Abstract
APE1 is a multifunctional protein that possesses several nuclease activities, including the ability to incise at apurinic/apyrimidinic (AP) sites in DNA or RNA, to excise 3′-blocking termini from DNA ends, and to cleave at certain oxidized base lesions in DNA. Pre-clinical and clinical data indicate a role for APE1 in the pathogenesis of cancer and in resistance to DNA-interactive drugs, particularly monofunctional alkylators and antimetabolites. In an effort to improve the efficacy of therapeutic compounds, such as temozolomide, groups have begun to develop high-throughput screening assays and to identify small molecule inhibitors against APE1 repair nuclease activities. It is envisioned that such inhibitors will be used in combinatorial treatment paradigms to enhance the efficacy of DNA-interactive drugs that introduce relevant cytotoxic DNA lesions. In this review, we summarize the current state of the efforts to design potent and selective inhibitors against APE1 AP site incision activity.
Keywords: APE1/APEX1/REF-1, Abasic endonuclease, DNA damage, Base excision DNA repair, Inhibitor, Cancer treatment
Introduction
It was not until after the elucidation of the DNA structure by Watson and Crick that the scientific community began to fully recognize the vulnerability of genetic material to spontaneous decomposition or reactions with exogenous or endogenous physical and chemical agents [1]. This realization prompted interest in the prospect that organisms had evolved corrective systems to preserve genome integrity to avert deleterious outcomes such as mutagenesis and cell death. The first evidence for the existence of “DNA repair” was described by Kelner [2] and Dulbecco [3], who discovered, independently although not deliberately, that the variability in survival of cells following ultraviolet irradiation was due to differences in exposure to visible light after treatment. It was later uncovered that there exists a light-dependent photoreactivation process (catalyzed by the enzyme photolyase) that removes cytotoxic photodamage (i.e., cyclobutane pyrimidine dimers) produced within the genome [4].
As the DNA repair field began to emerge, it was soon recognized that the most common forms of DNA damage are likely those that arise as spontaneous hydrolytic products due to the intrinsic instability of nucleic acid or as products of attack by endogenous chemical species, most notably reactive oxygen or nitrogen species formed during mitochondrial respiration [5]. One of the most frequent forms of DNA damage is the apurinic/apyrimidinic (AP) site, which has been estimated to arise at a rate of roughly 10,000 times per mammalian genome per day. AP sites, which pose a mutagenic and cytotoxic challenge to the cell [6], are products of both spontaneous and damage-induced base loss, and are also intermediates generated during the base excision DNA repair (BER) response (Fig. 1). In the early 1980s, two groups described a human enzyme from placenta and HeLa cell extracts that possessed the ability to incise at AP sites introduced into plasmid DNA by heat and acid [7, 8]. Almost 10 years later, the gene encoding the predominant human AP endonuclease was cloned and named APE [9], HAP1 [10] or APEX [11], now more commonly referred to as APE1 or APEX1.
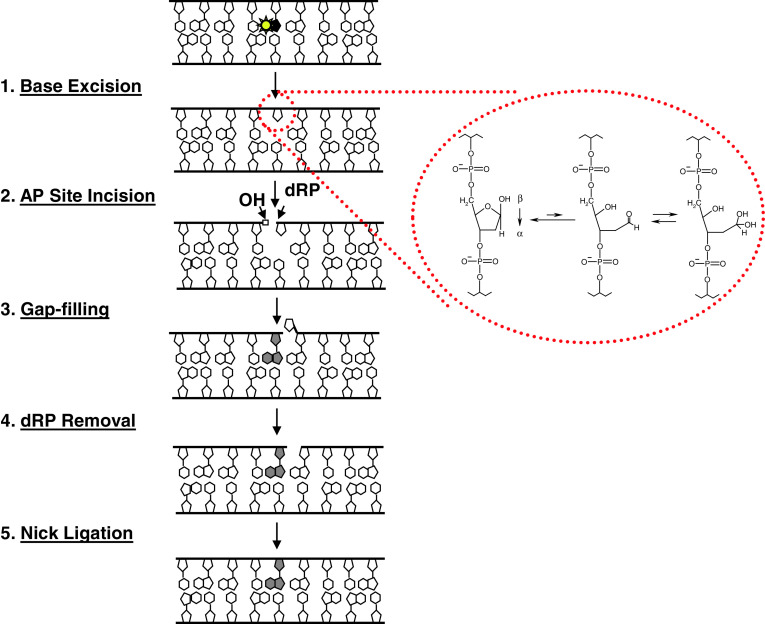
Fig. 1.
The base excision repair response. Left are the five major enzymatic steps of short-patch BER. Other sub-pathways of BER exist (e.g., long-patch), and are described in greater detail elsewhere [80, 81]. Typically, BER is initiated by removal of a damaged or inappropriate base (yellow star) through the action of a DNA glycosylase, which hydrolyzes the N-glycosidic bond that links the base to the sugar-phosphate backbone, generating an AP site. APE1, the major mammalian AP endonuclease, then cleaves the DNA backbone immediately 5′ to the abasic lesion. The subsequent steps of BER—gap-filling to replace the missing nucleotide, removal of the 5′-deoxyribose phosphate (dRP), and sealing of the final nick—are performed primarily by DNA polymerase β and a complex of XRCC1 and DNA ligase 3α, although back-up mechanisms exist [82]. As amplified within the red circle, the hydrolytic 2′-deoxyribose AP site exists primarily (at ~99%) in a ring-closed form (left), in one of two racemeric hemiacetal arrangements, α or β, which are in an equilibrium mixture. Reduction of the ring-closed AP site can produce a ring-opened aldehyde form (middle). This ring-opened form is susceptible to hydration, generating a hydrated aldehyde AP site (right) ([83] and references therein)
Since the discovery of the APE1 gene, significant research has been performed by a number of laboratories to characterize the structure–function mechanism and the biological roles of the protein (see reviews, such as [12–16]). More recently, the decades of information gathered on APE1, as well as the mounting evidence that DNA repair is a logical target for improving anticancer treatment paradigms involving DNA-interactive cytotoxins [17], has spurred investigators to initiate screens for small molecule inhibitors of DNA repair proteins in general and the APE1 nuclease in particular. This review summarizes the various nuclease activities of APE1, the principle behind the reported high-throughput screening assays, and the current picture of the identified endonuclease inhibitors.
Nuclease activities of APE1
APE1 is a ~35,500-Da protein comprised of a four-layered α/β-sandwich core domain (Fig. 2), with a disordered N-terminal extension (not seen in crystal structures to date). The globular core exhibits structural similarity to the divalent cation-dependent phosphoesterase superfamily of proteins, which includes nucleases, inositol (polyphosphate) and possibly protein phosphatases, and sphingomyelinases, and encompasses the endonuclease portion of APE1 [12, 18]. The unique N-terminal segment of ~50 amino acids appears to have been acquired via divergent evolution, and imparts several additional features to the mammalian protein, including nuclear targeting, transcriptional regulatory roles, and its so-called redox (or REF-1) function, which are reviewed in the accompanying articles of this Special Issue. The N-terminal portion of human APE1 may also serve as a docking platform for certain protein interactions (see for example [19–21]).
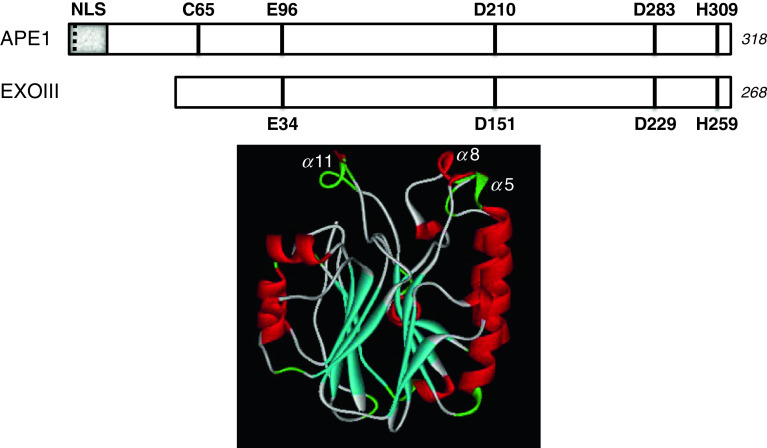
Fig. 2.
APE1 primary sequence conservation and 3-dimensional structure. Top is a linear, schematic comparison of the human APE1 and E. coli exonuclease III (Xth) proteins showing the conserved catalytic residues: E96, D210, D283 and H309 in APE1. The nuclear targeting region (shaded portion, NLS), the acetylation sites (K6 and K7, dashed line), and a critical redox regulatory cysteine residue (C65) is denoted within the unique N-terminal portion of the human protein [15, 16]. The amino acid length of the two proteins is indicated to the right in italics. Below is a ribbon diagram of the human APE1 protein. β-strands are shown in cyan and α-helices in red, with the four-layered α/β sandwich fold apparent. The proposed major groove DNA recognition loop, α8, is denoted, as is the minor groove recognition loop, α11. Coordinates were from 1BIX [84]
APE1 executes a Mg2+-promoted hydrolytic catalytic reaction that severs the phosphodiester bond to facilitate the BER response (Fig. 3, left). While the name “APE1” stems from the ability of the protein to incise at AP sites as an endonuclease, the enzyme was described early as a protein with phosphodiesterase activity for 3′-blocking groups (e.g., 3′-phosphoglycolate ester) in DNA (Fig. 3, right) [22]. In this capacity, APE1 has the ability to excise 3′-obstructive termini, which would normally be refractory to DNA repair synthesis and thus potentially genotoxic, to generate the needed 3′-hydroxyl priming group. In addition, APE1 has a weak (typically ≥100-fold less efficient than its AP endonuclease activity), albeit measurable, 3′–5′ exonuclease function that is influenced by reaction conditions, the configuration of the DNA substrate (i.e., sequence context and precise location of the 3′-target nucleotide), and the chemical composition of the terminal nucleotide [23–29]. For instance, APE1 exhibits a more robust excision activity on 3′ nucleotides in a single-strand break context than at a double-strand break end, and efficiently removes, both in vitro and in vivo, the chain-terminating nucleoside analog troxacitabine, which is currently in clinical trials for the treatment of refractory acute myelogenous leukemia and various solid tumors. APE1 also possesses less expected nuclease activities, such as an ability to incise 5′ to oxidative base modifications, e.g., 5,6-dihydro-2′-deoxyuridine, 5,6-dihydrothymidine, 5-hydroxy-2′-deoxyuridine, alpha-2′-deoxyadenosine and alpha-thymidine adducts, in a process known as nucleotide incision repair (NIR) [30].
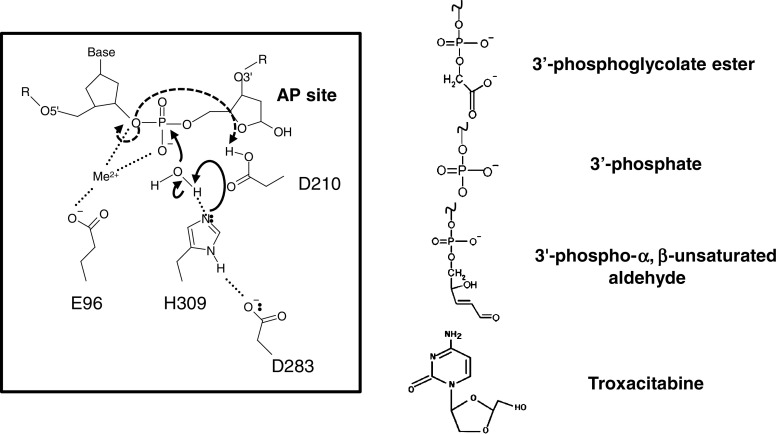
Fig. 3.
APE1 catalytic reaction mechanism and 3′-damage substrates. In the reaction shown (left), H309 acts as the general base and abstracts a proton from a water molecule to generate the active site nucleophile [85]. D283 forms a hydrogen bond with H309 to help stabilize the positive charge that develops upon proton abstraction. The metal ion bound by E96 interacts with the negatively charged phosphate group and aids in nucleophilic attack by the hydroxyl, and in one model also serves to stabilize the leaving group [84]. Alternatively, D210, whose pKa is potentially altered by extensive hydrogen bonding, plays the role of the Lewis base and protonates the 3′ leaving group [86]. Additional reaction mechanisms have been proposed, but are not depicted or described herein [75, 87–89]. To the right are 3′-damage substrates of APE1. Oxidation of C-4′ results in fragmentation of the deoxyribose, causing strand breakage and the formation of a 3′-phosphoglycolate ester or a 3′-phosphate; 3′-phosphate groups can also be generated by bifunctional DNA glycosylases [90]. The α,β-unsaturated aldehyde is the β-elimination product of a bifunctional DNA glycosylase. Troxacitabine is an L-stereoisomeric, chain-terminating nucleoside analog that is a primary substrate for APE1 excision activity [27]
Besides operating on DNA (see summary of APE1 nuclease substrates in Fig. 3), recent studies have found that APE1 can cleave at abasic sites or at certain sequence contexts in RNA molecules [21, 31, 32]. The former activity has been suggested to play a role in “cleansing” the RNA pool by removing damaged RNA templates, while the latter function may operate to regulate gene expression. Although the screening assays described next have focused on the identification of inhibitors of the AP site incision activity of APE1, it is likely that any inhibitor identified would inactivate all of the aforementioned nuclease functions. This conclusion stems largely from the fact that APE1 maintains a single nuclease active site, which is in part comprised of the critical, conserved amino acids, E96, D210 and H309 (Fig. 3) [12, 13]. However, it would be of interest to design inhibitor compounds, if feasible, that exhibit selective inactivation of a particular nuclease function. It is also possible that some inhibitors might be allosteric and as such might inhibit the various nuclease activities differentially or may exert effects on the redox or regulatory functions of this protein as well.
Screening strategy for inhibitors of APE1 incision activity
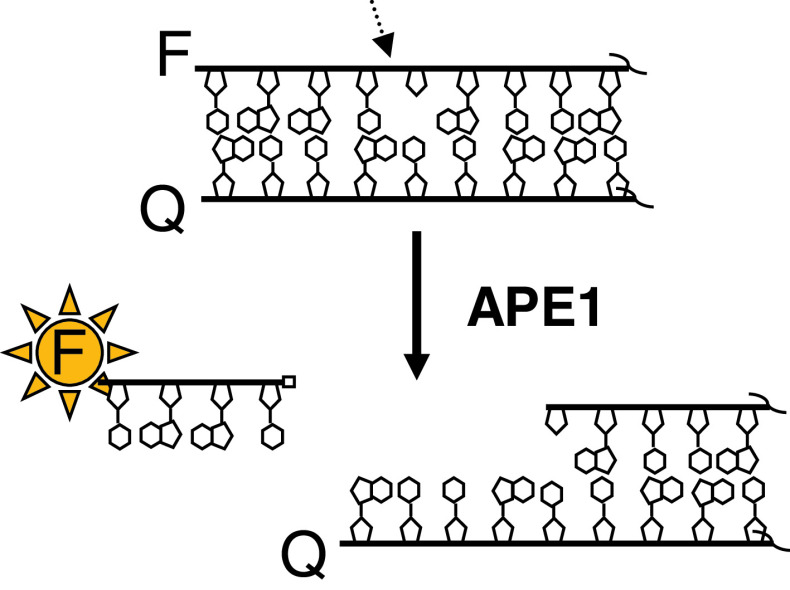
The strategy for high-throughput screening for inhibitors of APE1 AP site incision activity was first described by Madhusudan et al. [33], who designed a fluorescence-based (molecular beacon) assay system. In particular, a 5′-fluorescein-labeled, AP site-containing DNA strand was annealed to a 3′-dabcyl-labeled complementary oligonucleotide (Fig. 4). Positioning of the fluorophore and quench opposite one another resulted in a significantly reduced fluorescence relative to the 5′-fluorescein single-strand oligonucleotide alone. Since the AP site was located in close proximity to the 5′-terminal end, following APE1 cleavage, the short 5′-fluorescein-containing DNA product was spontaneously released from the duplex, resulting in increased fluorescence readout. Thus, inhibitors are revealed by a reduced (or background) fluorescence in high-throughput multi-well screens.
Fig. 4.
APE1 screening assay principle. A deoxyoligonucleotide containing an internal tetrahydrofuran abasic site analog [91] and a 5′ fluorophore is annealed to a complementary strand with a 3′ quencher to create a double-stranded DNA substrate. The close proximity of the fluorophore and the quencher results in a dampened signal upon light excitation. Following DNA backbone cleavage by APE1, a short deoxyoligonucleotide fluorophore-labeled product (typically around 4–6 nucleotides) is spontaneously released from the remaining DNA fragment possessing the quencher, causing the fluorophore emission to increase. F can be any fluorophore, and Q any compatible quench moiety. The APE1 incision site is indicated by the arrow. The right side of the duplex is not complete, as denoted by the squiggly lines
Additional groups have more recently described variations of this strategy, employing different sequence contexts (to optimize the signal to background ratio) or fluorogenic substrates (i.e., TAMRA-labeled, to shift the signal detection to a longer wavelength, thereby minimizing the interfering effect of compound autofluorescence), although each screening method operates under the same guiding principles (see, e.g., [34]). Indeed, the use of fluorogenic screening assays has gained broad support, and such strategies have been applied to a number of nucleic acid metabolizing enzymes, including DNA polymerases [35], ligases [36] and helicases [37]. Furthermore, recent advances in technology have allowed for the execution of APE1 inhibitor screens in very low reaction volumes (e.g., 4 μl in 1,536-well plates) and collection of data in real-time kinetic mode [34]. Recently, an alternative assay that monitors APE1 incision activity with potential high-throughput applicability was designed that employs an instrument for spectral cross-talk-free dual-color fluorescence cross-correlation spectroscopy, using a double-stranded DNA substrate labeled with Cy3 and IRD800 [38]. However, no inhibition measurement was demonstrated within this report, and the method relies on a complex equipment setup not presently suitable for testing a large number of small molecules.
Inhibitors of APE1 nuclease activities
APE1 is essential for animal viability, as deletion of both alleles leads to early stage embryonic lethality in mice [39], and more recent studies have indicated a vital role for this protein in human cell viability [40]. Approaches such as antisense and siRNA have demonstrated that reduced levels of APE1 protein increase the sensitivity of cells to DNA-damaging agents that generate BER substrates, e.g., oxidizing and alkylating compounds. However, in most cases, whether the hypersensitivity results from impairment in its nuclease function(s) or one of its other roles has not been explicitly addressed. That said, recent work, which will not be described in detail herein, has begun to delineate the unique contributions of the nuclease and redox regulatory functions of APE1 in cell viability and stress resistance [41, 42].
Several pre-clinical and clinical studies have validated APE1 as a rational anticancer drug target [43, 44]. For instance, APE1 expression levels and/or subcellular localization has been found to be altered in several tumor types, and, in some instances, is predictive of responsiveness to radio- or chemotherapy. In addition, studies have shown that reduced APE1 protein levels or impaired APE1 protein function increase cellular sensitivity to a range of direct and indirect DNA-interactive anticancer agents, including ionizing radiation, thiotepa, carmustine, temozolomide, and the antimetabolites troxacitabine and 5-fluorouracil [45–49]. This increased cellular sensitivity corresponds in many cases with elevated DNA damage, indicting the DNA metabolizing functions of the protein in resistance. Furthermore, as will be discussed next, many of the identified APE1 small molecule inhibitors, which were selected based on their ability to inhibit APE1 endonuclease activity and, thus, likely do not affect the other functions of APE1 (although this needs to be confirmed in most cases), have been shown to improve the efficacy of clinically relevant DNA-interactive drugs.
CRT0044876 (7-nitro-indole-2-carboxylic acid)
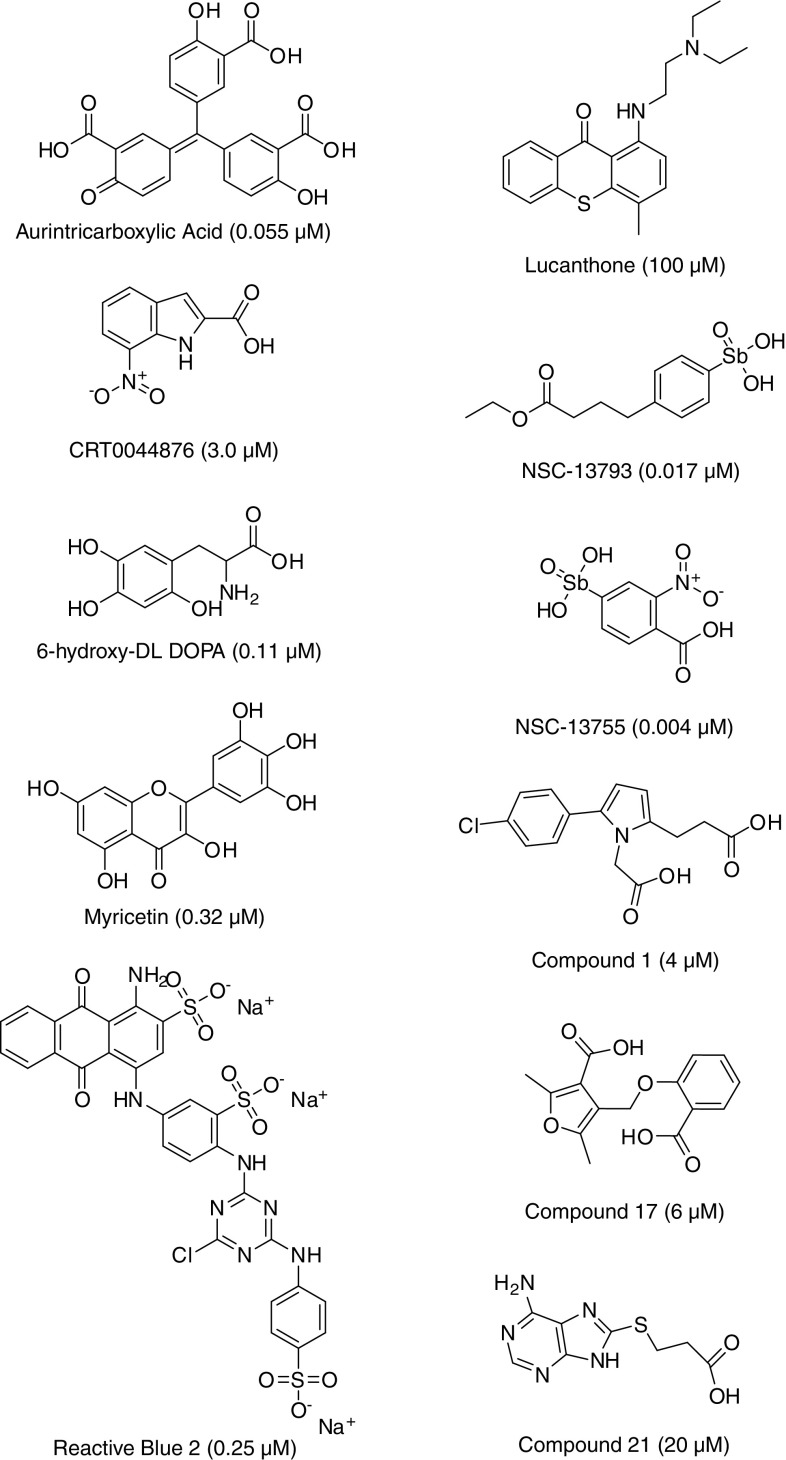
Using the fluorescein/dabcyl-based AP site incision assay described above, Madhusudan et al. [33] identified the first biochemical and biological inhibitor of APE1 from a screen of a collection of structurally diverse small molecules (5,000 in total). The top compound, CRT0044876 (Fig. 5), was shown to be a specific inhibitor of the E. coli exonuclease III family of AP endonucleases, which includes APE1, yet did not inhibit the repair activity of the structurally distinct E. coli AP endonuclease, endonuclease IV (EndoIV). Moreover, at non-toxic doses, CRT0044876 was found to potentiate the cell-killing effect of a number of DNA-damaging agents, including the laboratory chemical methyl methanesulfonate (MMS) and the clinically relevant alkylator temozolomide. CRT0044876 did not alter the cytotoxicity of agents that induced DNA damage not commonly repaired by BER, such as ultraviolet light, nitrogen mustard, and camptothecin, suggesting specificity for APE1 inhibition. However, since the initial report, the reproducibility of this compound has been brought into question [34, 43], and the lack of analogues reported for this singleton complicates the interpretation of any enzymatic or cellular activity that CRT0044876 may possess. While the reason for the apparent discrepancy in results is unclear, it may stem from differences in reaction conditions, the source of the small molecule, or possibly the amino acid composition or status of the recombinant protein. Further studies to better clarify the inconsistencies are needed, as investigators are actively employing CRT0044876 to evaluate the utility of APE1 inhibition in clinically relevant treatment paradigms (see, e.g., [50, 51]). It is also noteworthy that the nitro-aromatic feature of CRT0044876 is generally associated with toxicity issues [52], and may therefore limit the utility of this compound as a drug candidate.
Fig. 5.
Identified inhibitors of APE1 endonuclease activity. Shown are the identifiers, chemical features, and concentrations of compound reported to inhibit 50% of APE1 incision activity in vitro (in parentheses). IC50s were obtained from the following articles, [33, 34, 53, 57, 60, 66], and compounds 1, 17 and 21 are described in [57]
Since CRT0044876 has poor drug-like properties including aqueous solubility and membrane permeability (i.e., sharp drop-off between biochemical and cellular potency) and, thus, does not represent a suitable starting point for medicinal chemistry lead optimization, Madhusudan and colleagues have recently adopted a virtual screening strategy. In this approach, the CRT0044876 template, in addition to prototypical molecular scaffolds designed on the basis of the shape of the ligand binding site within APE1, have been used to perform a rapid structure-based similarity search of a large virtual library of drug-like molecules. ‘Hits’ from this search were subsequently subjected to the more computationally-costly process of docking-based evaluation, followed by detailed biochemical and cellular investigations. Several potent APE1 inhibitors have been isolated using this approach that appear to be promising hit to lead conversion candidates (Srinivasan Madhusudan, personal communication).
Arylstibonic acids
Since the compounds reported in the literature at the time, including CRT0044876, were generally weak (μM) inhibitors of APE1, Stivers and colleagues set out to identify more potent enzyme inactivators [53]. Using a standard fluorescein/dabcyl-based molecular beacon assay, they screened the 2,000-compound NCI Diversity Set of small molecules and identified aromatic nitroso, carboxylate, sulfonamide, and arylstibonic acid compounds with μM affinities for the protein. Since arylstibonic acids appeared frequently as inhibitors of APE1 incision activity in the initial screen, and thus potentially represented a useful scaffold for further diversification, an additional 37-member arylstibonic acid library was tested. Notably, two compounds (13755 and 13793; Fig. 5) were identified with IC50s of 4 and 17 nM, respectively; neither appeared to be a non-specific inhibitor via binding DNA, as they were negative in a conventional dye-displacement assay. However, despite the potent, presumably direct, inhibition observed in vitro, none of the tested antimony-containing small molecules exhibited an obvious effect on the survival of cells challenged with MMS. Whether these compounds are simply not taken up by cells, or do not target APE1 in vivo, needs to be more clearly resolved, but one of the more potent arylstibonic acids (NSC-13755) did display a significant inhibitory effect on total AP endonuclease activity of whole cell extracts, suggesting some specificity for APE1 [34].
As discussed by Seiple and colleagues [53], the very small size of these inhibitors, combined with their relatively high potency, indicates that they largely owe their activity to the arylstibonic core, akin to phosphonate derivatives. As such, this entire class of compounds can be expected to inhibit additional nucleic acid-interacting enzymes [54], making it difficult to develop them into selective agents. Further complicating the picture is the uncertainty surrounding the whole-organ toxicity potential of antimony-containing compounds, particularly as related to the interconversion between their trivalent and pentavalent forms. That said, examples exist of the continuing use and optimization of arsenic- and antimony-based therapeutic agents, particularly in cases of acute infections and cancers associated with poor prognosis [55, 56]. Whether such compounds can be modified in a way to direct specificity against APE1 is unclear.
Pharmacophore-guided inhibitors
Guided by the unique interactions of the APE1 active site with abasic DNA, Zawahir et al. [57] designed a set of pharmacophore models that were used to carry out a virtual screen of a 365,000 small molecule library. Identified hits were subsequently selected for further biochemical analysis based on pharmacophore fit value, pharmacophore mapping pattern, predicted binding orientation, and interactions of the compound within the APE1 active site. Employing a radiolabeled DNA substrate-based AP site incision assay, the authors found that the majority of inhibitors displayed IC50 values of ~3 to 20 μM against APE1, while showing no pronounced, non-specific inhibition of the unrelated AP endonuclease EndoIV or the general DNA phosphate backbone cleaving enzyme HIV-1 integrase. The most effective APE1 inhibitors maintained a structural platform that consisted of an optimum-sized central hydrophobic core linked to two negatively ionizable features, such as two carboxyl groups (see, e.g., compounds 1, 17, and 21 from the study; Fig. 5). However, as will be discussed below, not all inhibitors of APE1 possess these features, ruling out the exclusivity of the suggested pharmacophore and indicating that additional stabilizing interactions beyond those afforded by two negatively charged anchor points need to be considered during the design and optimization of direct APE1 inhibitors. Additional investigation of the top pharmacophore-based inhibitors, particularly to derive structure–activity relationship(s) and improve potency, is also necessary to further advance the overall strategy presented by Zawahir et al. [57]. Lastly, it is important to note that the biological efficacy of the compounds identified in this effort has yet to be described.
6-Hydroxy-DL-DOPA, Reactive Blue 2, and myricetin
Since the reported effectiveness of CRT0044876 had come into question, and since the biological value of the arylstibonic acids was not evident, Simeonov et al. [34] set out to identify novel APE1 inhibitors by screening the commercially available Library of Pharmacologically Active Compounds (LOPAC1280), a collection of well-characterized, drug-like molecules representing all major target classes. Using a series of optimized screening and validation assays (including an EndoIV counterscreen and a dye-displacement test), three compounds were identified as selective, direct inhibitors of APE1 incision activity, i.e., 6-hydroxy-DL-DOPA, Reactive Blue 2, and myricetin (Fig. 5). Each of these bioactives was also found to enhance the cytotoxic and genotoxic potency of the alkylating agent MMS. However, 6-hydroxy-DL-DOPA, Reactive Blue 2, and myricetin, which have well-characterized pharmacological profiles, are known to have additional cellular targets (see discussion within [34]) and are not easily amendable to chemical modification. Moreover, 6-hydroxy-DL-DOPA and some of its analogues suffer from instability in the presence of oxygen. We note that Levodova (i.e., L-DOPA), a naturally occurring dietary supplement and psychoactive drug used in the clinical treatment of Parkinson’s disease that maintains chemical features similar to 6-hydroxy-DL-DOPA, did not inhibit APE1 endonuclease activity in a conventional radiolabeled DNA substrate assay (unpublished observation).
Aurintricarboxylic acid
One of the most potent inhibitors of APE1 identified in the screen of the LOPAC1280 (see above) was aurintricarboxylic acid (ATA) [34], displaying an IC50 value in the tens of nanomolar range (Fig. 5). However, this compound appears to be a highly potent, pan-selective inhibitor of DNA- and RNA-processing enzymes [58], presumably due to its DNA-mimetic properties [59]. Thus, while ATA is a readily available compound that carries some utility as a control reagent, it is unattractive as a candidate agent for targeted combination therapy due to its promiscuity and therefore high probability for unpredictable off-target effects. Furthermore, ATA has been postulated to form a radical homopolymer in solution, casting doubt about its tractability. We stress that investigators should proceed with caution in ongoing and future screening efforts, as other compounds will likely be identified that are potent APE1 inhibitors simply due to DNA likeness, yet will turn out to be pan-selective inhibitors of nucleic acid-processing enzymes.
APE1 repair inhibitor 03
Employing a similar strategy to that outlined in Fig. 4, Bapat et al. [60] screened a library of 60,000 diverse small molecules for APE1 endonuclease inhibitors. After eliminating strong DNA-binders and insoluble compounds, four molecules, designated APE1 repair inhibitor 01 (AR01) (2-(4-(2,5-dimethyl-1Hprryol-1-yl)phenoxy acetic acid), AR02 (4-(2,6,8-trimethylquinolin-4-ylamino)phenol), AR03 (2,4,9-trimethylbenzo [b][1, 8] naphthyridin-5-amine) and AR06 (N-(3-chlorophenyl)-5,6-dihyro-4H-cyclopenta [d] isoxazole-3-carboxamide), were identified as the most promising of the chemically distinct inhibitors with IC50 values of ≤10 μM. Of these, AR03 (Fig. 5) was (1) validated using a radiolabeled oligonucleotide substrate, (2) shown to have only a partial effect on E. coli EndoIV AP site incision activity, and (3) found to inhibit total AP endonuclease activity of human whole cell extracts. Further characterization of AR03 demonstrated that this compound effectively kills cells at low micromolar concentrations (LD50 value of ~1 μM), promotes the accumulation of AP sites in chromosomal DNA of human cells, and potentiates the cytotoxicity of MMS and temozolomide in SF767 glioblastoma cells. While AR03 was argued to exhibit selectivity for APE1 and have potential therapeutic utility, its planar fused-ring structure is similar to that of recognized DNA-binders, a fact partially corroborated by AR03’s mild activity against Endo IV, and, as such, may warrant additional profiling work. Also, given the micromolar IC50 of this inhibitor, medicinal chemistry optimization will need to be performed in order to improve its potency and potentially enhance its selectivity. Nonetheless, AR03 does represent one of the better validated APE1 inhibitors to date.
Lucanthone
Lucanthone (also known as Miracil D) is an orally administered small molecule (a thiaxanthenone) that has been used in the treatment of human schistosomiasis. Its heterocyclic ring structure (Fig. 5) resembles that of actinomycin D, and like actinomycin D, the compound binds to DNA and inhibits RNA synthesis in bacteria [61]. While lucanthone has structural and biochemical similarities to actinomycin D, it has no hematological or gastrointestinal toxicity at clinically tolerated doses, thus making it an attractive therapeutic compound.
In the 1970s, lucanthone was shown to act as a radiosensitizer of cancer cells [62], possibly through the inhibition of post-radiation repair or as an inhibitor of topoisomerase II [63, 64]. Bases and colleagues [65] subsequently reported that exposure of cells to lucanthone resulted in an accumulation of abasic sites, implicating APE1 as a target of this small molecule. Luo and Kelley [66] soon thereafter demonstrated that lucanthone is a direct inhibitor of APE1 in vitro and enhances the cell-killing effect of the drug temozolomide in culture. Part of the inhibitory effect of lucanthone is undoubtedly a result of its ability to intercalate into DNA [61], but some appears to stem from a direct interaction with the APE1 protein (Mamta Naidu, personal communication). Nevertheless, due to the fact that the cellular consequences of lucanthone occur at doses much lower than required to inhibit APE1 nuclease activities in vitro, it has been postulated that the effects of the small molecule are mediated through inhibition of topoisomerase II or another cellular protein. Since lucanthone is currently in clinical development for the treatment of brain tumors due to its ability to readily cross the blood–brain barrier, further information regarding its mechanism of action, and in particular its possible cellular targets, are of great interest.
Methoxyamine
Methoxyamine (trademark TRC102) is small molecule that can be administered orally or intravenously, and is actively being pursued by TRACON Pharmaceuticals in Phase I clinical trials in the management of advanced cancers, such as malignant melanoma, primary brain tumors, and lung cancer (see further details at http://www.traconpharma.com). Methoxyamine forms a covalent alkoxyamine-adducted intermediate with the ring-open form of a deoxyribose residue, generating an AP site product that is refractory to APE1 cleavage, and hence, largely resistant to BER processing [67]. Thus, unlike the mechanism of action presumed for the molecules described earlier (Fig. 5), methoxyamine is an indirect inhibitor of APE1, as it is not known to bind physically to the protein. A sizeable collection of pre-clinical data, largely from the Gerson laboratory, indicates that methoxyamine significantly enhances the antitumor effect of alkylating agents such as carmustine and temozolomide, further supporting the idea that inhibition of AP site repair (i.e., APE1) would be advantageous in certain cancer therapy protocols [68, 69].
Departing thoughts
Many of the agents used in the clinic today to eradicate or manage cancer are cytotoxins that kill rapidly dividing cells by introducing lethal genetic damage [70]. Past and recent evidence has indicated the potential benefit of inhibiting DNA repair pathways in treatment paradigms that involve such DNA-interactive drugs. There has in fact been a renewed and enhanced effort to develop novel DNA repair inhibitors that may improve therapeutic agent efficacy, primarily due to the recent promising clinical results using inhibitors against the strand break response protein, PARP-1 [17, 71, 72]. APE1 is an attractive pharmaceutical target due to its central role in BER, and to the evidence that APE1 repair activity correlates with anticancer agent resistance. Thus, a number of groups have recently set out to develop small molecules that inhibit APE1 endonuclease activity directly, although the design of a potent and selective biological APE1 inhibitor remains elusive to date. Furthermore, a “dominant-negative” form of APE1, which binds substrate DNA with high affinity, but does not execute strand incision, consequently blocking the ensuing steps of BER, may have analogous utility in gene therapy modalities [49, 73].
Despite the lack of a pharmaceutical quality APE1 inhibitor, the recent reports detailed above represent an important step in that direction. In silico modeling indicates that many of the small molecules (Fig. 5), such as CRT0044876, 6-hydroxy-DL-DOPA, arylstibonic analogues and lucanthone, adhere in a similar manner to the hydrophobic pocket of APE1 (consisting of W280, F266 and L282) that forms the binding site for the abasic ring structure in the protein–DNA complex ([33, 34, 53], and unpublished observation). Indeed, all these compounds share a relatively small size and an aromatic hydrophobic central core, and, except for lucanthone, have a negative ionizable group, most frequently a carboxylate. Thus, most of the current APE1 inhibitors tend to resemble the nucleobase-phosphate unit of DNA (though exceptions have been noted, e.g., myricetin and lucanthone). It is conceivable that some degree of DNA “likeness” may need to be retained to achieve high potency. However, additional chemical features will also need to be present in order for the next-generation chemotypes to be APE1-selective. Structure-guided design for direct APE1 inhibitors should take into account not only the residues that define the abasic site binding pocket but also more distal features of the enzyme that are presumably responsible for recognition of the overall bend in DNA made possible by the abasic site [74, 75].
Looking forward, it is evident that synthetic chemical design of analogs is necessary to create more powerful structure–activity relationship profiles, and ultimately, high-affinity selective compounds. In addition, characterization of the binding mode of one or more of the current inhibitors by X-ray crystallography would expedite the rational design of APE1 inhibitors with improved potency. Furthermore, experiments beyond simple potentiation of toxicity in cell culture are needed, such as extending the use of prioritized small molecules into relevant tissue or animal cancer models to explore possible side-effects and further validate APE1 as a pharmacological target for anticancer therapies. While it seems evident that we are at the early stages of direct APE1 inhibitor design and some time away from formal clinical trials, it is important to keep in mind that inhibition of BER is likely to be most advantageous when coupled with a relevant DNA-interactive drug, such as the alkylator temozolomide, commonly used in the treatment of brain cancers [76]. Of course, there are additional considerations—such as delivery of the inhibitor to the site of the tumor and the utility of the current animal models as pre-clinical assessment tools—that fall outside the scope of this review.
It is worth pointing out that, in addition to direct inhibitors of APE1 nuclease activities, inhibitors against the other functions of APE1 may also prove clinically valuable. For example, inhibition of the REF-1 function of APE1 may be a novel therapeutic approach in the management of asthma and airway fibrosis [77], or an anti-proliferative or anti-angiogenesis strategy in the treatment of solid tumors [78, 79]. A recent report by Tell et al. [21] described an interaction between APE1 and nucleophosmin 1 (NPM1), which appears to provide an additional mechanism for controlling the localization, substrate specificity, and activity of APE1. Thus, small molecule inhibitors of this interaction might serve as an alternative valuable modulator of the repair enzyme. The readers of this article are encouraged to review the other manuscripts of this Special Issue, which touch upon several aspects of APE1 biochemistry/biology and DNA damage responses.
Acknowledgments
We thank Drs. Mamta Naidu (Brookhaven National Laboratory, USA) and Srinivasan Madhusudan (University of Nottingham, UK) for sharing unpublished observations and for constructive input to the manuscript. This work was supported by the Intramural Research Program of the National Institute on Aging, NIH; the Molecular Libraries Initiative of the NIH Roadmap for Medical Research; the Intramural Research Program of National Human Genome Research Institute, NIH; and grant 1 R03 MH086444-01 to D.M.W. III.
References
- 1.Friedberg EC. A brief history of the DNA repair field. Cell Res. 2008;18:3–7. doi: 10.1038/cr.2007.113. [DOI] [PubMed] [Google Scholar]
- 2.Kelner A. Effect of visible light on the recovery of streptomyces griseus conidia from ultra-violet irradiation injury. Proc Natl Acad Sci USA. 1949;35:73–79. doi: 10.1073/pnas.35.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dulbecco R. Reactivation of ultra-violet-inactivated bacteriophage by visible light. Nature. 1949;163:949. doi: 10.1038/163949b0. [DOI] [PubMed] [Google Scholar]
- 4.Rupert CS. Enzymatic photoreactivation: overview. Basic Life Sci. 1975;5A:73–87. doi: 10.1007/978-1-4684-2895-7_11. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 7.Kane CM, Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J Biol Chem. 1981;256:3405–3414. [PubMed] [Google Scholar]
- 8.Shaper NL, Grafstrom RH, Grossman L. Human placental apurinic/apyrimidinic endonuclease. Its isolation and characterization. J Biol Chem. 1982;257:13455–13458. [PubMed] [Google Scholar]
- 9.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robson CN, Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim Biophys Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- 12.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 13.Wilson DM, III, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 14.Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, He H, Kelley MR, Georgiadis M. Redox regulation of DNA repair: implications for human health and cancer therapeutic development. Antioxid Redox Signal. 2009;12:1247–1269. doi: 10.1089/ars.2009.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 18.Dlakic M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 19.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., III Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D’Ambrosio C, Scaloni A, Quadrifoglio F, Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson DM, III, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 24.Suh D, Wilson DM, III, Povirk LF. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadi MZ, Ginalski K, Nguyen LH, Wilson DM., III Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J Mol Biol. 2002;316:853–866. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- 26.Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- 27.Chou KM, Kukhanova M, Cheng YC. A novel action of human apurinic/apyrimidinic endonuclease: excision of L-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J Biol Chem. 2000;275:31009–31015. doi: 10.1074/jbc.M004082200. [DOI] [PubMed] [Google Scholar]
- 28.Chou KM, Cheng YC. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J Biol Chem. 2003;278:18289–18296. doi: 10.1074/jbc.M212143200. [DOI] [PubMed] [Google Scholar]
- 29.Wilson DM., III Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol. 2003;330:1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- 30.Gros L, Ishchenko AA, Ide H, Elder RH, Saparbaev MK. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berquist BR, McNeill DR, Wilson DM., III Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J Mol Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, Dianov GL, Hickson ID. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, Wilson DM., III Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS ONE. 2009;4:e5740. doi: 10.1371/journal.pone.0005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorjsuren D, Wilson DM, III, Beard WA, McDonald JP, Austin CP, Woodgate R, Wilson SH, Simeonov A. A real-time fluorescence method for enzymatic characterization of specialized human DNA polymerases. Nucleic Acids Res. 2009;37:e128. doi: 10.1093/nar/gkp641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Ballin JD, la-Maria J, Tsai MS, White EJ, Tomkinson AE, Wilson GM. Distinct kinetics of human DNA ligases I, IIIalpha, IIIbeta, and IV reveal direct DNA sensing ability and differential physiological functions in DNA repair. DNA Repair (Amst) 2009;8:961–968. doi: 10.1016/j.dnarep.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhary S, Sommers JA, Brosh RM., Jr Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J Biol Chem. 2004;279:34603–34613. doi: 10.1074/jbc.M401901200. [DOI] [PubMed] [Google Scholar]
- 38.Lee W, Lee YI, Lee J, Davis LM, Deininger P, Soper SA. Cross-talk-free dual-color fluorescence cross-correlation spectroscopy for the study of enzyme activity. Anal Chem. 2010;82:1401–1410. doi: 10.1021/ac9024768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fung H, Demple B. A vital role for ape1/ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 41.Izumi T, Brown DB, Naidu CV, Bhakat KK, MacInnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y, Guo C, Fishel ML, Wang ZY, Vasko MR, Kelley MR. Role of APE1 in differentiated neuroblastoma SH-SY5Y cells in response to oxidative stress: use of APE1 small molecule inhibitors to delineate APE1 functions. DNA Repair (Amst) 2009;8:1273–1282. doi: 10.1016/j.dnarep.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): From mechanistic insights to druggable target in cancer. Cancer Treat Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, Kolstoe DD. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res. 2002;8:3008–3018. [PubMed] [Google Scholar]
- 46.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 47.Bobola MS, Finn LS, Ellenbogen RG, Geyer JR, Berger MS, Braga JM, Meade EH, Gross ME, Silber JR. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res. 2005;11:7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- 48.Lam W, Park SY, Leung CH, Cheng YC. Apurinic/apyrimidinic endonuclease-1 protein level is associated with the cytotoxicity of L-configuration deoxycytidine analogs (troxacitabine and beta-L-2′, 3′-dideoxy-2′, 3′-didehydro-5-fluorocytidine) but not D-configuration deoxycytidine analogs (gemcitabine and beta-D-arabinofuranosylcytosine) Mol Pharmacol. 2006;69:1607–1614. doi: 10.1124/mol.105.021527. [DOI] [PubMed] [Google Scholar]
- 49.McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM., III Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol Cancer Res. 2009;7:897–906. doi: 10.1158/1541-7786.MCR-08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koll TT, Feis SS, Wright MH, Teniola MM, Richardson MM, Robles AI, Bradsher J, Capala J, Varticovski L. HSP90 inhibitor, DMAG, synergizes with radiation of lung cancer cells by interfering with base excision and ATM-mediated DNA repair. Mol Cancer Ther. 2008;7:1985–1992. doi: 10.1158/1535-7163.MCT-07-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo Y, Kinsella TJ. Essential role of DNA base excision repair on survival in an acidic tumor microenvironment. Cancer Res. 2009;69:7285–7293. doi: 10.1158/0008-5472.CAN-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rickert DE (ed) (1985) Toxicity of nitroaromatic compounds. Hemisphere, Washington, D.C.
- 53.Seiple LA, Cardellina JH, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmacol. 2008;73:669–677. doi: 10.1124/mol.107.042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim H, Cardellina JH, Akee R, Champoux JJ, Stivers JT. Arylstibonic acids: novel inhibitors and activators of human topoisomerase IB. Bioorg Chem. 2008;36:190–197. doi: 10.1016/j.bioorg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frezard F, Demicheli C, Ribeiro RR. Pentavalent antimonials: new perspectives for old drugs. Molecules. 2009;14:2317–2336. doi: 10.3390/molecules14072317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma P, Perez D, Cabrera A, Rosas N, Arias JL. Perspectives of antimony compounds in oncology. Acta Pharmacol Sin. 2008;29:881–890. doi: 10.1111/j.1745-7254.2008.00818.x. [DOI] [PubMed] [Google Scholar]
- 57.Zawahir Z, Dayam R, Deng J, Pereira C, Neamati N. Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem. 2009;52:20–32. doi: 10.1021/jm800739m. [DOI] [PubMed] [Google Scholar]
- 58.Bina-Stein M, Tritton TR. Aurintricarboxylic acid is a nonspecific enzyme inhibitor. Mol Pharmacol. 1976;12:191–193. [PubMed] [Google Scholar]
- 59.Gonzalez RG, Haxo RS, Schleich T. Mechanism of action of polymeric aurintricarboxylic acid, a potent inhibitor of protein–nucleic acid interactions. Biochemistry. 1980;19:4299–4303. doi: 10.1021/bi00559a023. [DOI] [PubMed] [Google Scholar]
- 60.Bapat A, Glass LS, Luo M, Fishel ML, Long EC, Georgiadis MM, Kelley MR (2010) Novel small molecule inhibitor of Ape1 endonuclease blocks proliferation and reduces viability of glioblastoma cells. J Pharmacol Exp Ther (in press) [DOI] [PMC free article] [PubMed]
- 61.Hirschberg E, Weinstein IB, Gersten N, Marner E, Finkelstein T, Carchman R. Structure-activity studies on the mechanism of action of miracil D. Cancer Res. 1968;28:601–607. [PubMed] [Google Scholar]
- 62.Bases R. Enhancement of x-ray damage in HeLa cells by exposure to lucanthone (Miracil D) following radiation. Cancer Res. 1970;30:2007–2011. [PubMed] [Google Scholar]
- 63.Bases R, Leifer A, Rozycki H, Blake C, Jr, Neubort S. Effects of lucanthone on the sedimentation properties of DNA from HeLa cells. Cancer Res. 1977;37:2177–2181. [PubMed] [Google Scholar]
- 64.Bases RE, Mendez F. Topoisomerase inhibition by lucanthone, an adjuvant in radiation therapy. Int J Radiat Oncol Biol Phys. 1997;37:1133–1137. doi: 10.1016/s0360-3016(97)00113-2. [DOI] [PubMed] [Google Scholar]
- 65.Mendez F, Goldman JD, Bases RE. Abasic sites in DNA of HeLa cells induced by lucanthone. Cancer Invest. 2002;20:983–991. doi: 10.1081/cnv-120005914. [DOI] [PubMed] [Google Scholar]
- 66.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 67.Rosa S, Fortini P, Karran P, Bignami M, Dogliotti E. Processing in vitro of an abasic site reacted with methoxyamine: a new assay for the detection of abasic sites formed in vivo. Nucleic Acids Res. 1991;19:5569–5574. doi: 10.1093/nar/19.20.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu L, Gerson SL. Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr Opin Investig Drugs. 2004;5:623–627. [PubMed] [Google Scholar]
- 69.Fishel ML, He Y, Smith ML, Kelley MR. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res. 2007;13:260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 70.Thurston DE. Chemistry and pharmacology of anticancer drugs. Boca Raton, FL: CRC Press; 2007. [Google Scholar]
- 71.Chalmers AJ. The potential role and application of PARP inhibitors in cancer treatment. Br Med Bull. 2009;89:23–40. doi: 10.1093/bmb/ldp005. [DOI] [PubMed] [Google Scholar]
- 72.Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur J Cancer. 2010;46:9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 73.McNeill DR, Wilson DM., III A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 74.Barsky D, Foloppe N, Ahmadia S, Wilson DM, III, MacKerell AD., Jr New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res. 2000;28:2613–2626. doi: 10.1093/nar/28.13.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 76.Villano JL, Seery TE, Bressler LR. Temozolomide in malignant gliomas: current use and future targets. Cancer Chemother Pharmacol. 2009;64:647–655. doi: 10.1007/s00280-009-1050-5. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen C, Teo JL, Matsuda A, Eguchi M, Chi EY, Henderson WR, Jr, Kahn M. Chemogenomic identification of Ref-1/AP-1 as a therapeutic target for asthma. Proc Natl Acad Sci USA. 2003;100:1169–1173. doi: 10.1073/pnas.0437889100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, Nyland RL, Borch RF, Qiao X, Georgiadis MM, Kelley MR. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1. Antioxid Redox Signal. 2008;10:1853–1867. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zou GM, Karikari C, Kabe Y, Handa H, Anders RA, Maitra A. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: therapeutic implications in tumor angiogenesis. J Cell Physiol. 2009;219:209–218. doi: 10.1002/jcp.21666. [DOI] [PubMed] [Google Scholar]
- 80.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilde JA, Bolton PH, Masumder A, Manoharan M, Gerlt JA. Characterization of the equilibrating forms of the aldehydic abasic site in duplex DNA by 17O NMR. J Am Chem Soc. 1989;111:1894–1896. [Google Scholar]
- 84.Gorman MA, Morera S, Rothwell DG, de La FE, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mundle ST, Delaney JC, Essigmann JM, Strauss PR. Enzymatic mechanism of human apurinic/apyrimidinic endonuclease against a THF AP site model substrate. Biochemistry. 2009;48:19–26. doi: 10.1021/bi8016137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erzberger JP, Wilson DM., III The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J Mol Biol. 1999;290:447–457. doi: 10.1006/jmbi.1999.2888. [DOI] [PubMed] [Google Scholar]
- 87.Lowry DF, Hoyt DW, Khazi FA, Bagu J, Lindsey AG, Wilson DM., III Investigation of the role of the histidine-aspartate pair in the human exonuclease III-like abasic endonuclease, Ape1. J Mol Biol. 2003;329:311–322. doi: 10.1016/s0022-2836(03)00382-6. [DOI] [PubMed] [Google Scholar]
- 88.Mundle ST, Fattal MH, Melo LF, Coriolan JD, O’Regan NE, Strauss PR. Novel role of tyrosine in catalysis by human AP endonuclease 1. DNA Repair (Amst) 2004;3:1447–1455. doi: 10.1016/j.dnarep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 89.Lipton AS, Heck RW, Primak S, McNeill DR, Wilson DM, III, Ellis PD. Characterization of Mg2+ binding to the DNA repair protein apurinic/apyrimidic endonuclease 1 via solid-state 25 Mg NMR spectroscopy. J Am Chem Soc. 2008;130:9332–9341. doi: 10.1021/ja0776881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilson DM., III Processing of nonconventional DNA strand break ends. Environ Mol Mutagen. 2007;48:772–782. doi: 10.1002/em.20346. [DOI] [PubMed] [Google Scholar]
- 91.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]