Abstract
Phylogeographic methods enable inference of the geographical history of genetic lineages. Recent examples successfully explore the patterns of human migration and the origins and spread of viral pandemics. Nevertheless, longstanding disagreement exists over the use and validity of certain phylogeographic inference methodologies. In this paper, we highlight three distinct frameworks for phylogeographic inference to give a taste of this disagreement. Each of the three approaches presents a different viewpoint on phylogeography, most fundamentally how we view the relationship between the inferred history of the sample and the history of the population the sample is embedded in. Satisfactory resolution of this relationship between history of the tree and history of the population remains a challenge for all but the most trivial models of phylogeographic processes. Intriguingly, we believe that some recent methods that entirely side-step inference about the history of the population will eventually help the field toward this goal.
Emerging Pathways of Phylogeographic Inference
The influence of phylogeography is spreading throughout biology. Amongst other examples, phylogeographic techniques have enabled us to infer the origins of mice [1], modern humans [2; 3], and man’s “best friend”, the domesticated dog [4]. Phylogeographic analyses also allow public health officials to understand the origin and spread of emerging infectious diseases [5; 6; 7; 8]. In spite of these successes, disagreement and confusion lingers over the most effective ways to learn about phylogeographic processes from geo-spatially identified molecular sequence data. Major points of contention include: how to model the phylogeographic spread of the represented taxa under study, what statistical frameworks provide effective inference tools and how to best reconcile population frameworks with geographic information? Over the past few years, these questions have been touched upon extensively in other reviews, and we do not belabor points already made [9; 10; 11; 12]. Instead, we highlight several newer phylogeographic approaches: a Bayesian approach to nested clade phylogeographic analysis (NCPA) [13; 14], stochastic-process driven spatial diffusion models mapping viral outbreaks [15; 16], and several recent population genetic approaches to phylogeography [17; 18; 19]. After this, we draw parallels between these methods in an attempt to shed light on their similarities and differences. We hope that by exposing a general audience to a cross-section of methods, new perspectives will be shed on longstanding issues in phylogeography.
A Comparative Approach
For almost a decade, supporters of NCPA [14] and supporters of model-based phylogeographic methods [20] have argued over the merits of each respective method. Similar to previous parley in phylogenetics, this debate has generated several positive outcomes; most importantly, investigators now vigorously question assumptions when analyzing geographic, demographic, and evolutionary data [21]. Nevertheless, this long-standing discussion has introduced considerable confusion [22; 23]. Thankfully, the debate appears to be coming to an end [20]. In addition, a new model-based approach (highlighted below) overcomes many points of contention over NCPA, making the debate increasingly moot at this point. For those not familiar with the debate between NCPA and model-based approaches, we provide a synopsis in Box 1.
Box 1: NCPA Debate Background
Nested clade phylogeographic analysis (NCPA) originated in a single-locus formulation in the landmark paper by Templeton [26] where he addresses the construction of a haplotype tree [28], the nesting of the clades on the haplotype tree [31; 32], and the use of a permutation test and inference key to interpret significant results [68; 26]. The debate began when Knowles and Maddison [69] showed that single-locus NCPA has a high false-positive rate. Later, Panchal and Beaumont [27; 36] found similar high false-positive rates.
In response to these claims, Templeton [35; 70] modified his original NCPA, introduced a multi-locus “cross-validated” version, and conducted an extensive empirical validation study of single-locus NCPA. Nevertheless, supporters of NCPA and those against it soon released a flurry of papers both denouncing the method [34; 71; 72] and defending it [73; 74; 75]. Following this, both sides began to question the philosophical basis of both methods [9; 76]. Within the past six months, Templeton [77; 14] and those criticizing his method [20] continue this back-and-forth debate.
The debate has focused around two general points: i) does single-locus and multi-locus NCPA have an inherently high false-positive rate, and does this preclude its use? And ii) do model-based methods or NCPA provide a more appropriate way to analyze phylogeographic data? One decade ago, the answer to the latter question was NCPA, since model-based methods for phylogeography were not widely available and, arguably, only NCPA could explicitly incorporate geographic features. Today, however, this situation has changed. Spatial diffusion and population genetics approaches are gaining attention; the former does currently incorporate important features, like physical distance and known geographic barriers, and the latter has the potential to do so. As such, we personally believe Beaumont et al. [20] resolve both questions posed above in favor of model-based methods. Nevertheless, we encourage others to read over the relevant publications to form their own opinion.
Before discussing this newer approach, we give a short background to NCPA, a technique that, for many years, stood alone in explicitly modeling geography in an evolutionary context. The method originated in previous efforts to jointly analyze phenotypic and phylogenetic data in a comparative framework [24; 25] and follows the same general three-step scheme [26; 13; 27; 9]. First, a haplotype tree or network is constructed from molecular data using a variety of approaches [28; 29; 30]. Second, clades on the haplotype tree are nested typically following the stepwise guidelines of Templeton [31; 32]. Finally, using these nested clades, a permutation test assesses the significance of the geographical spread.
Significant ambiguity occurs in all three stages of the NCPA pipeline [9]. During the first and second stages, alternative methods can lead to different, and possibly better, haplotype tree inference and nesting structures [33; 13]. More troubling, in the third stage, determining the level of significance for a particular clade does not typically take into account multiple testing, likely leading to a high false-positive rate [34; 9] (see Box 1). Templeton [35] has introduced a multi-locus “cross-validated” remedy for the apparent high false-positive rate, although even this revision still does not provide a complete solution [20; 36]. To be fair, we note that Templeton [14] disagrees with the ambiguous label for NCPA and its apparent high false-positive rate; but, extensive research suggests otherwise (Box 1).
In addition to these issues, the pipeline nature of NCPA leads to overconfidence in the final result [13]. This phenomenon is well known in the statistics and model-based molecular evolution literature. Multiple sequence alignment provides an excellent example where conditioning on a unknown multiple sequence alignment can lead to biased conclusions and overconfidence [37; 38]. Fortunately, a joint Bayesian approach provides a formal and straightforward fix to pipelined analyses [39; 40; 41]. Recently, Brooks et al. [13] and Manolopoulou (I. Manolopoulou, PhD thesis, University of Cambridge, 2008) adopt this simultaneous paradigm to combine the three stages of NCPA. For the first stage, Manolopoulou introduces a tractable procedure to infer a haplotype tree from molecular sequence data that takes into account possible homoplasy. She then uses approximate Bayesian computation to achieve computational efficiency for this part. For the second stage of NCPA, Brooks et al. [13] describe a multivariate clustering model applicable to both phenotypic and phylogeographic data. Brooks et al. [13] do not provide an analog of the third stage of NCPA, but nevertheless suggest that posterior probabilities of various qualitative events provide a solid alternative to the inference key of NCPA. To demonstrate the model, Manolopoulou applies both methods to three empirical examples. In general, the two methods give similar conclusions, but the Bayesian approach makes formal the intuitive results from NCPA. This Bayesian framework should excite those fond of the comparative method since it maintains the spirit of the comparative method, but corrects many of the statistical issues plaguing previous efforts. We look forward to many new developments from this approach.
A Spatial Diffusion Approach
As an alternative to the comparative method and its Bayesian extensions, we now highlight recent developments towards model-based approaches that take a probabilistic perspective on spatial diffusion [15; 16]. Although much of this work is implemented as part of a comprehensive statistical inference package that witnessed fruitful advances in demographic models [42; 43], we immediately want to emphasize that, unlike spatial-coalescent approaches [44], phylogenetic diffusion models do not claim to infer population-based spatial histories. Instead, they aim to uncover the ancestral history of a particular sample of molecular sequences, namely when and where the direct ancestors relating the sample existed. Nevertheless, such models can extract important information about the processes that underlie geographic structure in genetic data. For example, the inferred movement of the direct ancestors reflects movement of or between historical populations. Noteably, limiting oneself to the ancestors, the spatial diffusion approach can accommodate geographical context more explicitly by modeling the diffusion as trait evolution on a phylogeny.
Phylogenies naturally lend themselves to exploration of population structure and to assessment of significance in various ways [45]. More challenging however is the analytical reconstruction of how this structure took shape throughout the phylogenetic history. It is almost a law of statistical inquiry that challenging problems are initially probed by heuristic approaches. Parsimony methods represent model-free heuristic approaches that achieved enormous popularity in reconstructing such ancestral histories [46], be they of nucleotides, discrete organism traits or geographical locations [47]. Probabilistic approaches to ancestral reconstruction employ continuous-time Markov chain (CTMC) models to infer discrete realizations in continuous time. Although conceptually simple, they can capture considerable complexity of the discrete transition process among many states, such as geographic locations, albeit at the expense of an increased number of instantaneous rate parameters that make up the CTMC rate matrix. An obvious criticism is that a single observation does not hold the information required to estimate all parameters in the rate matrix, since we do not observe the intermediate states of this process. Nonetheless, this should be no reason to abandon probabilistic approaches all together.
Others have reviewed ancestral reconstruction methods and highlighted the advantages of likelihood-based statistical methods [48; 49]. Maximum likelihood methods and, to a greater extent, Bayesian inference accommodate important sources of uncertainty that accompany the estimation of character evolution [49]. A recent example of a Bayesian approach to island biogeography, in which specific Canary islands represent the character state data, has recently demonstrated great potential when applied to many groups of organisms evolving on independent phylogenies [50]. For a single character, however, a probabilistic approach that diligently accounts for phylogenetic and parameter uncertainty can yield particularly high variance estimates.
Recent developments in Bayesian phylogeography have made great strides towards efficient statistical inference of spatial diffusion [15]. In general, Bayesian approaches are more robust against over-parametrization via priors and this can be exploited most efficiently in discrete phylogeography by specifying informative priors on the rate parameters. For example, prior distributions based upon distance could formalize the prior expectation of a higher organismal lineage exchange between adjacent island groups in the Canary island biogeography [50]. Such distance-informed priors have recently been explored in viral epidemiology [15]. Arguably a more important advance, presented in the same work, is the ability to reduce the set of rate parameters to a limited number that provides a parsimonious description of the spatial diffusion process. This has been achieved by a Bayesian stochastic search variable selection procedure [51], which augments the rate matrix with indicator variables and imposes a prior distribution on the total sum of non-zero rate parameters. Parametrization of the truncated Poisson prior distribution proposed in Lemey et al. [15] fulfills the wish for statistical efficiency and is motivated by the believe that most pairwise exchange rates are probably not required to explain the diffusion process along the phylogeny. As such, it has been used to construct a formal Bayes factor test to establish epidemiological linkage in viral phylogeographic histories [15], removing the need to resort to ad hoc procedures [47].
It is a basic assumption of discretized dispersal that ancestral organisms at any point along the phylogeny reside in the locations from which samples were drawn. Spatial diffusion in continuous space presents an attractive alternative for more realistic phylogeographic inference. Following Brownian motion models for quantitative traits, bivariate Brownian random walks have been used for phylogeographic analysis in a likelihood approach [52]. This basic model has recently been greatly expanded and introduced into a Bayesian statistical framework [16]. To overcome the limiting assumption of constant diffusion rate throughout the phylogeny, rate heterogeneity is accomplished by borrowing ideas from uncorrelated relaxed clock models [53]. In our opinion, this represents a much-needed extension because it might prove difficult to find any real life example of spatial diffusion that adheres to a homogeneous Brownian process. For example, mountains or valleys, bodies of water and the environmental tolerances of organisms complicate the situation.
In particular, these relaxed random walks admit several diffusion processes commonly harnessed to model animal movement in ecological contexts [54]. These include independent Cauchy- and, more generally, Student-t-distributed increments along each branch of the uncertain phylogeny. These distributions emit infinite variance and derive motivation from Lévy flight models [55; 56]. Further, the relaxed walks do not require a strict enforcement of power-law tail behavior that remains contentious in some animal movement studies [57]. Encouragingly, even for relatively simple epidemic expansions, relaxed random walks yield reconstructions with increased statistical efficiency. We believe this can add considerable credibility to spatial reconstructions in continuous space, in particular because Brownian trait estimates have often been found too variable for practical use [58].
The continuous and discrete stochastic processes furnish complementary models that expand our ability to infer phylogeographic processes. They are both implemented in the “Bayesian evolutionary analysis sampling trees” (BEAST) software package that has matured into a rich resource of stochastic evolutionary models [59]. Focusing entirely on time-measured phylogenies and offering different flavors of demographic models, such an integrated approach appears well suited for biogeographical analyses. The first phylogeographic steps with this framework were made in the field of viral epidemiology, in particular for rapidly evolving RNA viruses [8; 16]. As general tools however, these models can also prove useful in the analysis of genetic data from more slowly evolving organisms. To illustrate this, we provide a phylogeographic reconstruction in continuous space for the freshwater snail Biomphalaria glabrata, a major vector of schistosomiasis in the New World [60] (see Box 2). This box also opens a discussion on visualizing inferred evolutionary histories through both time and space.
Box 2: Visualization Tools
We revisit the phylogeography of the freshwater snail Biomphalaria glabrata that has limited dispersal abilities and occupies fragmented habitats in South America and the Caribbean islands [60]. Mavarez et al. [60] originally obtained sequence data from populations of B. glabrate sampled at the scale of the current geographic distribution of the species to study its phylogeography. As illustrated in Figure I, we apply the recently developed continuous diffusion model to 17 concatenated nuclear internal transcribed spacer-2 (484 bp) gene sequences and the partial mitochondrial large ribosomal subunit (374 bp) generated in the original study. The reconstruction confirms a clear separation into Northern (Venezuela and Lesser Antilles) and Southern clades (Brazil), in line with the Amazon river constituting an important barrier to gene flow [60]. Because the equatorial forest of the Amazon basin does not provide appropriate habitats for this species, it might therefore represent an additional barrier to gene flow [78]. The considerable divergence among the sampled haplotypes has led to the suggestion that B. glabrata constitutes a species complex [60]. As suggested by the Lesser Antilles haplotypes clustering in the same monophyletic clade [79; 80], there also appears to be significant isolation by distance at a more restricted geographic scale.
With this illustration, we also touch upon the developing visualization tools that complement the Bayesian inference. Phylogenies have previously been projected in virtual globe software [81], but we would like to argue for directing these efforts towards model-based estimates. If probabilistic approaches are able to rigorously capture the uncertainty in phylogeographic inference, it is critical that visualization tools are able to fully present this uncertainty for appropriate interpretation. For discrete ancestral reconstruction this still remains an open issue; while location probability distributions for ancestral nodes can be readily obtained, the phylogenetic uncertainty adds a level of complexity that does not easily lend itself to straightforward visual summaries. Phylogeographic estimates through time and continuous space are more naturally streamlined into rich visualization [16]. Not only does this model offer the ability to draw inference about location realizations and various summary statistics at arbitrary points in time, researchers can now explore the results in an interactive fashion (for several examples, we refer to www.phylogeography.org). Finally, more work is needed to introduce these summaries in more dedicated visualization software [82], or to accommodate them in feature-rich, multi-purpose geographic information systems software [83].
A Population Genetics Approach
By far the most popular statistical approaches to phylogeography rely on the structured-coalescent [61; 44; 62; 9]. In general, these methods assume that evolutionary trees are random draws from some underlying population-level process [9]. Essentially, population-level processes fossilize their histories as evolutionary trees that we indirectly view through molecular sequence and other data [61; 10]. These processes include selection, migration, population size changes, and recombination.
Nearly all population-level approaches embed evolutionary trees into a structured-coalescent framework to make inference [63]. Herein lies the strength and limitations of these methods. If constructed appropriately, structured-coalescent methods allow us to infer population-level information from a relatively small sample. Unfortunately, constructing these methods in an appropriate manner requires extensive time, computation, and ingenuity. The task is not trivial. Nearly a decade ago, Felsenstein et al. [64] maintained that any implementation of structured-coalescent methods, and related models, took nearly two years to develop. Nielsen and Beaumont et al. [9] suggest that things remain the same. As such, the field still does not have a way to handle even simple recombination between linked loci in a structured-coalescent framework.
Nevertheless, population genetic approaches for ancestral inference are flourishing. In the past few years, numerous highly significant contributions have appeared. As examples, Liu and Pearl [65] and Heled and Drummond [18] fully take into account gene tree uncertainty and species tree uncertainty through Bayesian statistical modeling [66; 67]. Focusing on gene-flow and migration, Hey [17] extends his previous work to incorporate migration between multiple species. Also focusing on gene-flow, Beerli and Palczewski [19] adopt thermodynamic integration to compute Bayes factors assessing panmixia and migration in a structured-coalescent framework [44]. And the list goes on. Approximate Bayesian computation allows empirical investigators to assess complex demographic histories, conditional likelihood methods allow for the approximation of the coalescent with recombination, and efficient calculation allows for full assessment of it [9].
In sum, much as in the previous two approaches, the application of population genetics to phylogeography remains a vibrant field of research. Highly significant work continues to be created, but numerous, biologically-relevant, methods need to implemented, adapted, and created. This route to phylogeography is far from a dead-end.
Three Routes To the Same Destination
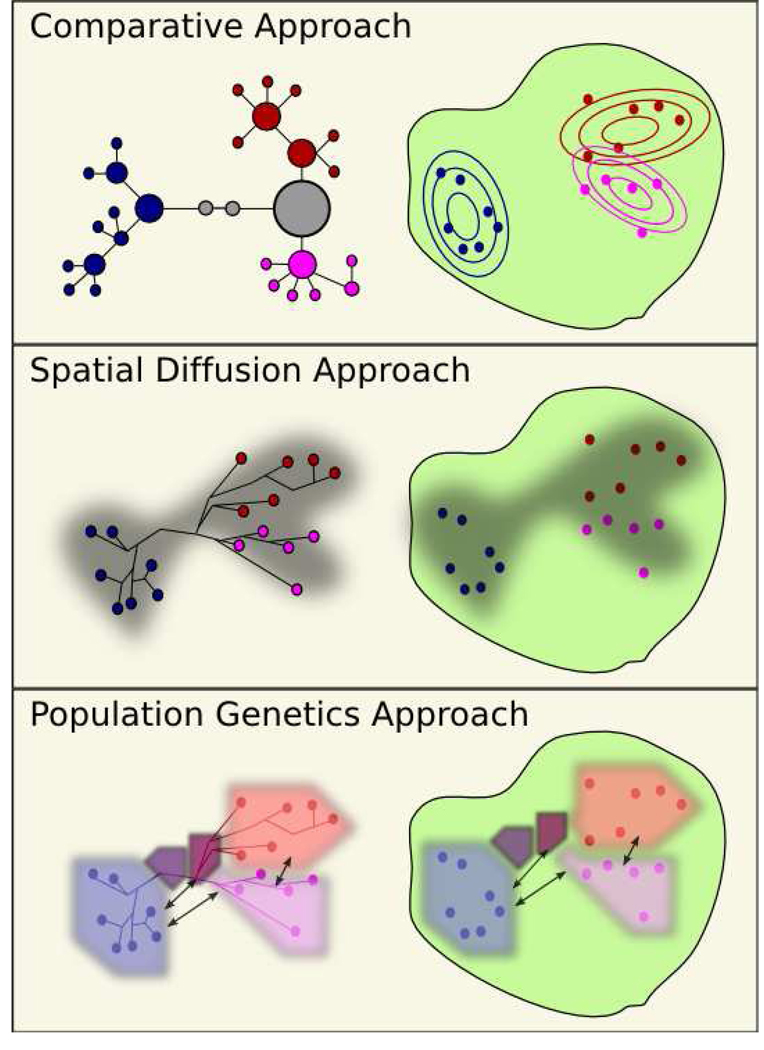
With the expansion of phylogeography in new Bayesian directions involving NCPA and spatial diffusion, the field might appear to be fragmenting. But, we believe all three approaches address the same basic question we stated at the beginning of this article: what are the most effective ways to learn about phylogeographic processes from geo-spatially identified molecular sequence data? Essentially, we believe all three frameworks produce effective answers, if one knows what questions to ask. Under the comparative approach, if one wants to assess geographical spread and molecular data, without modeling this spread explicitly, the method of Manolopoulou will be the most appropriate. If one wants to model rates of spread and take into account geographical features, the method of Lemey et al. [16] will be most appropriate. If one wants information about population size and migration, but considers population to be a coarse feature, the population genetics approach will be most appropriate. Figure 1 provides a schematic of these three phylogeographical frameworks.
Figure 1.
Three approaches to phylogeography. In all three windows, the data consist of three species (red, purple and blue) sampled across an island (green). Comparative Approach: In this window, the haplotype tree is displayed on the left; concentric circles on the island represent the geographical distribution of the three species. Spatial Diffusion Approach: In this window, the phylogenetic tree, superimposed upon the geographical location of the samples, is displayed on the left. We color the phylogenetic tree black to reinforce that we do not infer population information. The gray area behind the phylogenetic tree, and on the island, represents a high probability region contour of the locations of the ancestors of the sampled taxa. Population Genetics Approach: This window is based upon Hey (2010). The five colored polygons represent the ancestral populations and their respective population sizes. Arrows represent strong migrations between the populations. Currently, the idealized population polygons abstract away all geographic features to keep the models tractable.
One might wonder whether the advantages of all three approaches can be combined into a single framework. We hesitate to speculate on this particular question, and simply leave it as open and tractable and encourage its exploration (see Box 3 for other open issues on the verge of solution). We do note that with a shift towards likelihood-based statistical inference, ad hoc procedures are no longer the only or preferable methods available. Instead, garnering novel insight into phylogeographic processes reduces to envisioning stochastic process-driven models that incorporate the relevant aspects of the research question at hand and then developing the tools necessary to efficiently fit these models to data. The limiting aspects of this approach are chiefly creativity in modeling and effective strategies to reduce the high-dimensional model fit into a human-interpretable form.
Box 3: Some Open Issues Approaching Solution
Availability of Geo-Coded Sequence Data
The inclusion of geographical data into molecular phylogenetics increases the size and complexity of datasets. As a benefit, investigators can learn the relationships between several disparate aspects, gaining a better understanding of biological diversity and history. The downside to these complex datasets, however, is reproducibility and validity. In the past, investigators could reproduce and validate inferences since the models and datasets were readily available. In the modern climate, however, inferences are more difficult to reproduce because the geographic data is rarely provided with the molecular sequences and many modeling assumptions go unreported. One way to avoid this difficultly is to require geo-coded sequence submission and a central online repository where investigators can upload their computational scripts. Some journals have begun to require these uploads for publications, but the push needs to be stronger.
Incorporating Geographic Features and Niche Modeling
The structured-coalescent and discretized spatial diffusion approaches generate models in which researchers can inject a limited amount of information about geographic or niche features. This injection involves the definition of prior distributions on migration rates between populations or geographic locations. For these forms of discrete Markov chain models, O’Brien et al. [84] offer advice on how to make summary statistics about the diffusion process robust to some of the model misspecification that results from ignoring additional features. However, it is unclear how effective informed prior specification will remain when researchers attempt to learn from the observed data which geographic or niche features significantly affect the migration or diffusion process. In these situations, directly modeling the geographic features as hard or soft barriers in a continuous diffusion framework appears as a tenable solution. Ranking features by their barrier strength or testing which strengths do not significantly differ from zero produces a sound statistical framework. Looming over us is the modeling challenge of efficiently computing transition probabilities along the tree of migration or diffusion in the presence of multiple, possibly irregular barriers.
Next-Generation Sequencing
With the advent of next-generation sequencing technologies, biologists can now obtain de novo genomic samples from entire biological communities. Still, these metagenomic studies are in their infancy. In particular, the analysis and design of metagenomic studies is unrefined and rudimentary. For model-based methods to move into this area, advances in computational efficiency will be vital. Moreover, model-based methods will need to account for the data-collection technology, as well as the phylogeographic history of a sample under study. Currently, the lack of theoretical tools here is hindering scientific progress.
To Tree or Not To Tree
The relationship between the inferred phylogenetic tree and its embedded population has been described extensively over the past 30 years [85; 62; 9]. Nevertheless, the relationship between the two remains poorly understood, as evidenced by the three distinct approaches highlighted in this paper. Put simply, we still poorly understand whether the inferred evolutionary history remains a nuisance or fundamental entity [86]. Resolving this issue will likely require extensive work in the upcoming decade.
Figure I.
Freshwater snail phylogeny (maximum clade credibility tree) obtained using Bayesian phylogeographic inference in continuous space. The maps in the virtual globe visualization are based on satellite pictures made available in Google Earth (http://earth.google.com). A) Projection of the complete phylogeny with a red-white color (on-line) or grayscale (in print) gradient informing the relative age of the branches. The location uncertainty for each node is visualized by a polygon representing a 80% credibility contour. B) A detailed view of the Northern phylogeographic clade, as indicated by the rectangle in panel A. The orientation of the view is indicated by matching arrows in both panels. Taxa habitat (freshwater) is an important geographic feature to begin modeling directly.
Acknowledgments
We thank Chris Simon, Allen Rodrigo, Brian C. Carstens, H. Lisle Gibbs, Laura S. Kubatko, Peter Beerli and Ioanna Manolopoulou for their comments and suggestions. We also thank the National Evolutionary Sythnesis Center (NSF #EF-0423641) for fostering our collaboration. Alexei J. Drummond contributed greatly to discussions on an earlier version of this paper. EWB is partially supported by the National Science Foundation under Agreement No. 0635561. PL is supported by a postdoctoral fellowship from the Fund for Scientific Research (FWO) Flanders. MAS is partially supported by the United States National Institutes of Health (R01 GM086887), the National Science Foundation (DMS 0856099) and the Marsden Fund, New Zealand. The research leading to this article has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013) / ERC Grant agreement #260864.
Glossary
- Ancestral history
any information about the direct ancestors relating a sample of molecular sequences. This term can refer, for example, to inferred sequence composition or phenotype, such as geography, and is often associated with a time-scale.
- Approximate Bayesian computation (ABC)
a simulation technique used to draw statistical inference based on data summaries, often employed when the full data likelihood is impractical to compute.
- Bayes factor
the ratio of marginal likelihoods of a given data set comparing two competing models that naturally incorporates uncertainty about unknown parameters in both models.
- Bayesian stochastic search variable selection
a framework that estimates the posterior probability that a particular explanatory variable should be included in a model. The method is most commonly used in Bayesian inference of linear regression.
- BEAST
an open-source MCMC package for the analysis of several Bayesian evolutionary models for molecular sequences and associated traits, such as geography.
- Brownian diffusion
a stochastic process on the real number line or, in the work described in this article, on a geographic surface, in which increments are independent and normally-distributed with mean zero and a variance that scales linearly with duration.
- Continuous-time Markov chain
a stochastic process on a discrete state-space or, in the work described in this article, a set of locations that is memoryless and whose waiting times between transitions are exponentially-distributed.
- Comparative approach
a framework with which to relate observed phenotype information to an evolutionary history. Nested clade phylogeographic analysis assumes geography to be a phenotypic trait and falls under this category.
- Model-based approach
a method in which a fully specified probabilistic model describes how the observed data are generated. Unknown parameters can characterize this model, and statistical inference proceeds through estimating and testing these parameters.
- Nested clade phylogeographic analysis
a resampling based approach to infer geographic information from haplotype trees and networks.
- Phylogeography
the interdisciplinary field that studies the evolutionary history and the geographic spread of biological populations or taxa.
- Population genetics
as used in this article, a framework that uses sample molecular sequences to make statements about a population under study. The coalescent is the most widely used population genetics framework. Extensions of the coalescent to phylogeography typically focus on migration rates between multiple populations fixed in space.
- Spatial diffusion model
the application of independent stochastic processes to describe changes in geographic phenotypes on a two-dimensional surface. Under the discrete framework described in this article, one divides the surface into discrete regions and models movement between regions as a continuous-time Markov chain; under the continuous diffusion framework, one subdivides these regions until they become infinitesimal and considers generalizations of Brownian diffusion.
- Structured coalescent
an extension of the basic coalescent model to multiple populations. This method focuses on ancestral population sizes and migration rates between populations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Searle J, et al. Of mice and (Viking?) men: phylogeography of British and Irish house mice. Proceedings of the Royal Society B: Biological Sciences. 2009;276:201–207. doi: 10.1098/rspb.2008.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fagundes N, Ray N, Beaumont M, Neuenschwander S, Salzano F, Bonatto S, Excoffier L. Statistical evaluation of alternative models of human evolution. Proceedings of the The National Academy of Sciences, USA. 2007;104 doi: 10.1073/pnas.0708280104. 1761417619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 4.von Holdt B, et al. Genome-wide snp and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biek R, Drummond A, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- 6.Biek R, Henderson J, Waller L, Real CRL. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceedings of the The National Academy of Sciences, USA. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith G, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 8.Lemey P, Suchard M, Rambaut A. Reconstructing the initial global spread of a human influenza pandemic: a Bayesian spatial-temporal model for the global spread of H1N1. PLoS Current Influenza. 2009 doi: 10.1371/currents.RRN1031. RRN1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen R, Beaumont M. Statistical inferences in phylogeography. Molecular Ecology. 2009;18:1034–1047. doi: 10.1111/j.1365-294X.2008.04059.x. [DOI] [PubMed] [Google Scholar]
- 10.Knowles L. Statistical phylogeography. Annual Review of Ecology, Evolution, and Systematics. 2009;40:593–612. [Google Scholar]
- 11.Avise J. Phylogeography: retrospect and prospect. Journal of Biogeography. 2009;36:3–15. [Google Scholar]
- 12.Hickerson M, Carstens B, Cavender-Bares J, Crandall K, Graham C, Johnson J, Rissler L, Victoriao P, Yoder A. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Molecular Phylogenetics and Evolution. 2010;54:291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Brooks S, Manolopoulou I, Emerson B. Assessing the effect of genetic mutation: a Bayesian framework for determining population history from DNA sequence data. In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics 8. Oxford University Press; 2007. pp. 1–26. [Google Scholar]
- 14.Templeton A. Coalescent-based, maximum likelihood inference in phylogeography. Molecular Ecology. 2010;19:431–435. doi: 10.1111/j.1365-294X.2009.04514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemey P, Rambaut A, Drummond A, Suchard M. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemey P, Rambaut A, Welch J, Suchard M. Phylogeography takes a relaxed random walk in continuous space and time. Molecular Biology and Evolution. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hey J. Isolation with migration models for more than two populations. Molecular Biology and Evolution. 2010 doi: 10.1093/molbev/msp296. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heled J, Drummond A. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010 doi: 10.1534/genetics.109.112532. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaumont M, et al. In defence of model-based inference in phylogeography. Molecular Ecology. 2010;19:436–446. doi: 10.1111/j.1365-294X.2009.04515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camargo A, Heyer W, de Sá R. Phylogeography of the frog Leptodactylus validus (Amphibia: Anura): patterns and timing of colonization events in the Lesser Antilles. Molecular Phylogenetics and Evolution. 2009;53:571–579. doi: 10.1016/j.ympev.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Phillipsen I, Metcalf A. Phylogeography of a stream-dwelling frog (Pseudacris cadaverina) in southern California. Molecular Phylogenetics and Evolution. 2009;53:152–170. doi: 10.1016/j.ympev.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Gante H, Micael J, Oliva-Paterna F, Doadrio I, Dowling T, Judite Alves M. Diversification within glacial refugia: tempo and mode of evolution of the polytypic fish Barbus sclateri. Molecular Ecology. 2009;18:3240–3255. doi: 10.1111/j.1365-294X.2009.04264.x. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Phylogenies and the comparative method. The American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- 25.Harvey P, Pagel D. The comparative method in evolutionary biology. Oxford University Press; 1991. [Google Scholar]
- 26.Templeton A. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Molecular Ecology. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 27.Panchal M, Beaumont M. The automation and evaluation of nested clade phylogeographic analysis. Evolution. 2007;61:1466–1480. doi: 10.1111/j.1558-5646.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- 28.Templeton A, Crandall K, Sing C. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. III. Cladogram estimation. Genetics. 1992;134:659–669. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 30.Posada D, Crandall K. Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology and Evolution. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- 31.Templeton A, Boerwinkle E, Sing C. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophilia. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Templeton A, Boerwinkle E, Sing C. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics. 1993;134:659–669. doi: 10.1093/genetics/134.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cassens I, Mardulyn P, Milinkovitch M. Evaluating intraspecific “network” construction methods using simulated sequence data: do existing algorithms outperform the global maximum parsimony approach? Systematic Biology. 2005;54:363–372. doi: 10.1080/10635150590945377. [DOI] [PubMed] [Google Scholar]
- 34.Knowles L. Why does a method that fails continue to be used? Evolution. 2008;62:2713–2717. doi: 10.1111/j.1558-5646.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 35.Templeton A. A maximum likelihood framework for cross validation of phylogeographic hypotheses. In: Wasser S, editor. Evolutionary Theory and Processes: Modern Horizons. Kluwer Academic Publishers; 2004. pp. 209–230. [Google Scholar]
- 36.Panchal M, Beaumont M. Evaluating nested clade phylogenetic analysis under models of restricted gene flow. Systematic Biology. 2010;59:415–432. doi: 10.1093/sysbio/syq022. [DOI] [PubMed] [Google Scholar]
- 37.Wong K, Suchard M, Huelsenbeck J. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]
- 38.Redelings B, Suchard M. Robust inferences from ambiguous alignments. In: Rosenberg M, editor. Sequence alignment: methods, models, concepts, and strategies. University of California Press; 2009. pp. 209–271. [Google Scholar]
- 39.Suchard M, Weiss R, Sinsheimer J. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Bio Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 40.Redelings B, Suchard M. Joint Bayesian estimation of alignment and phylogeny. Syst Biol. 2005;54:401–418. doi: 10.1080/10635150590947041. [DOI] [PubMed] [Google Scholar]
- 41.Novák A, Miklós I, Lyngsø R, Hein J. StatAlign: an extendable software package for joint Bayesian estimation of alignments and evolutionary trees. Bioinformatics. 2008;24:2403–2404. doi: 10.1093/bioinformatics/btn457. [DOI] [PubMed] [Google Scholar]
- 42.Drummond A, Rambaut A, Shapiro B, Pybus O. Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 43.Minin V, Bloomquist E, Suchard M. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Molecular Biology and Evolution. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proceedings of the The National Academy of Sciences, USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarate S, Pond S, Shapshak P, Frost S. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. Virology. 2007;81:6643–6641. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swofford D, Maddison W. Parsimony, character-state reconstructions and evolutionary inferences. In: Mayden R, editor. Systematics, Historical Ecology, and North American Freshwater Fishes. Stanford University Press; 1992. pp. 186–223. [Google Scholar]
- 47.Wallace R, HoDac H, Lathrop R, Fitch W. A statistical phylogeography of influenza A H5N1. Proceedings of the The National Academy of Sciences, USA. 2007;104:4473–4478. doi: 10.1073/pnas.0700435104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cunningham C, Omland K, Oakley T. Reconstructing ancestral character states: a critical reappraisal. Trends in Ecology and Evolution. 1998;13:361–366. doi: 10.1016/s0169-5347(98)01382-2. [DOI] [PubMed] [Google Scholar]
- 49.Ronquist F. Bayesian inference of character evolution. Trends in Ecology and Evolution. 2004;19:475–481. doi: 10.1016/j.tree.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Sanmartin I, van der Mark P, Ronquist F. Inferring dispersal: a Bayesian approach to phylogeny-based island biogeography, with special reference to the Canary Islands. Journal of Biogeography. 2008;35:428–449. [Google Scholar]
- 51.Kuo L, Mallick B. Variable selection for regression models. Sankhya B. 1998;60:65–81. [Google Scholar]
- 52.Lemmon A, Lemmon E. A likelihood framework for estimating phylogeographical history on a continuous landscape. Systematic Biology. 2008;57:544–561. doi: 10.1080/10635150802304761. [DOI] [PubMed] [Google Scholar]
- 53.Drummond A, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paradis E, Baillie S, Sutherland W. Modeling large-scale dispersal distances. Ecological Modelling. 2002;151:279–292. [Google Scholar]
- 55.Viswanathan G, Afanasyev V, Buldyrev S, Murphy E, Prince P, Stanley H. Lévy flight search patterns of wandering albatrosses. Nature. 1996;381:413–415. doi: 10.1038/nature06199. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds A, Rhodes C. The Lévy flight paradigm: random search patterns and mechanisms. Ecology. 2009;90:877–887. doi: 10.1890/08-0153.1. [DOI] [PubMed] [Google Scholar]
- 57.Edwards A, et al. Revisiting Lévy flight search patterns of wandering albatrosses, bumblebees and deer. Nature. 2007;449:1044–1048. doi: 10.1038/nature06199. [DOI] [PubMed] [Google Scholar]
- 58.Schluter D, Price T, Ludwig D. Likelihood of ancestral states in adaptive radiation. Journal of Organic Evolution. 1997;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- 59.Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mavárez J, Steiner C, Pointer J, Jarne P. Evolutionary history and phylogeography of the schistosome-vector freshwater snail Biomphalaria glabrata based upon nuclear and mitochondrial DNA sequences. Heredity. 2002;89:266–272. doi: 10.1038/sj.hdy.6800128. [DOI] [PubMed] [Google Scholar]
- 61.Avise J. Phylogeography: the history and formation of species. Harvard University Press; 2000. [Google Scholar]
- 62.Hey J, Machado C. The study of structured populations-a new hope for a difficult and divided science. Nature Reviews Genetics. 2003;4:535–543. doi: 10.1038/nrg1112. [DOI] [PubMed] [Google Scholar]
- 63.Kingman J. The coalescent. Stochastic Processes and their Applications. 1982;13:235–248. [Google Scholar]
- 64.Felsenstein J, Kuhner M, Yamato J, Beerli P. Likelihoods on coalescents: a Monte Carlo sampling approach to inferring parameters from populations samples of molecular data. In: Seillier-Moiseiwitsch F, editor. Statistics in Molecular Biology, vol. 33 of IMS Lecture Notes. Institute of Matematical Statistics; 1999. pp. 163–184. [Google Scholar]
- 65.Liu L, Pearl D. Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Systematic Biology. 2007;56:504514. doi: 10.1080/10635150701429982. [DOI] [PubMed] [Google Scholar]
- 66.Edwards S. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- 67.Degnan J, Rosenberg N. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology and Evolution. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 68.Templeton A, Routman E, Phillips C. Separating population structure from population history - a cladisitic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander. Ambystoma tigrinum. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knowles L, Maddison W. Statistical phylogeography. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 70.Templeton A. Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology. 2004;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- 71.Petit R. The coup de grâce for nested clade phylogeographic analysis. Molecular Ecology. 2008;17:516–518. doi: 10.1111/j.1365-294X.2007.03589.x. [DOI] [PubMed] [Google Scholar]
- 72.Beaumont M, Panchal M. On the validity of nested clade phylogeographic analysis. Molecular Ecology. 2008;17:2563–2565. doi: 10.1111/j.1365-294X.2008.03786.x. [DOI] [PubMed] [Google Scholar]
- 73.Garrick R, Dyer R, Beheregaray L, Sunnucks P. Babies and bathwater: a comment on the premature obituary for nested clade phylogeograhical analysis. Molecular Ecology. 2008;17:1401–1403. doi: 10.1111/j.1365-294X.2008.03675.x. [DOI] [PubMed] [Google Scholar]
- 74.Templeton A. Nested clade analysis: an extensively validated method for strong phylogeographic inference. Molecular Ecology. 2008;17:1877–1880. doi: 10.1111/j.1365-294X.2008.03731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Templeton A. Why does a method that fails continue to be used? The answer. Evolution. 2009;63:807–812. doi: 10.1111/j.1558-5646.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Templeton A. Statistical hypothesis testing in intraspecific phylogeography: nested clade phylogeographic analysis vs. approximate Bayesian computation. Molecular Ecology. 2009;18:319–331. doi: 10.1111/j.1365-294X.2008.04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Templeton A. Coherent and incoherent inference in phylogeography and human evolution. Proceedings of the The National Academy of Sciences, USA. 2010 doi: 10.1073/pnas.0910647107. In Press, doi:10.1073/pnas.0910647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paraense W. A survey of planorbid molluscs in the Amazonian region of Brazil. Memorias do Instituto Oswaldo Cruz. 1983;78:343–361. doi: 10.1590/s0074-02761983000300011. [DOI] [PubMed] [Google Scholar]
- 79.Mavárez J, Amarista M, Pointer J, Jarne P. Fine-scale population structure and dispersal of Biomphalaria glabrata, the intermediate snail host of Schistosoma mansoni, in Venezuela. Molecular Ecology. 2002;11:879–899. doi: 10.1046/j.1365-294x.2002.01486.x. [DOI] [PubMed] [Google Scholar]
- 80.Mavárez J, Pointer J, David P, Delay B, Jarne P. Genetic differentiation, dispersal and mating system in the schistosome-transmitting freshwater snail Biomphalaria glabrata. Heredity. 2002;89:258–265. doi: 10.1038/sj.hdy.6800127. [DOI] [PubMed] [Google Scholar]
- 81.Janies D, Hill A, Guralnick R, Habib F, Waltari E, Wheeler W. Genomic analysis and geographic visualization of the spread of avian influenza (H5N1) Systematic Biology. 2007;56:321–329. doi: 10.1080/10635150701266848. [DOI] [PubMed] [Google Scholar]
- 82.Parks D, Porter M, Churcher S, Wang S, Blouin C, Whalley J, Brooks S, Beiko R. GenGIS: a geospatial information system for genomic data. Genome Research. 2009;19:1896–1904. doi: 10.1101/gr.095612.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kidd D, Ritchie M. Phylogeographic information systems: putting the geography into phylogeography. Journal of Biogeography. 2006;33:1851–1865. [Google Scholar]
- 84.O’Brien J, Minin V, Suchard M. Learning to count: robust estimates for labeled distances between molecular sequences. Molecular Biology and Evolution. 2009;26:801–814. doi: 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annual Review of Genetics. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 86.Smouse P. To tree or not to tree. Molecular Ecology. 1998;7:399–412. [Google Scholar]