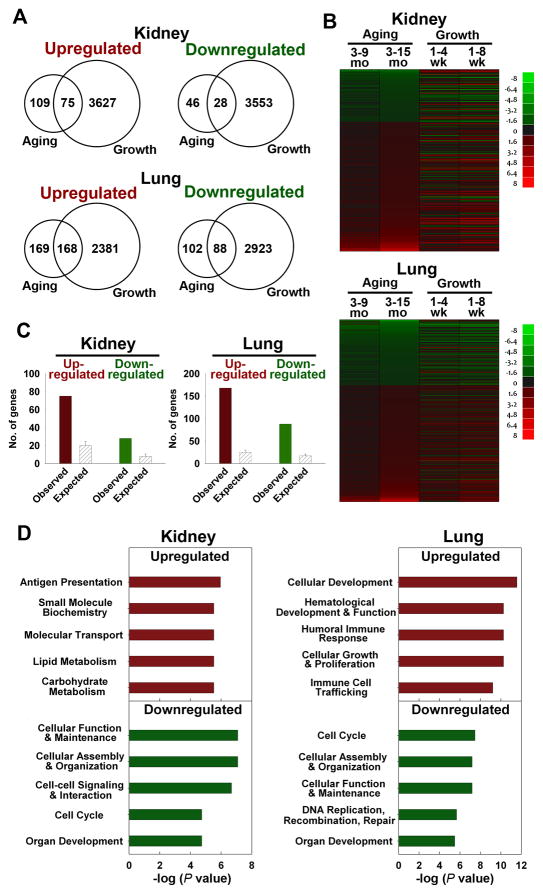
We also compared the overlap between the genes that changed significantly during aging (P < 0.0077, FDR < 10%, ≥1.5-fold) and genes that changed significantly during juvenile growth (P < 0.05, FDR < 5%, ≥1.5-fold). 40% (103/258) and 49% (256/527) of the genes that changed during aging had also changed significantly during earlier juvenile growth deceleration in the kidney and the lung, respectively (Fig. 3A, Venn diagram; Fig. 3B, heatmap; Supplemental Table 2). The observed overlap between aging and juvenile growth is far more than expected by chance (Fig. 3C, P < 0.001 in each organ, Chi-square test). Furthermore, when the genes that showed significant changes in both aging and growth were analyzed by IPA, one of the most overrepresented biological functions in the age-downregulated genes was cell cycle (Fig. 3D), suggesting that the downregulation of cell-cycle-related genes during juvenile growth continued in the aging process, in both organs studied. We also asked whether a specific part of the cell cycle pathway was downregulated in both aging and growth. However, bioinformatic analysis using IPA did not point to a specific phase of the cell-cycle. For example, in lung, Cdk6 and E2f2, which are involved in G1/S progression and Ccnb1 (cyclin B1), which is involved in G2/M progression, are downregulated during aging (3 to 15 mo). Similarly, in liver Cdkn2a, which inhibits G1/S progression, is upregulated and Cdc25b, which activates Cdk1 and is required for G2/M progression, is downregulated. Similarly, both E2f2 and Cdk1 are downregulated in the kidney during aging.
Fig 3. Many genes up- or downregulated during adult aging showed similar regulation during juvenile growth.
(A) Venn diagrams showing the number of genes significantly upregulated and downregulated during aging (3 vs 15 mo) and juvenile growth (1 vs 8 wk) in mouse kidney and lung. Significant changes were defined by: aging, P < 0.0077, FDR < 10%, and ≥ 1.5-fold; juvenile growth, P < 0.05, FDR < 5%, and ≥ 1.5-fold. (B) Heat maps were constructed from microarray data using JMP software version 8 for genes that show significant changes during aging (3 vs 15 mo, P < 0.0077, FDR < 10%, and ≥ 1.5-fold) in either kidney or lung. Green represents down-regulated genes, and red represents up-regulated genes compared to the baseline (3 mo for aging, 1 wk for postnatal growth). The color intensity corresponds to the magnitude of the change from baseline (log2 [value at later time point/value at baseline time point]). (C) Observed and expected overlap between aging and juvenile growth in kidney or lung. The solid bars represent the observed number of genes concordantly regulated during aging and juvenile growth (red, upregulated; green, downregulated) in kidney or lung. The hatched bars represent the number of concordantly regulated genes that would be expected by chance (mean ± SD). The observed overlap between organs was significantly greater than the overlap expected by chance (Pearson’s Chi-square test, all P < 0.001). (D) Pathway analyses of genes commonly regulated during aging and juvenile growth (kidney, upregulated: 75 genes, downregulated: 28 genes; lung, upregulated: 168 genes, downregulated: 88 genes). Upregulated (red) and downregulated (green) groups were analyzed separately, using Ingenuity Pathway Analysis (IPA) 7.1. The 5 most overrepresented molecular, cellular, or physiological functions are shown.
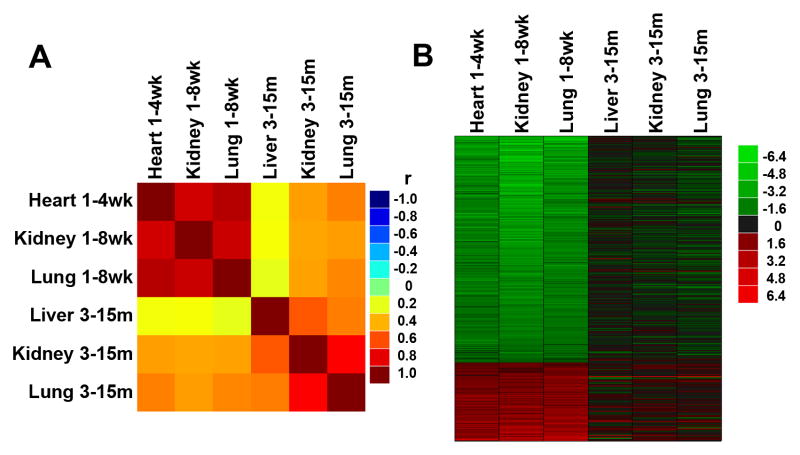
As an alternative test of our hypothesis, we focused on changes in gene expression that occur as juvenile growth slows, and asked whether these changes continued into adult life. We recently identified a genetic program comprising 316 genes that changed significantly and in the same direction in multiple organs of mice and rats (Lui et al. 2010). There is evidence that this genetic program contributes to the declining cell proliferation that occurs in these organs as somatic growth slows and the animal approaches adult body size (Lui et al. 2010). When correlation analysis was performed between the aging dataset and the juvenile dataset for these 316 genes, a substantial similarity between aging and juvenile growth was again found (Fig. 4A, correlation map, Fig. 4B, heatmap). The correlations involving the aging liver were statistically significant (P < 0.001) but again tended to be weaker (r = 0.20–0.24, ρ = 0.17–0.24) than those involving the aging kidney and lung (r = 0.42–0.50, ρ = 0.43–0.54, all P < 0.0001, Table 4). Of these 316 genes, there were 42, 84, and 118 genes that continued to change with P < 0.05 in the same direction in the aging liver, kidney, and lung, respectively. Furthermore, pathway analysis suggested that, particularly in the lung and kidney, one of the major biological functions for these genes that continue to decline during aging is the cell cycle (Fig. 5). This finding supports the hypothesis that some of the genetic changes in early life that slow cell proliferation and somatic growth continue into adult life to slow down cell proliferation in aging tissues.
Fig 4. Changes in gene expression occurring during postnatal growth deceleration continued during aging.
The analysis involved 316 genes that showed coordinate changes in genes expression in multiple organs of mice and rats during juvenile growth (Lui et al. 2010). (A) Correlation analysis was performed between temporal changes in gene expression during juvenile growth (1vs 8 wk) and aging (3 vs 15 mo) in various organs of mice. The correlation coefficients (r values) are depicted in a color map. Yellow - red, positive correlation; green, no correlation (r = 0); cyan - blue, negative correlation. (B) Heat maps were constructed from microarray data using JMP software version 8 for the same set of 316 genes. Green represents downregulated genes, and red represents upregulated genes compared to the baseline (3 mo for aging, 1 wk for postnatal growth). The color intensity corresponds to the magnitude of the change from baseline (log2 [value at later time point/value at baseline time point]). For genes that were downregulated from 1 to 8wk, there was a strong tendency for continued decline during aging. Similarly, genes upregulated during juvenile life tended to continue that upregulation during aging.
Table 4.
Correlation between aging and postnatal growth (growth genes)
| Gene set analyzed: all genes that showed uniform regulation (P < 0.05, ≥2-fold in multiple organs, both mouse and rat) during ju venile growth (n=316) | ||||
| Comparison between: | Pearson’s Correlation | Spearman’s Correlation | ||
| Heart, 1 to 4wk | vs | Liver, 3 to 15 mo | r = 0.23 | ρ = 0.24 |
| P = <0.0001 | P = <0.0001 | |||
| Kidney, 3 to 15 mo | r = 0.44 | ρ = 0.47 | ||
| P = <0.0001 | P = <0.0001 | |||
| Lung, 3 to 15 mo | r = 0.50 | ρ = 0.52 | ||
| P = <0.0001 | P = <0.0001 | |||
| Pearson’s Correlation | Spearman’s Correlation | |||
| Kidney, 1 to 8wk | vs | Liver, 3 to 15 mo | r = 0.24 | ρ = 0.20 |
| P = <0.0001 | P = 0.0003 | |||
| Kidney, 3 to 15 mo | r = 0.42 | ρ = 0.43 | ||
| P = <0.0001 | P = <0.0001 | |||
| Lung, 3 to 15 mo | r = 0.44 | ρ = 0.46 | ||
| P = <0.0001 | P = <0.0001 | |||
| Pearson’s Correlation | Spearman’s Correlation | |||
| Lung, 1 to 8wk | vs | Liver, 3 to 15 mo | r = 0.20 | ρ = 0.17 |
| P = <0.0001 | P = 0.002 | |||
| Kidney, 3 to 15 mo | r = 0.43 | ρ = 0.46 | ||
| P = <0.0001 | P = <0.0001 | |||
| Lung, 3 to 15 mo | r = 0.49 | ρ = 0.54 | ||
| P = <0.0001 | P = <0.0001 | |||
Fig 5. Pathway analysis of genes that showed persistent up- or downregulation durng juvenile growth and aging.
Ingenuity Pathway Analysis (IPA 7.1) was applied to genes that showed coordinate up- or downregulation during juvenile growth in multiple organs of mice and rats and also persistent changes in expression during aging (P < 0.05) in liver (hatched bars), kidney (solid grey bars), or lung (solid black bars). The 5 most overrepresented molecular, cellular, or physiological functions are shown. Cell cycle, DNA replication, and cellular growth and proliferation were amongst the most overrepresented functions in kidney and lung, suggesting that genes implicated in postnatal growth that continue to decline during aging were involved in cell proliferation.
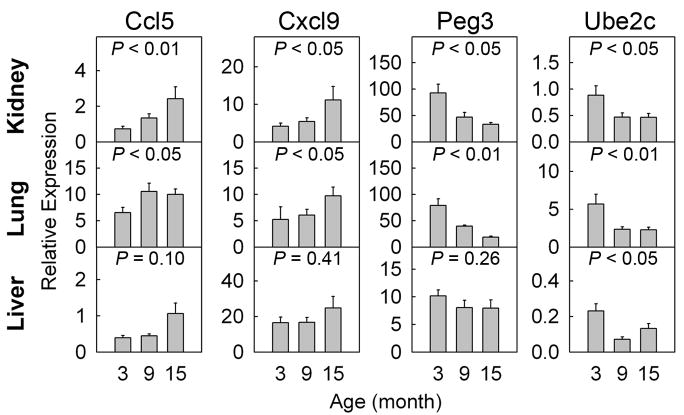
To verify the microarray findings, changes in gene expression during aging were assessed by real-time PCR. For this purpose, we studied genes that showed consistent changes in multiple organs both during postnatal growth and aging. Two of these genes, Ube2c and Peg3, were downregulated during growth and aging in multiple organs. Ube2c encodes a ubiquitin-conjugating enzyme that catalyses the ubiquitination and degradation of mitotic cyclin A and B. This enzyme has been reported to be growth stimulatory (Lin et al. 2006) and overexpressed in a number of cancer cell lines and tumors (Okamoto et al. 2003). Similarly, Peg3 encodes a zinc finger transcription factor (Kuroiwa et al. 1996) that is reported to be growth stimulatory (Lui et al. 2010), and involved in the “stemness” of muscle satellite cells (Seale et al. 2004; Nicolas et al. 2005). Two other genes, Ccl5 and Cxcl9, are chemokines that showed consistent increases in gene expression with age. Ccl5 has been reported to be secreted by senescent fibroblasts and implicated in benign prostatic hyperplasia of the aging male (Eyman et al. 2009), whereas Cxcl9, also known as monokine induced by gamma-interferon (MIG), was found to be elevated in the circulation of aged individuals (Shurin et al. 2007), and has been proposed to contribute to the Th1-mediated proinflammatory response and increased susceptibility of the elderly to autoimmune diseases. The expression patterns observed with real-time PCR generally agreed with those observed using microarray. Using real-time PCR, we found statistically significant changes in the same direction as shown by microarray for all four genes in kidney and lung, although in liver changes for only one of the four genes reached statistical significance (Fig. 6).
Fig 6.
Temporal changes in gene expression (mean± SEM) of Ccl5, Cxcl9, Peg3, and Ube2c in kidney, lung, and liver of 3, 9, and 15 month old mice were measured by real-time PCR and normalized to 18S RNA. P values (ANOVA) in each panel show overall change in expression with age. The trend observed in all three organs was consistent with the expression microarray, but did not reach statistical significance in the liver for 3 genes.
Discussion
In this study, we used microarray to analyze the changes in gene expression that occur during aging in mouse liver, lung, and kidney and compared these changes to those occurring during juvenile growth deceleration. We found that in each organ, hundred of genes were changing significantly during aging, from 3 months to 15 months. Pathway analyses suggested that a wide variety of biological functions were involved, underscoring the complex nature of the aging process.
We first asked whether the molecular genetic changes occurring during aging are unique to each organ or whether there are common changes occurring in multiple organs. To address this question, we compared the changes in gene expression occurring during aging in liver, kidney, and lung using microarray. The analysis indicated that the biological functions affected by aging differ among different organs. For instance, lipid metabolism is altered in liver, while immune functions were strongly overrepresented in the kidney. However, there were also similarities among the organs, as indicated by correlation analysis. Thus the findings suggest that the genetic changes occurring during aging have substantial organ-specific but also some shared components. Our findings are consistent with previous studies showing both tissue-specific (Weindruch et al. 2002) and common changes in gene expression during aging (Zahn et al. 2007; Schumacher et al. 2008).
The primary purpose of this study was to explore the possibility that changes in gene expression that are involved in restraining juvenile growth in early postnatal life persist into adulthood and therefore might contribute to the aging process. In mammals, the proliferation rate in multiple tissues is rapid in early life but slows with age as the animal approaches adult body size. This decline in growth velocity is common to many different organs. We previously showed evidence that this growth deceleration is driven by a genetic program that involves the downregulation of many growth-promoting genes (Lui et al. 2010). The program itself appears to be dependent on cell proliferation, as temporarily stalling proliferation by inducing hypothyroidism or tryptophan deficiency will delay progression of the program (Finkielstain et al. 2009; Lui et al. 2010). Thus somatic growth appears to be limited by a negative feedback loop in which proliferation induces widespread changes in gene expression that then limit further proliferation. In some organs, the adult parenchymal cells stay in a quiescent state, but retain the ability to proliferate for normal cellular turnover (Michalopoulos and DeFrances 1997; Bergmann et al. 2009). In contrast, in organs considered post-mitotic, the parenchymal cells no longer divide but adult stem cells in the organ allow for tissue repair (Relaix et al. 2005; Little 2008). In either case, a low level of cell proliferation is present in adult organs. We therefore hypothesized that the juvenile growth-limiting program continues to progress in adulthood, driven by low-level proliferation, and that this progression further limits the proliferative capacity in different organs. With age, this dwindling proliferative capacity might impair the ability of the organs to replace dying cells for tissue maintenance and regeneration following injury, thus contributing to the aging process. If this hypothesis is correct, we would expect genes that showed declining expression in early life to continue the decline during aging. However, the decline during adulthood would be expected to occur more gradually; because the changes in gene expression are driven by proliferation, one would predict that rapid proliferation in early life would cause rapid progression of this genetic program, but the much lower rate of proliferation in adult organs would lead to much slower progression of the program.
Consistent with this hypothesis, we found significant correlation between the changes in gene expression that occur during juvenile growth deceleration and those that occur during aging. For instance, within an organ, either lung or kidney, 40–50% of the genes that show a change in expression during aging also changed significantly (P < 0.05, ≥1.5-fold) in the same direction during growth. Thus, changes in gene expression that occur during aging frequently originate during juvenile growth. Similarly, many of the genes that showed a significant change during juvenile growth deceleration continue to change during aging, generally with a smaller magnitude. Bioinformatic analysis of these genes that showed persistent changes in expression indicated that cell-cycle-related genes are the most strongly overrepresented. This finding is consistent with previous studies based on expression microarray that showed increased expression with age of genes involved in cell-cycle arrest (Zahn et al. 2007; de Magalhaes et al. 2009). Taken together, these data support the hypothesis that changes in gene expression during juvenile growth deceleration persist into adulthood and therefore the aging process might be due in part to persistent gradual progression of the genetic program that limits growth in juvenile life. However, additional functional studies will be required to determine whether these continued changes in expression actually contribute to a progressive decline in proliferative capacity in aging organs and to the other varied structural and functional changes associated with aging. Compared to the kidney and lung, changes in gene expression in the aging liver showed weaker correlation with juvenile organs. Whether this difference might reflect greater proliferative capacity of the adult liver compared to kidney and lung, thus allowing regeneration after partial hepatectomy, remains to be determined.
The hypothesis that the genetic program that limits juvenile growth continues to progress into adulthood and contributes to aging is consistent with the developmental theory of aging, which states that some developmental mechanisms inadvertently affect postreproductive life (de Magalhaes and Church 2005; Fontana et al. 2010). In particular, there is prior evidence that body growth and aging are linked (Finch 1976; de Magalhaes and Faragher 2008). For example, many mutations that cause premature aging in mice and humans, such as disruption of ATR (Ruzankina et al. 2007), interfere with proliferation and body growth (de Magalhaes and Faragher 2008). This connection between growth and aging is further supported by the observations that caloric restriction (Fernandes et al. 1976; Rollo 2002) and deficiency of growth hormone and insulin-like growth factor-I (Flurkey et al. 2002; Shimokawa et al. 2003; Holzenberger et al. 2004; Cohen et al. 2009) delay organismal aging (Rollo 2002; de Magalhaes and Faragher 2008). Caloric restriction and GH/IGF-1 deficiency both inhibit proliferation and consequently may conserve proliferative capacity for later life, thereby slowing the aging process (de Magalhaes and Faragher 2008). This proposed effect of prior growth on aging might be explained by a cell-cycle counter that limits proliferation (de Magalhaes and Faragher 2008). The findings in the current study suggest a possible underlying mechanism at the molecular level. Recent studies indicate that growth-inhibiting conditions slow progression of the growth-limiting genetic program during juvenile life (Lui et al. 2010). Therefore, caloric restriction and GH/IGF-I deficiency, which also inhibit growth, may similarly slow progression of the growth-limiting genetic program and consequently delay the loss of proliferative capacity associated with aging. However, the effects of caloric restriction and GH/IGF-I deficiency on aging might involve mechanisms not directly related to proliferative capacity, such as a slowing of tissue metabolism, decreased oxidative stress, and/or suppression of the mTOR pathway (Beckman and Ames 1998; Carter et al. 2002; Merry 2002; Rollo 2002; de Magalhaes and Faragher 2008; Fontana et al. 2010).
This hypothesis, linking deceleration of juvenile growth and aging, might also help explain the general, though imperfect, positive correlation between body size and longevity among different mammalian species (R2 = 0.34, n = 1701) (Austad and Fischer 1991; Austad 2005). In general, small mammals, such as mice, have a shorter lifespan (Rollo 2002) than do large mammals such as humans and elephants (de Magalhaes et al. 2005). In small mammals, the diminutive body size is achieved by a rapid suppression of proliferation (Bogin 1999; Chang et al. 2008) and hence somatic growth, occurring over weeks, whereas in large mammals a greater body size is achieved by a more gradual suppression of somatic growth, occurring over years (Bogin 1999). If the mechanisms responsible for the rapid growth suppression of small mammals continue to progress into adulthood, one would expect relatively rapid loss of regenerative capacity in adult tissues of the small mammal. In contrast, in the large mammal, in which the growth-decelerating mechanisms act more gradually, a continuation of these mechanisms would be expected to produce a slower loss of proliferative capacity in the adult, thus perhaps contributing to the slower progression of aging and longer lifespan. Thus, aging might represent, in part, a tradeoff for body size control, with small mammals paying the greatest price for their rapid suppression of proliferation. This concept is further supported by the observation that, among mammalian species of similar adult body size, those that grow more slowly and for a more prolonged period of time tend to have a greater lifespan (Bowen and Atwood 2004; de Magalhaes et al. 2007). Similarly, mammalian and avian species that undergo rapid embryonic growth tend to have a shorter lifespan (Ricklefs 2006). However, other possible explanations have been proposed for the relationship between body size and longevity, for example, the higher basal metabolic rate and/or more rapid generation of reactive oxygen species in smaller mammals may accelerate aging compared to larger mammals (Beckman and Ames 1998). However, when corrected for body size, interspecific differences in basal metabolic rate do not correlate with lifespan (Harvey et al. 1991; de Magalhaes et al. 2007).
The evolutionary basis for aging is unclear. One possible explanation is that deterioration in body functions can occur in later adulthood because natural selection pressure decreases with age (Ricklefs 1998; de Magalhaes and Church 2005). Alternatively, the antagonistic pleiotropy theory proposes the existence of genes that have harmful effects in later life but are perpetuated because they have beneficial effects in early life, when natural selection pressure is stronger (Williams G.C. 1957). Our current study supports the antagonistic pleiotropy theory and implicates a specific biological function – growth suppression – in this paradigm. A potent growth-suppressing genetic program appears to have evolved to regulate somatic growth and thus target an adult size that is optimal for survival. However, the continuation of this program into adulthood may lead to a gradual loss of proliferative capacity, eventually contributing to organismal aging. Therefore, the gradual progressive loss of regenerative capacity in the aging mammal may be in part a lingering consequence of the evolutionary need to limit body size.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Austad SN. Diverse aging rates in metazoans: targets for functional genomics. Mech Ageing Dev. 2005;126:43–49. doi: 10.1016/j.mad.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Baylis C, Corman B. The aging kidney: insights from experimental studies. J Am Soc Nephrol. 1998;9:699–709. doi: 10.1681/ASN.V94699. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogin B. Evolutionary perspective on human growth. Annu Rev Anthropol. 1999;28:109–153. doi: 10.1146/annurev.anthro.28.1.109. [DOI] [PubMed] [Google Scholar]
- Bowen RL, Atwood CS. Living and dying for sex. A theory of aging based on the modulation of cell cycle signaling by reproductive hormones. Gerontology. 2004;50:265–290. doi: 10.1159/000079125. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Burton DG. Cellular senescence, ageing and disease. Age (Dordr) 2009;31:1–9. doi: 10.1007/s11357-008-9075-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CS, Ramsey MM, Sonntag WE. A critical analysis of the role of growth hormone and IGF-1 in aging and lifespan. Trends Genet. 2002;18:295–301. doi: 10.1016/S0168-9525(02)02696-3. [DOI] [PubMed] [Google Scholar]
- Chang M, Parker EA, Muller TJ, Haenen C, Mistry M, Finkielstain GP, Murphy-Ryan M, Barnes KM, Sundaram R, Baron J. Changes in cell-cycle kinetics responsible for limiting somatic growth in mice. Pediatr Res. 2008;64:240–245. doi: 10.1203/PDR.0b013e318180e47a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, Masliah E, Dillin A. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Church GM. Genomes optimize reproduction: aging as a consequence of the developmental program. Physiology(Bethesda) 2005;20:252–259. doi: 10.1152/physiol.00010.2005. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33:D537–D543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Faragher RG. Cell divisions and mammalian aging: integrative biology insights from genes that regulate longevity. Bioessays. 2008;30:567–578. doi: 10.1002/bies.20760. [DOI] [PubMed] [Google Scholar]
- Eyman D, Damodarasamy M, Plymate SR, Reed MJ. CCL5 secreted by senescent aged fibroblasts induces proliferation of prostate epithelial cells and expression of genes that modulate angiogenesis. J Cell Physiol. 2009;220:376–381. doi: 10.1002/jcp.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes G, Yunis EJ, Good RA. Influence of diet on survival of mice. Proc Natl Acad Sci USA. 1976;73:1279–1283. doi: 10.1073/pnas.73.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. The regulation of physiological changes during mammalian aging. Q Rev Biol. 1976;51:49–83. doi: 10.1086/409053. [DOI] [PubMed] [Google Scholar]
- Finkielstain GP, Forcinito P, Lui JC, Barnes KM, Marino R, Makaroun S, Nguyen V, Lazarus JE, Nilsson O, Baron J. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology. 2009;150:1791–1800. doi: 10.1210/en.2008-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Harrison DE. The Snell dwarf mutation Pit1(dw) can increase life span in mice. Mech Ageing Dev. 2002;123:121–130. doi: 10.1016/s0047-6374(01)00339-6. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD, Rees JA. Mammalian Metabolism and Life Histories. The American Naturalist. 1991;137:556–566. [Google Scholar]
- Hayashi Y, Hiyoshi T, Takemura T, Kurashima C, Hirokawa K. Focal lymphocytic infiltration in the adrenal cortex of the elderly: immunohistological analysis of infiltrating lymphocytes. Clin Exp Immunol. 1989;77:101–105. [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Kappeler L, De Magalhaes FC. IGF-1 signaling and aging. Exp Gerontol. 2004;39:1761–1764. doi: 10.1016/j.exger.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A. Stem cell: balancing aging and cancer. Int J Biochem Cell Biol. 2005;37:935–941. doi: 10.1016/j.biocel.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li LL, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, Barton SC, Ishino F, Surani MA. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- Lin J, Raoof DA, Wang Z, Lin MY, Thomas DG, Greenson JK, Giordano TJ, Orringer MB, Chang AC, Beer DG, Lin L. Expression and effect of inhibition of the ubiquitin-conjugating enzyme E2C on esophageal adenocarcinoma. Neoplasia. 2006;8:1062–1071. doi: 10.1593/neo.05832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH. Tracing the life of the kidney tubule- re-establishing dogma and redirecting the options. Cell Stem Cell. 2008;2:191–192. doi: 10.1016/j.stem.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Lui JC, Finkielstain GP, Barnes KM, Baron J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol. 2008;295:R189–R196. doi: 10.1152/ajpregu.00182.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Forcinito P, Chang M, Chen W, Barnes KM, Baron J. Coordinated postnatal down-regulation of multiple growth-promoting genes: evidence for a genetic program limiting organ growth. FASEB J. 2010 doi: 10.1096/fj.09-152835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry BJ. Molecular mechanisms linking calorie restriction and longevity. Int J Biochem Cell Biol. 2002;34:1340–1354. doi: 10.1016/s1357-2725(02)00038-9. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Misra V, Lee H, Singh A, Huang K, Thimmulappa RK, Mitzner W, Biswal S, Tankersley CG. Global expression profiles from C57BL/6J and DBA/2J mouse lungs to determine aging-related genes. Physiol Genomics. 2007;31:429–440. doi: 10.1152/physiolgenomics.00060.2007. [DOI] [PubMed] [Google Scholar]
- Mote PL, Grizzle JM, Walford RL, Spindler SR. Influence of age and caloric restriction on expression of hepatic genes for xenobiotic and oxygen metabolizing enzymes in the mouse. J Gerontol. 1991;46:B95–100. doi: 10.1093/geronj/46.3.b95. [DOI] [PubMed] [Google Scholar]
- Nicolas N, Marazzi G, Kelley K, Sassoon D. Embryonic deregulation of muscle stress signaling pathways leads to altered postnatal stem cell behavior and a failure in postnatal muscle growth. Dev Biol. 2005;281:171–183. doi: 10.1016/j.ydbio.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Ozaki T, Miyazaki K, Aoyama M, Miyazaki M, Nakagawara A. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res. 2003;63:4167–4173. [PubMed] [Google Scholar]
- Pellettieri J, Sanchez AA. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Phillips JI. Inflammatory plasma cell infiltration of the urinary bladder in the aging C57BL/Icrfa(t) mouse. Invest Urol. 1981;19:75–78. [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE. Evolutionary theories of aging: confirmation of a fundamental prediction, with implications for the genetic basis and evolution of life span. Am Nat. 1998;152:24–44. doi: 10.1086/286147. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE. Embryo development and ageing in birds and mammals. Proc Biol Sci. 2006;273:2077–2082. doi: 10.1098/rspb.2006.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollo CD. Growth negatively impacts the life span of mammals. Evol Dev. 2002;4:55–61. doi: 10.1046/j.1525-142x.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, van dPI, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van SH, Niedernhofer LJ, van IW, Bartke A, Spindler SR, Hoeijmakers JH, van der Horst GT, Garinis GA. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Ishibashi J, Holterman C, Rudnicki MA. Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev Biol. 2004;275:287–300. doi: 10.1016/j.ydbio.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Shimokawa I, Higami Y, Tsuchiya T, Otani H, Komatsu T, Chiba T, Yamaza H. Life span extension by reduction of the growth hormone-insulin-like growth factor-1 axis: relation to caloric restriction. FASEB J. 2003;17:1108–1109. doi: 10.1096/fj.02-0819fje. [DOI] [PubMed] [Google Scholar]
- Shurin GV, Yurkovetsky ZR, Chatta GS, Tourkova IL, Shurin MR, Lokshin AE. Dynamic alteration of soluble serum biomarkers in healthy aging. Cytokine. 2007;39:123–129. doi: 10.1016/j.cyto.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Singh P, Coskun ZZ, Goode C, Dean A, Thompson-Snipes L, Darlington G. Lymphoid neogenesis and immune infiltration in aged liver. Hepatology. 2008;47:1680–1690. doi: 10.1002/hep.22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JV, Heilbronn LK, Ravussin E. Energy restriction and aging. Curr Opin Clin Nutr Metab Care. 2004;7:615–622. doi: 10.1097/00075197-200411000-00005. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Dhahbi JM. Conserved and tissue-specific genic and physiologic responses to caloric restriction and altered IGFI signaling in mitotic and postmitotic tissues. Annu Rev Nutr. 2007;27:193–217. doi: 10.1146/annurev.nutr.27.061406.093743. [DOI] [PubMed] [Google Scholar]
- Stocker E, Heine WD. Regeneration of liver parenchyma under normal and pathological conditions. Beitr Pathol. 1971;144:400–408. [PubMed] [Google Scholar]
- Timchenko NA. Aging and liver regeneration. Trends Endocrinol Metab. 2009;20:171–176. doi: 10.1016/j.tem.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Kayo T, Lee CK, Prolla TA. Gene expression profiling of aging using DNA microarrays. Mech Ageing Dev. 2002;123:177–193. doi: 10.1016/s0047-6374(01)00344-x. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution. 1957;11:398–411. [Google Scholar]
- Yui J, Chiu CP, Lansdorp PM. Telomerase activity in candidate stem cells from fetal liver and adult bone marrow. Blood. 1998;91:3255–3262. [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, Lakatta EG, Boheler KR, Xu X, Mattson MP, Falco G, Ko MS, Schlessinger D, Firman J, Kummerfeld SK, Wood WH, III, Zonderman AB, Kim SK, Becker KG. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.