Abstract
Although chemotherapy is an important therapeutic strategy for cancer treatment, it fails to eliminate all tumor cells due to intrinsic or acquired drug resistance, which is the most common cause of tumor recurrence. Emerging evidence suggests an intricate role of cancer stem cells (CSCs) and epithelial-mesenchymal transition (EMT)-type cells in anticancer drug resistance. Recent studies also demonstrated that microRNAs (miRNAs) play critical roles in the regulation of drug resistance. Here we will discuss current knowledge regarding CSCs, EMT and the role of regulation by miRNAs in the context of drug resistance, tumor recurrence and metastasis. A better understanding of the molecular intricacies of drug resistant cells will help to design novel therapeutic strategies by selective targeting of CSCs and EMT-phenotypic cells through alterations in the expression of specific miRNAs toward eradicating tumor recurrence and metastasis. A particular promising lead is the potential synergistic combination of natural compounds that affect critical miRNAs, such as curcumin or epigallocatechin-3-gallate (EGCG) with chemotherapeutic agents.
Keywords: drug resistance, cancer, miRNA, EMT, cancer stem cells, natural compounds, curcumin
1. Introduction
Chemotherapy is an important therapeutic option for most cancer patients; however, drug resistance causes the failure of chemotherapy even after combination chemotherapy. Therefore, increasing the drug sensitivity is a key step towards improved treatment of cancer patients. Resistance to anticancer drug therapy is generally classified in two categories: intrinsic (de novo) and acquired resistance (Szakacs et al., 2006). Intrinsic resistance would make the therapy ineffective prior in therapy-naïve patients because the tumor cells have already a resistant phenotype at achievable doses of anti-cancer drugs. While tumor(cell)s often show initial sensitivity to anti-cancer drugs, acquired resistance develops during the treatment, which leads to tumor recurrence and further progression. Although the mechanisms responsible for multidrug resistance have been investigated intensely over the past 50 years the clinical causes of multidrug resistance are still very incompletely understood (Borst et al., 2007; Broxterman et al., 2009). The most cited mechanisms for the acquisition of multidrug resistance are the expression of energy-dependent transporters that eject anti-cancer drugs from cells, insensitivity to drug-induced apoptosis and the induction of drug-detoxification mechanisms (Gottesman, 2002). For instance, three ATP-binding cassette (ABC) drug transporters, namely ABCB1 (MDR1, Pgp, P-glycoprotein), ABCG2 (BCRP, breast cancer resistant protein) and ABCC1 (MRP1, multidrug resistance associated protein) have been associated frequently with drug resistant phenotypes in experimental systems (Broxterman et al., 1996; Gottesman, 2002; Szakacs et al., 2006); however, there is currently no treatment strategy to override these transporters for therapeutic benefit (Kolitz et al., 2010). Furthermore, a number of proteins, including K-ras, COX-2, cyclin D1, Bcl-2, and Survivin, play critical roles in drug resistance to conventional chemotherapeutics (Bardelli and Siena, 2010; Gottesman, 2002; Liu et al., 2010a; Lopez-Chavez et al., 2009). In addition, major cell survival signaling pathway receptors and downstream proteins have been reported to be involved in drug resistance such as the EGFR (epidermal growth factor receptor), FGFR (fibroblast growth factor receptor), PDGFR (platelet derived growth factor receptor) and IGFR (insulin-like growth factor recptor), PTEN (phosphatase and tensin homolog on chromosome 10), ERK (extracellular signal-regulated kinase), MAPK (mitogen-activated protein kinase), MEK (MAP/ERK kinase), Akt, mTOR (mammalian target of rapamycin), NF-κB (nuclear factor-kappa B) and Notch (Haagenson and Wu, 2010; Hendrickson and Haluska, 2009; Hopper-Borge et al., 2009; Jiang and Liu, 2008; Kono et al., 2009; Lin et al., 2010; LoPiccolo et al., 2008; Mehta and Osipo, 2009; Wang et al., 2008; Wang et al., 2009a; Wang et al., 2010d; Wang et al., 2010c). Moreover, recent studies have shown that cancer stem cells (CSCs) and epithelial-mesenchymal transition (EMT)-type cells could play critical roles in drug resistance (Konopleva et al., 2009; Todaro et al., 2007; Voulgari and Pintzas, 2009; Wang et al., 2009b). Thus, the molecular knowledge of drug resistance related to CSCs and EMT is now considered an important focus for cancer research. Finally, recent studies have demonstrated that microRNAs (miRNAs) are involved in the regulation of drug resistance (Sarkar et al., 2010) and the role of miRNA in CSCs and EMT regulation is just beginning to emerge. Gaining further insight in these new concepts would likely be helpful not only in the discovery of new drugs but also in the design of novel therapeutic strategies for the treatment of human cancer with better outcome. The following sections will summarize what we know about these new concepts in drug resistance of human cancers, and how such knowledge might be useful to design novel therapeutic strategies by selective targeting of EMT, CSCs, and miRNAs towards achieving better treatment outcome by preventing tumor recurrence and metastatic progression.
2. EMT and drug resistance
Emerging evidence suggest a molecular and phenotypic association between chemo-resistance and the acquisition of the EMT-like phenotype of cancer cells. It is now widely accepted that epithelial cells can acquire mesenchymal phenotype by a fundamental and complex processes. The processes of EMT is a unique process by which epithelial cells undergo remarkable morphologic changes characterized by a transition from their epithelial cobblestone phenotype to an elongated fibroblastic phenotype (mesenchymal phenotype) accompanied by increased motility and invasion (Thiery et al., 2009). During the acquisition of EMT characteristics, cells loose epithelial cell-cell junctions, undergo actin cytoskeleton reorganization and decrease in the expression of proteins that promote cell-cell contact such as E-cadherin and γ-catenin, and gain in the expression of mesenchymal markers such as vimentin, fibronectin, α-smooth muscle actin (SMA), N-cadherin as well as increased activity of matrix metalloproteinases (MMPs) like MMP-2, MMP-3 and MMP-9, associated with an invasive phenotype (Thiery and Sleeman, 2006).
Recent studies have shown an intimate relationship between drug resistance and the EMT phenotype. For instance, an epithelial, but not mesenchymal gene signature has been associated with sensitivity to the EGFR inhibitor erlotinib-mediated growth inhibition in lung cancer cells (Yauch et al., 2005). These results were confirmed in other types of tumors like head and neck squamous cell carcinoma and hepatocellular carcinoma as well as for the treatment of cancer with other EGFR inhibitors such as gefitinib and cetuximab (Frederick et al., 2007; Fuchs et al., 2008; Voulgari and Pintzas, 2009). The process of EMT has also been shown to be important in conferring drug resistance to cancer cells against conventional therapeutics including taxol, vincristine and oxaliplatin (Sabbah et al., 2008). Recent studies showed a link between EMT and gemcitabine-resistant pancreatic cancer cells, oxaliplatin-resistant colorectal cancer cells, lapatinib-resistant breast cancer and paclitaxel-resistant ovarian carcinoma cells (Arumugam et al., 2009; Kajiyama et al., 2007; Konecny et al., 2008; Shah et al., 2007; Yang et al., 2006). For example, paclitaxel-resistant ovarian cancer cells showed phenotypic changes consistent with EMT with decreased expression of the epithelial adhesion molecule, E-cadherin and an increase in mesenchymal markers such as vimentin, α-SMA and fibronectin (Kajiyama et al., 2007). In parallel, tamoxifen-resistant breast cancer cells demonstrated altered morphological characteristic of cells similar to EMT with altered β-catenin phosphorylation (Hiscox et al., 2006; Kim et al., 2009). Recently, Rho et al. reported phenotypic changes such as a spindle-cell shape and increased pseudopodia formation, suggesting the presence of EMT, in gefitinib-resistant lung cancer cells, which was further documented by a decrease in the expression of E-cadherin and an increase in the expression of vimentin (Rho et al., 2009). Our studies have also shown that gemcitabine-resistant (GR) pancreatic cancer cells acquired EMT characteristics (Shah et al., 2007; Wang et al., 2009c). Moreover, we found that down-regulation of Notch signaling led to partial reversal of the EMT phenotype, resulting in mesenchymal-epithelial transition (MET), which was associated with decreased expression of vimentin, ZEB1, Slug, Snail and NF-κB (Wang et al., 2009c). These results suggest that the increased Notch signaling is mechanistically associated with chemoresistance and EMT characteristics of pancreatic cancer cells. Very interestingly, some reports indicated that mesenchymal-like, basal breast cancers are initially more sensitive to chemotherapy than epithelial-like luminal breast cancers (Carey et al., 2007; Liedtke et al., 2008; Paik et al., 2006). However, it was discussed that basal, mesenchymal-like breast cancers possibly would be more prone to develop drug resistance because patients with basal cancers have worst prognoses even though they have initial responses. It has been indicated that mesenchymal-like cancers might be more sensitive to DNA damaging agents such as doxorubicin, whereas epithelial-like cancers are more sensitive to targeted therapies, such as EGFR and HER2 antagonists (Singh and Settleman, 2010). Clearly, further investigations need to identify and characterize the mechanisms of EMT in drug resistance. The discovery of the precise mechanisms that govern the acquisition of EMT characteristics in cancer cells would likely be useful for devising better targeted therapeutic approaches in combination with conventional therapeutics. Collectively, these studies clearly provide strong evidence linking chemo-resistance to EMT. A comprehensive list of EMT involvement in chemoresistance is presented in Table 1.
Table 1.
A comprehensive list of EMT involved in chemoresistance
| Chemotherapeutic Drug | Characteristics | Cell or tissue | References |
|---|---|---|---|
| Cetuximab | Epithelial cell lines were significantly more susceptible to this agent. | Human hepatoma cells | (Fuchs et al., 2008) |
| Erlotinib | Epithelial gene signature is associated with sensitivity to erlotinib. | Human hepatoma cells; Lung cancer | (Yauch et al., 2005) |
| Gefitinib | Epithelial cell lines were found to be significantly more susceptible to this agent; gefitinib-resistant cancer cells became EMT-type cells. | Head and neck squamous cell carcinoma; non-small cell lung carcinoma | (Frederick et al., 2007; Rho et al., 2009) |
| Gemcitabine | Gemcitabine-resistant cells have EMT features; EMT cells have high expression of Notch; EMT contributes to drug resistance | Pancreatic cancer cells | (Arumugam et al., 2009; Shah et al., 2007; Wang et al., 2009c) |
| Lapatinib | Lapatinib-resistant cell lines exhibited EMT features. | Breast cancer | (Konecny et al., 2008) |
| Oxaliplatin | Chronic oxaliplatin resistance induced EMT | Colorectal cancer cells | (Yang et al., 2006) |
| Paclitaxel | Paclitaxel-resistant cells showed EMT characteristics. | Ovarian carcinoma cells | (Kajiyama et al., 2007) |
| Tamoxifen | Tamoxifen resistance promotes EMT-like behavior and involved regulation of beta-catenin phosphorylation and Pin1 expression | Breast cancer cells | (Hiscox et al., 2006; Kim et al., 2009) |
| Doxorubicin | Doxorubicin activates TGFβ signaling and EMT | Human and murine breast cancer cells | (Bandyopadhyay et al., 2010) |
The obvious question that has to be answered in order to be able to rationally design novel treatment strategies is how EMT is mechanistically involved in drug-resistance. A body of literature currently suggests that the relative resistance of EMT tumor cells is related to their cancer stem cell-like features. Then it appears that elimination of these cells is essential for overcoming drug resistance (Singh and Settleman, 2010; Voulgari and Pintzas, 2009). Emerging evidence has implicated EMT with the conversion of early stage tumors into invasive malignancies accompanied by increased cell motility and invasion, and these processes are consistent with the acquisition of “cancer stem-like cell” phenotype also known as “stemness (Thiery et al., 2009; Thiery and Sleeman, 2006). Therefore, we will discuss the role of CSCs in drug resistance in the following section.
3. Cancer stem cell and drug resistance
Current cancer therapeutic strategies based on tumor regression may target and kill differentiated tumor cells, which constitute the bulk of the tumor, while sparing the rare CSC population. CSCs constitute a mostly small (Dirks, 2010) subset of cancer cells that possess the ability to self-renew and generate the diverse differentiated cell populations that comprise the cancer mass (Gupta et al., 2009a; Frank et al., 2010). A growing body of evidence now supports the concept that cancers are diseases driven by subpopulation of self renewing CSCs, which have been identified and isolated from tumors of the hematopoietic system, breast, lung, prostate, colon, brain, head and neck, and pancreas (Tang et al., 2007). The CSCs possess the capacity for self-renewal and have the ability to drive continued expansion of the population of malignant cells with invasive and metastatic propensity. For example, the CSCs are able to self-renew, differentiate, and regenerate to phenotypic cells of the original tumor when implanted into the severe combined immunodeficient mouse.
Recently, CSCs have been blamed for playing a critical role in drug resistance and cancer metastasis, which may explain why it is difficult to completely eradicate cancer and why recurrence is a real threat to eradicate tumors completely. It has been reported that CSCs in mammary tumors may contribute to cisplatin and paclitaxel resistance in vitro and in vivo (Shafee et al., 2008; To et al., 2010). Similarly, CSCs in colorectal cancers are also believed to be responsible for resistance to chemotherapeutic drugs (Cammareri et al., 2010; Dylla et al., 2008; Fang et al., 2010; Ong et al., 2010; Todaro et al., 2007). In acute myelogenous leukemia (AML), CD34+CD38− human primary AML stem cells residing in the endosteal region of the bone marrow are chemotherapy resistant (Ishikawa et al., 2007; Saito et al., 2010). Chronic myelogenous leukemia reveals the presence of a CD34− cell population with intrinsic resistance to imatinib (Lemoli et al., 2009). In glioblastoma, a population of CD133+ cancer stem cells showed significant resistance to chemotherapeutic agents including temozolomide, carboplatin, paclitaxel and etoposide (Liu et al., 2006). In human hepatocellular carcinoma, CSCs showed significantly higher viability following treatment with doxorubicin or methotrexate (Zhang et al., 2010). In small-cell lung carcinoma, a small population of uPAR+ stem-like cells showed high clonogenic activity and co-expression of CD44 and MDR1 (Gutova et al., 2007), fitting with a drug resistant profile (Toole and Slomiany, 2008). Another group confirmed that small-cell lung CSCs, which are CD133+/ABCG2+ cells, are resistant to cisplatin (Wang et al., 2010b). Recently, osteosarcoma CSCs are reported to be associated with metastasis and drug resistance. Osteosarcoma CSCs showed highly invasive and drug-resistant properties and were enriched for CXCR4 and ABCG2 (Adhikari et al., 2010). Hermann et al. have reported that human pancreatic CSCs are exclusively tumorigenic and highly resistant to chemotherapy (Hermann et al., 2007). They also found a distinct subpopulation of CSCs in the invasive front of pancreatic tumors and depletion of this “migratory CSC pool” virtually abrogated the metastatic phenotype of pancreatic tumors without affecting their tumorigenic potential (Hermann et al., 2007). Another study has shown that pancreatic CSCs are gemcitabine-resistant (Hong et al., 2009). Recently, it has been reported that ovarian CSCs are markedly resistant to carboplatin and paclitaxel (Liu et al., 2010b; Shi et al., 2010b). Another group confirmed that ovarian CSCs are resistant to cisplatin in vitro and in vivo due to high expression of ABCG2 (Hu et al., 2010). Prostate stem cells showed innate resistance to arsenic-induced cytolethality, which was in part due to higher expression of ABCC1 (Tokar et al., 2010). Prostate CSCs are resistant to chemotherapeutics such as cisplatin, paclitaxel, doxorubicin and methotrexate (Liu et al., 2010c). However, an intriguing study on colorectal cancer, suggested that an ABCB1 transporter expressing subpopulation of differentiated cells protected the CSCs against irinotecan (Emmink et al., 2010). A comprehensive list of CSCs involvement in chemoresistance is presented in Table 2.
Table 2.
A list of CSCs involved in chemoresistance
| Cancer stem cells | CSCs surface markers | Chemotherapeutic Drug | Characteristics | References |
|---|---|---|---|---|
| acute myeloid leukemia | CD34+/CD38- | Ara-C | CSCs are chemotherapy resistant | (Ishikawa et al., 2007; Saito et al., 2010) |
| Breast cancer | CD29hi/CD24med CD44hi/CD24low | Cisplatin; paclitaxel | CSCs contribute to cisplatin and paclitaxel resistance | (Shafee et al., 2008; To et al., 2010) |
| Chronic myelogenous leukemia | CD34- | Imatinib | CSCs are resistant to imatinib | (Lemoli et al., 2009) |
| Colon cancer | CD133+ | Oxaliplatin; 5-fluorouracil; cyclophosphami de; irinotecan | CSCs are resistant to oxaliplatin and 5-FU due to induction of interleukin-4; resistant to cyclophosphamide due to production of Aldehyde dehydrogenase 1 activity. | (Dylla et al., 2008; Fang et al., 2010; Ong et al., 2010; Todaro et al., 2007) |
| Glioblastoma | CD133+ | temozolomide, carboplatin, paclitaxel, and etoposide | CSCs showed significant resistance to these chemotherapeutic agents | (Liu et al., 2006) |
| Hepatocellular carcinoma | Hoechst+ | Doxorubicin; methotrexate | CSCs showed significantly higher viability following treatment with doxorubicin or methotrexate | (Zhang et al., 2010) |
| Osteosarcoma | CD117+ Stro-1+ | Doxorubicin | CSCs showed drug-resistant properties and were enriched for CXCR4 and ABCG2 | (Adhikari et al., 2010) |
| Ovarian cancer | CD44+/CD24-CD133+/CD117+ Hoechst+ | Carboplatin and paclitaxol; cisplatin | CSCs markedly resistant to carboplatin and paclitaxol. CSCs resisted cisplatin due to high expression of ABCG2 | (Hu et al., 2010; Shi et al., 2010b) |
| Pancreatic cancer | CD44+ CD44+/CD24- | Gemcitabine | CSCs showed dramatic drug resistance to gemcitabine | (Hermann et al., 2007; Hong et al., 2009) |
| Prostate cancer | CD117+ABCG 2+ | Arsenite; casplatin, paclitaxel, adriamycin, methotrexate | CSCs showed resistance to arsenic-induced cytolethality due to higher expression of ABCC1 | (Liu et al., 2010c; Tokar et al., 2010) |
| Small cell lung cancer | CD133+/ABCG 2+ uPAR+ | 5-FU; cisplatin and etoposide | CSCs demonstrated multi-drug resistance. | (Gutova et al., 2007; Wang et al., 2010b) |
The mechanisms causing CSCs drug resistance are still poorly understood and overexpression of drug transporters and DNA repair enzymes, may only partly explain the entire resistance spectrum (Styczynski and Drewa, 2007; Zhou et al., 2001; Patrawala et al., 2005) and regulation by miRNAs is one emerging level of complexity. Another mechanism might be that CSCs accumulate mutations over time as a consequence of a long-term exposure to drug, which then confer a drug resistance phenotype acquired by the daughter cancer cells (Reya et al., 2001).
4. miRNAs and drug resistance
Recent evidence suggests that microRNAs (miRNAs), which are single-stranded 19–25 nucleotide short RNAs, play important roles in the regulation of drug resistance (Sarkar et al., 2010). It is well documented that miRNAs elicit their regulatory effects in post-transcriptional regulation of genes by binding to the 3′ untranslated region (3′UTR) of target messenger RNA (mRNA), leading to translational repression or target mRNA cleavage (Croce and Calin, 2005). Some miRNAs have oncogenic activity while others have tumor suppressor activity. Oncogenic miRNAs are up-regulated in cancer and contribute to its pathology through various mechanisms such as targeting tumor suppressor genes. These oncogenic miRNAs for example include miR-21, miR-17-92, miR-155, miR-221 and miR-222. In contrast to the oncogenic miRNAs, other miRNAs are considered to have tumor suppressor activity and are down-regulated in cancer. These miRNAs include let-7, miR-15, miR-16, miR-17-5p, miR-29, miR-34, miR-124a, miR-127, miR-143, miR-145 and miR-181 (Croce, 2009). The list of these miRNAs is still increasing.
Specific miRNAs have altered expression in drug-resistant tumor cells (Galluzzi et al., 2010; Garofalo et al., 2009; Iorio et al., 2009; Iorio and Croce, 2009; Kotani et al., 2009; Kovalchuk et al., 2008; Miller et al., 2008; Vere White et al., 2009; Wang et al., 2010a; Xia et al., 2008; Zhao et al., 2008; Zhu et al., 2008). For example, the expression of three miRNAs (miR-192, miR-424 and miR-98) was significantly increased while the expression of three other miRNAs (miR-194, miR-200b and miR-212) was decreased in docetaxel-resistant NSCLC cells (Rui et al., 2010). Recently, the expression of miR-140 was found to be associated with chemo-sensitivity to 5-fluorouracil (5-FU) and methotrexate in osteosarcoma. Blocking endogenous miR-140 sensitized resistant cancer cells to 5-FU treatment, whereas overexpression of miR-140 made tumor cells more resistant to 5-FU, suggesting that miR-140 could be a novel target to develop a therapeutic strategy to overcome drug resistance (Song et al., 2009). In breast cancer cells, miR-21 was overexpressed, causing upregulation of MDR1 (Bourguignon et al., 2009). MiR-34a is down-regulated in drug resistant prostate cancer cells and ectopic over-expression of miR-34a resulted in growth inhibition and attenuated chemoresistance to camptothecin (Fujita et al., 2008). Another group showed that miR-34a was down-regulated in doxorubicin and verapamil resistant MCF-7 breast cancer cells (Chen et al., 2010). Interestingly, Ji et al reported that pancreatic cancer stem cells are enriched with tumor-initiating cells or CSCs with loss of miR-34 (Ji et al., 2009b), suggesting that miR-34 could be involved in pancreatic CSCs self-renewal and leading to drug resistance. Ectopic miR-1 expression sensitized lung cancer cells to doxorubicin, suggesting that up-regulation of miR-1 has the potential as a target for therapy against lung cancers (Nasser et al., 2008). The expression of miR-200b was significantly down-regulated in docetaxel-resistant NSCLC cells (Rui et al., 2010). Recently, many studies have shown that the miR-200 family regulates EMT associated with drug resistance. One study showed that miR-200 expression regulates EMT in bladder cancer cells and reverses resistance to EGFR inhibitor therapy (Adam et al., 2009). Another study reported that miR-200c restored microtubule-binding chemotherapeutic agents in breast and ovarian cancer cells (Cochrane et al., 2009). We have also shown that re-expression of miR-200 family resulted in increased cell sensitivity to gemcitabine (Li et al., 2009b). A comprehensive list of miRNAs implicated in chemoresistance is presented in Table 3.
Table 3.
A list of miRNAs involved in chemoresistance
| miRNA | Characteristics | Target genes | Cell or tissue | References |
|---|---|---|---|---|
| miR-1 | Expression sensitize lung cancer cells to doxorubicin | Pim1, FoxP1, HDAC4, MET | Lung cancer cells | (Nasser et al., 2008) |
| miR-15/16 | Sensitized cancer cells to anti-cancer drugs | Bcl-2 | Gastric cancer cells | (Xia et al., 2008) |
| miR-21 | mediated chemoresistance, inhibited PDCD4, | PDCD4, LRRFIP1, PTEN, TPM1, TIMP3 | Breast cells, glioblastoma cells, pancreatic cancer cells | (Ali et al., 2010; Bourguignon et al., 2009; Li et al., 2009a) |
| miR-27 | Upregulated in multidrug-resistant cancer cells | MDR | Ovarian cancer cells, cervix cancer cells | (Zhu et al., 2008) |
| miR-34 | Mediated suppression of self-renewal, down-regulated in drug resistant cells; over-expression resulted in attenuated chemoresistant to camptothecin | Notch, Bcl2, HMGA2, SIRT1 | Prostate, gastric, breast, pancreatic cancer. | (Chen et al., 2010; Fujita et al., 2008; Ji et al., 2009b) |
| miR-98 | Up-regulated in docetaxel-resistant cells | Not detected | NSCLC cells | (Rui et al., 2010) |
| miR-125b | Rendered cells resistant to androgen withdrawal | CDK6, CDC25A | Prostate cancer | (Vere White et al., 2009) |
| miR-128b | Down-regulated in resistant cells. Over-expression increased cell sensitivity. | MLL, AF4, MLL-AF4, AF4-MLL | Acute lymphocytic leukemia cells | (Kotani et al., 2009) |
| miR-140 | Upregulated and associated with chemosensitivity to 5-FU and methotrexate | P53, p21 | Osteosarcoma, colon cancer cells | (Song et al., 2009) |
| miR-181 | Enhanced cisplatin-induced apoptosis; Enhanced resistance of HCC cells to the anticancer drug doxorubicin | TIMP-3, MMP-2, MMP-9 | NSCLC cells Hepatocellular carcinomas | (Galluzzi et al., 2010; Wang et al., 2010a) |
| miR-192 | Up-regulated in docetaxel-resistant cells | Not detected | NSCLC cells | (Rui et al., 2010) |
| miR-200 | Down-regulation in drug-resistant cells, regulated EMT, increased cell sensitivity to anti-cancer drug | ZEB1, ZEB2, TUBB3, ERRFI, | Breast, prostate, lung, bladder, ovarian, pancreatic cancer cells | (Adam et al., 2009; Cochrane et al., 2009; Li et al., 2009b; Rui et al., 2010) |
| miR-205 | Increased sensitivity to gefitinib and lapatinib | HER3 | Breast cancer cells | (Iorio et al., 2009; Iorio and Croce, 2009) |
| miR-214 | Induced cisplatin resistance by targeting PTEN | PTEN | Ovarian cancer cells | (Yang et al., 2008) |
| miR-215 | Elevated in cells that exhibit slow proliferating rate and chemoresistance. Over-expression decreased sensitivity to methotrexate and tomudex. | P21, p53 | colon and osteosarcoma cells | (Song et al., 2010) |
| miR-221/222 | Re-expression sensitized cancer cells to glucocorticoids and TRAIL; Up-regulated in 4-hydroxytamoxifen resistant cells. Knock-down sensitized to tamoxifen, | CDKN1B, ERα, P27, | Acute lymphocytic leukemia cells, breast cancer cells, NSCLC cells | (Garofalo et al., 2009; Kotani et al., 2009; Miller et al., 2008; Zhao et al., 2008) |
| miR-342 | Down-regulated in the 4-hydroxytamoxifen resistant cells | Not detected | Breast cancer cells | (Miller et al., 2008) |
| miR-424 | Up-regulated in docetaxel-resistant cells | Not detected | NSCLC cells | (Rui et al., 2010) |
| miR-451 | Increased sensitivity to Doxorubicin | MDR1 | Breast cancer cells, ovarian, cervix cancer cells | (Kovalchuk et al., 2008; Zhu et al., 2008) |
| miR-489 | Down-regulated in the 4-hydroxytamoxifen resistant cells | Not detected | Breast cancer cells | (Miller et al., 2008) |
| miR-630 | Decreased cisplatin-induced apoptosis | P27, p53 | NSCLC cells | (Galluzzi et al., 2010) |
Collectively, these reports suggest a role of miRNAs in drug resistance. Further in-depth research is needed in order to fully understand this role and to find novel ways to regulate miRNAs for highly innovative treatment strategies. Below, we will further discuss the interrelationships between CSCs, EMT and miRNAs in the context of drug resistance.
5. Connection between EMT, CSCs and miRNA
EMT cells have cancer stem cell-like features and CSCs exhibit mesenchymal phenotype under most circumstances. Aberrant miRNA expression has been correlated with tumor development, cancer progression, the presence of CSCs and the acquisition of an EMT phenotype. Therefore, in the following paragraphs, we will summarize the relationships between EMT, CSCs and miRNA.
5.1. EMT and CSCs
A relationship between EMT and CSCs has recently emerged with evidence suggesting that EMT cells have cancer stem cell-like features and CSCs exhibit a mesenchymal-like phenotype. Mani et al. initially disclosed that immortalized human mammary epithelial cells (HMLEs) undergoing EMT are CSCs-like as characterized by their CD44high/CD24low phenotype (Mani et al., 2008). Ectopic expression of Twist or Snail induced EMT in HMLEs, which acquired a fibroblastoid mesenchymal appearance, down-regulated epithelial markers such as E-cadherin and up-regulated mesenchymal markers such as N-cadherin, vimentin, and fibronectin (Mani et al., 2008). Importantly, these EMT-type cells had the properties of CSCs, such as self-renewal and the capacity to form mammospheres. Serially transplanted tumors derived from BRCA1 mammary tumors had features of EMT and gave rise to cell lines that contained a distinct CD44+/CD24− population (Wright et al., 2008). Other evidence showed that mouse mammary stem cells exhibit a mesenchymal phenotype (Mani et al., 2008; Radisky and LaBarge, 2008) and TGF-β induced EMT and mammosphere-forming capabilities in human mammary epithelial cells (Morel et al., 2008). Consistently, mammosphere-forming activity is abrogated in breast CSCs after the EMT is shut-down (Shimono et al., 2009). Alignment of EMT with the CSCs signature was also found in cells derived from a breast cancer lung metastasis (DiMeo et al., 2009). Brabletz et al. studied the colorectal cancer progression and found that tumor cells at the tumor–host interface express EMT-associated genes as well as stemness-associated genes (Brabletz et al., 2005), suggesting a relationship between EMT and CSCs in colorectal cancer. More importantly, it is well-known that many signaling pathways, such as the Wnt, Notch and Hedgehog that regulate EMT, also drive CSCs self-renewal and maintenance (Blick et al., 2010; Hollier et al., 2009; Radisky and LaBarge, 2008).
5.2. Regulation of EMT by microRNA
Recently, studies have shown that miRNAs regulate EMT through the regulation of E-cadherin and other molecules such as ZEB and vimentin (Bracken et al., 2009; Gibbons et al., 2009; Gregory et al., 2008b; Wellner et al., 2009). For example, the transcription factors ZEB1 and ZEB2, which repress E-cadherin, have been confirmed as direct targets for repression by miR-200. Over-repression of miR-200 had dramatic effects on the expression of ZEB. Moreover, a striking negative correlation was found between ZEB and miR-200 expression in many human cancer cell lines (Gregory et al., 2008a; Korpal et al., 2008; Park et al., 2008). MiR-205 was also reported to be down-regulated during EMT (Gregory et al., 2008a) and also inhibits ZEB1 and ZEB2 expression and was specifically expressed in a panel of epithelial but not mesenchymal-like breast cancer lines (Gregory et al., 2008a). Ma et al. found that miR-10b is likely to represent an important component of a larger Twist-regulated EMT gene expression program (Ma et al., 2007). We found that miR-200a, miR-200b, and miR-200c were down-regulated in gemcitabine resistant (GR) pancreatic cancer cells, which show the acquisition of EMT phenotype (Li et al., 2009b). Re-expression of the miR-200 family up-regulated the epithelial marker E-cadherin and down-regulated mesenchymal markers, including ZEB1 and vimentin. Members of the let-7 family are also down-regulated in EMT-type GR cells (Li et al., 2009b). Furthermore, re-expression of miR-200b in PDGF-D over-expressing EMT-type cells resulted in the reversal of EMT with the down-regulation of ZEB1, ZEB2 and slug expression (Kong et al., 2009). Recently, miR-661 was found in Snai1-induced EMT-type cells and required for efficient invasion of breast cancer cells by destabilizing two of its predicted mRNA targets, the cell-cell adhesion protein Nectin-1 and the lipid transferase StarD10, resulting in the down-regulation of epithelial markers (Vetter et al., 2010). Expression of miR-30 in mesenchymal anaplastic thyroid carcinoma-derived cells reduced their invasive potential and induced mesenchymal-epithelial transition (MET) by regulating the expression of MET marker proteins (Braun et al., 2010). Expression of miR-203 was also found to be involved in EMT. Stable knockdown of ZEB1 in mesenchymal Panc1 cells results in the up-regulation of the miR-200 family members miR-141 and miR-200c, as well as the stemness-inhibiting miR-203 (Wellner et al., 2009). Over-expression of ZEB1 as well as of other EMT-inducers, Snail1 and ZEB2, suppresses the activity of the putative miR-203 promoter in pancreatic cancer cell lines (Wellner et al., 2009). Altogether, these data suggest that miRNAs play critical roles in the acquisition of EMT.
5.3. Regulation of CSCs by microRNA
Recent studies have shown involvement of several miRNAs in the regulation of CSCs. For example, miR-200c strongly suppressed the ability of normal mammary stem cells to form mammary ducts and tumor formation driven by human breast CSCs in vivo (Shimono et al., 2009). The miR-200 family miRNAs were found to be strongly suppressed in CD44+/CD24-lineage human breast cancer cells. The miR-125b recently was found to be necessary for stem cell fission to bypass the normal G1/S checkpoint and make stem cells insensitive to chemotherapy. Further research on the mechanism demonstrated that miR-125b regulates the proliferation of glioma stem cells through the cell cycle regulated proteins CDK6 and CDC25A (Shi et al., 2010a). Recently, miRNAs was also reported in pancreatic CSCs. Pancreatic CSCs have been described as CD24+/CD44+ subpopulation. Stable knockdown of ZEB1 in Panc1 cells led to a reduction of this subpopulation, correlating with reduced CD24 mRNA levels in stable ZEB1-knockdown clones. Knockdown of ZEB1 also resulted in decreased expression of stem cell factors such as Sox2, Bmi1 and p63. ZEB1 has been shown to directly control the transcription of the miR-200, miR-203 and miR-183 genes (Wellner et al., 2009). In parallel, miR-34 was reported to be involved in pancreatic CSCs self-renewal, potentially via the direct modulation of downstream targets Bcl-2 and Notch, suggesting that miR-34 may play an important role in pancreatic CSCs self-renewal and/or cell fate determination (Ji et al., 2009b). Recently, Yu et al. found that re-expression of miR-30 in breast cancer stem-like cells inhibits their self-renewal capacity by reducing Ubc9 (ubiquitin-conjugating enzyme 9), and inducing apoptosis through silencing ITGB3 (integrin β3). Furthermore, over-expression of miR-30 in breast CSCs xenografts reduced tumorigenesis and lung metastasis in mice, whereas blocking miR-30 expression enhanced tumorigenesis and metastasis. These results suggest that miR-30 could be one of the important miRNAs in regulating the stem-like features of breast cancer (Yu et al., 2010). More recently, Wong et al. reported increased levels of the miR-17-19b at a higher frequency in leukemia stem cells, consistent with reduced differentiation and increased proliferation, which was also associated with reduced expression of p21 (Wong et al., 2010). These results suggest that miR-17-19b quantitatively regulates leukemia stem cell self-renewal in part by modulating the expression of p21. The miR-181 is important for the maintenance of hepatic cancer stem cells by posttranslational down-regulation of two hepatic transcriptional regulators of differentiation and an inhibitor of Wnt/β-catenin signaling (nemo-like kinase [NLK]) (Ji et al., 2009a). It is likely that in the near future the link between miRNAs and CSCs will find further experimental support.
6. Targeting the EMT, CSCs, miRNA connection to increase drug sensitivity
Recently, experimental evidence revealed that EMT, CSCs and miRNAs are involved in anti-cancer drug resistance, indicating that specific targeting those miRNAs involved in the regulation of EMT may lead to the elimination of CSCs or EMT-type cells that are typically drug-resistant and are believed to be the “root cause” of tumor recurrence (Figure 1).
Figure 1.
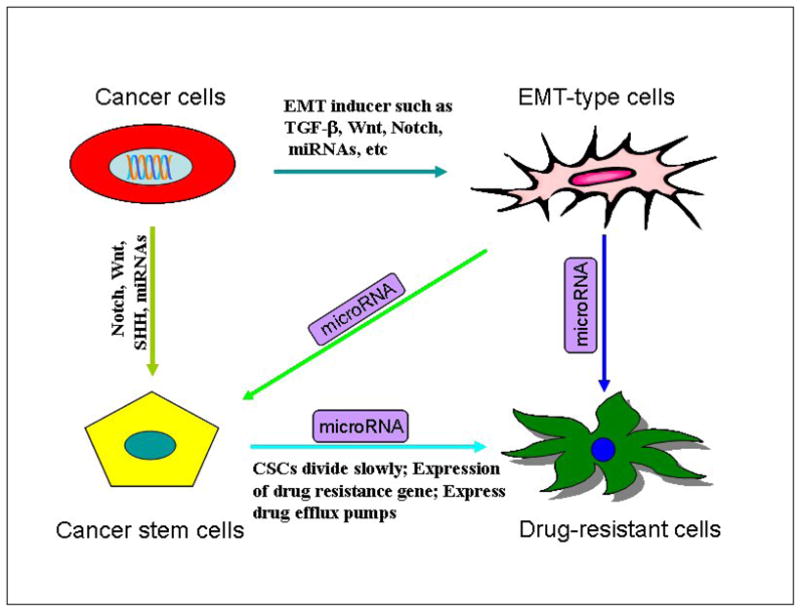
The connection between EMT, cancer stem cells, and miRNA. EMT cells have cancer stem cell-like features, and CSCs exhibit mesenchymal phenotype. Aberrant miRNA expression has been correlated with the formation of CSCs and the acquisition of EMT phenotype. miRNAs affect and connect CSCs through regulation of EMT.
6.1. Targeting EMT to increase drug sensitivity
Targeting EMT opens a new window for therapeutic applications. For instance, it was reported that E-cadherin-negative NSCLC patients had a worse overall survival after erlotinib treatment compared to E-cadherin-positive patients. Therefore, erlotinib is currently used in the epithelial early stage and in advanced stage metastatic pancreatic tumors (Thomson et al., 2005). Lupeol, a triterpene found in vegetables and fruits, has been reported to impair head and neck squamous cell carcinoma (HNSCC) cell invasion by reversal of the NF-κB-dependent EMT. Moreover, lupeol exerted a synergistic effect with cisplatin, resulting in chemosensitization of HNSCC cell lines with high NF-κB activity as documented by in vitro and in vivo studies (Lee et al., 2007). The cysteine protease inhibitor cystatin C has been proven to inhibit the acquisition of EMT and invasion stimulated by TGF-beta in breast cancer cell by preventing actin cytoskeletal rearrangements and E-cadherin down-regulation (Sokol et al., 2005). Very interestingly, dasatinib, a Src kinase inhibitor, has been found to be more effective in inhibiting growth of breast cancer cells with EMT features (Finn et al., 2007). All these data suggests that drug combinations using conventional or targeted therapies together with targeting the EMT-related mechanisms need to be considered for winning the battle against drug resistant cancer cells.
6.2. Targeting CSCs to increase drug sensitivity
Recent studies have suggested that eradication of CSCs is an important goal towards the cure of cancer by overcoming drug resistance. Salinomycin was found to kill selectively CSCs by an as yet unknown mechanism and when compared to paclitaxel, salinomycin reduced the proportion of CSCs by more than 100-fold and inhibited mammary tumor growth in mice, which was associated with increased epithelial differentiation of tumor cells (Gupta et al., 2009b). Interestingly, salinomycin was found to overcome ABC transporter-mediated drug resistance in human leukemia stem cell-like cells (Fuchs et al., 2010; Riccioni et al., 2010). In addition, two other compounds, nigericin and abamectin which are identified as inhibitors of CSCs growth are also PGP inhibitors (Riccioni et al., 2010). In another study, it was found that metformin, a standard drug for diabetes, inhibits cellular transformation and selectively kills CSCs in breast cancer. Moreover, the combination of metformin and doxorubicin killed both CSCs and non-stem cancer cells (Hirsch et al., 2009). Recently, metformin was also found to act synergistically with the anti-HER2 monoclonal antibody trastuzumab to suppress self-renewal and proliferation of CSCs in HER2-positive carcinomas (Vazquez-Martin et al., 2010). Jin et al. have reported a therapeutic strategy using an activating monoclonal antibody directed to the adhesion molecule CD44, a key regulator of leukemic stem cells, in mice transplanted with human acute myelogenous leukaemia (Jin et al., 2006). A recent study has shown that gene insertion into stem cells followed by direct specific delivery into the tumor in animal models is feasible (Aboody et al., 2008). In summary, targeting CSCs appears to be a promising therapeutic strategy. However, more innovative research in this area is warranted for the discovery of agents that selectively kill CSCs.
6.3. Targeting miRNA to increase drug sensitivity
As discussed above miRNAs are critically involved in CSCs and EMT regulation and the associated drug resistance phenotype. Therefore, targeting miRNAs for cancer therapy is an emerging field for treatment optimization aiming at restoring the sensitivity of drug-resistant cells to chemotherapy. For instance, miR-214 induced cisplatin resistance through targeting the PTEN in human ovarian cancer (Yang et al., 2008) and re-expression of miR-200 increased the sensitivity to microtubule-targeting chemotherapeutic agents (Cochrane et al., 2009). Over-expression of miR-221 and miR-222 by transfection made breast cancer cells more resistant to tamoxifen, while knock-down of miR-221 and miR-222 sensitizes breast cancer cells to tamoxifen-induced apoptosis (Zhao et al., 2008). In addition to breast cancer, miR-221 and miR-222 also caused TRAIL-sensitive NSCLC cells to become resistant to TRAIL (Garofalo et al., 2009; Garofalo et al., 2008). Song et al. recently reported that endogenous miR-215 was elevated in colon CSCs that exhibit slow proliferating rate and chemoresistance (Song et al., 2010). Suppression of miR-21 led to an increased cytotoxicity of a semisynthetic podophyllotoxin derivative (VM-26) against glioblastoma cells, suggesting that over-expression of miR-21 could contribute to drug resistance in glioblastoma (Li et al., 2009a). These examples suggest that targeting specific miRNAs could be useful for restoring drug sensitivity and preventing tumor recurrence.
6.4. Using natural agents to increase drug sensitivity
Because CSCs and EMT-type cells play important roles in drug resistance, novel inhibitors of EMT or agents that could either reverse the EMT phenotype or kill CSCs or EMT-type cells would be a novel strategy for the treatment of cancers. For example as mentioned earlier, salinomycin and metformin reduced the proportion of CSCs in breast cancer (Gupta et al., 2009; Hirsch et al., 2009). Several studies have also shown that anti-sense oligonucleotides can block the function of miRNAs (Hutvagner et al., 2004; Hutvagner and Simard, 2008; Meister et al., 2004; Orom et al., 2006; Orom and Lund, 2007); however, such strategies have limitation for the treatment of human cancer because of the lack of appropriate in vivo delivery systems. To overcome such limitations, recently researchers considered “natural agents” to target EMT or CSCs through miRNAs. Indeed, it has been shown that natural agents including curcumin, isoflavone, 3,3′-diinodolylmethane (DIM), indole-3-carbinol (I3C), epigallocatechin-3-gallate (EGCG) and others could regulate the EMT and miRNAs (Li et al., 2010b; Li et al., 2010a; Melkamu et al., 2010; Sun et al., 2008; Tsang and Kwok, 2010). DIM and isoflavone treatments could increase the level of miR-200 family in MiaPaCa-2 cells (EMT-type cells) (Li et al., 2009b) and the morphology of MiaPaCa-2 cells changed from elongated fibroblastoid-type to epithelial cobblestone-like appearance and it seems that the cell-cell contact was increased after the treatments (Li et al., 2009b). Moreover, we found that pretreatment of MiaPaCa-2 cells with DIM or isoflavone also increased the sensitivity of MiaPaCa-2 cells to gemcitabine, suggesting that miR-200 re-expression by DIM or isoflavone treatment could partially increase their sensitivity to gemcitabine through miR-200 mediated reversal of EMT status (Li et al., 2009b). Consistent with our findings, Sun et al reported that curcumin, another “natural agent” could up-regulate the miR-22 expression and suppress the expression of its target genes SP1 transcription factor and estrogen receptor 1 (Sun et al., 2008). Curcumin and piperine separately and in combination could inhibit breast cancer stem cell self-renewal but did not cause toxicity to differentiated cells (Kakarala et al., 2009). I3C down-regulated miR-21 and up-regulated miR-21 target genes PTEN (Melkamu et al., 2010) and also curcumin down-regulated the miR-21 and up-regulated the miR-200 in pancreatic cancer, leading to increased sensitivity to gemcitabine (Ali et al., 2010). Another natural agent, EGCG, was found to upregulate the expression of miR-16 in human tumor cells (Tsang and Kwok, 2010), suggesting that EGCG could increase drug sensitivity through up-regulation of miR-16. The isothiocyanate from broccoli, sulforaphane, was shown to synergize with the kinase inhibitor sorafenib in eradicating pancreatic CSCs-like cells (Rausch et al., 2010). Considering the relatively non-toxic nature of “natural agents”, targeting miRNAs, EMT-type cells and CSCs by these agents combined with conventional chemotherapeutics could be a novel and safer approach for achieving better treatment outcome. However, further in-depth preclinical and clinical studies are warranted in order to appreciate the value of “natural agents” in overcoming drug resistance.
7. Conclusion
In this review, we attempted to summarize the role of EMT, CSCs and their regulation by miRNAs in drug resistance. We discuss recent studies which demonstrate that EMT, CSCs and miRNAs could play a critical role in the regulation of anti-cancer drug sensitivity and resistance and that targeting EMT, CSCs and miRNAs is an emerging novel treatment strategy. Evidence is beginning to suggest that “natural agents” could be useful for the regulation of miRNA-mediated inhibition of cancer growth, reversal of EMT phenotype and elimination of drug-resistant CSCs, all of which might result in increasing drug sensitivity of human cancers and perhaps in combination with conventional therapy achieve better treatment outcome for cancer patients.
Acknowledgments
The authors’ work cited in this review was funded by grants from the National Cancer Institute, NIH (5R01CA131151, 5R01CA083695, 1R01CA132794, 1R01CA101870) to F.H.S. and Department of Defense Postdoctoral Training Award W81XWH-08-1-0196 (Zhiwei Wang) and also partly supported by a subcontract award (F.H.S.) from the University of Texas MD Anderson Cancer Center through a SPORE grant (5P20-CA101936) on pancreatic cancer awarded to James Abbruzzese. We also sincerely thank both Puschelberg and Guido foundation for their generous contributions to our research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboody KS, Najbauer J, Danks MK. Stem and progenitor cell-mediated tumor selective gene therapy. Gene Ther. 2008;15:739–752. doi: 10.1038/gt.2008.41. [DOI] [PubMed] [Google Scholar]
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, Bar-Eli M, Dinney C. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari AS, Agarwal N, Wood BM, Porretta C, Ruiz B, Pochampally RR, Iwakuma T. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010;70:4602–4612. doi: 10.1158/0008-5472.CAN-09-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Wang L, Agyin J, Tang Y, Lin S, Yeh IT, De K, Sun LZ. Doxorubicin in combination with a small TGFβ inhibitor: a potential novel therapy for metastatic breast cancer in mouse models. Plos One. 2010;5:e10365. doi: 10.1371/journal.pone.0010365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- Blick T, Hugo H, Widodo E, Waltham M, Pinto C, Mani SA, Weinberg RA, Neve RM, Lenburg ME, Thompson EW. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:235–252. doi: 10.1007/s10911-010-9175-z. [DOI] [PubMed] [Google Scholar]
- Borst P, Jonkers J, Rottenberg S. What makes tumors multidrug resistant? Cell Cycle. 2007;6:2782–2787. doi: 10.4161/cc.6.22.4936. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Khew-Goodall Y, Goodall GJ. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci. 2009;66:1682–1699. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010 doi: 10.1038/onc.2010.169. Epub 24 May 2010. [DOI] [PubMed] [Google Scholar]
- Broxterman HJ, Gotink KJ, Verheul HM. Understanding the causes of multidrug resistance in cancer: a comparison of doxorubicin and sunitinib. Drug Resist Updat. 2009;12:114–126. doi: 10.1016/j.drup.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Broxterman HJ, Lankelma J, Pinedo HM. How to probe clinical tumour samples for P-glycoprotein and multidrug resistance-associated protein. Eur J Cancer. 1996;32A:1024–1033. doi: 10.1016/0959-8049(96)00045-7. [DOI] [PubMed] [Google Scholar]
- Cammareri P, Scopelliti A, Todaro M, Eterno V, Francescangeli F, Moyer MP, Agrusa A, Dieli F, Zeuner A, Stassi G. Aurora-a is essential for the tumorigenic capacity and chemoresistance of colorectal cancer stem cells. Cancer Res. 2010;70:4655–4665. doi: 10.1158/0008-5472.CAN-09-3953. [DOI] [PubMed] [Google Scholar]
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Zhao ZW, Zhou HY, Liu YJ, Yang HJ. Systematic analysis of microRNA involved in resistance of the MCF-7 human breast cancer cell to doxorubicin. Med Oncol. 2010;27:406–415. doi: 10.1007/s12032-009-9225-9. [DOI] [PubMed] [Google Scholar]
- Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks P. Invitation to a second round. Nature. 2010;466:40–41. doi: 10.1038/466040a. [DOI] [PubMed] [Google Scholar]
- Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan LW, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3:1–13. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmink BL, van Houdt WJ, Vries RG, Hoogwater FJ, Jimenez CR, Clevers H, Borel Rinkes IH, Kranenburg O. Differentiated colorectal cancer cells protect tumor-initiating cells from irinotecan. Proc. Am. Assoc. Cancer Res; April 17–21, 2010; Washington DC. 2010. Abstract 4294. [Google Scholar]
- Fang DD, Kim YJ, Lee CN, Aggarwal S, McKinnon K, Mesmer D, Norton J, Birse CE, He T, Ruben SM, Moore PA. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–1275. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, Slamon DJ. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr, Raben D. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol Cancer Ther. 2007;6:1683–1691. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M, Tanabe KK. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391–2399. doi: 10.1158/0008-5472.CAN-07-2460. [DOI] [PubMed] [Google Scholar]
- Fuchs D, Daniel V, Sadeghi M, Opelz G, Naujokat C. Salinomycin overcomes ABC transporter-mediated multidrug and apoptosis resistance in human leukemia stem cell-like KG-1a cells. Biochem Biophys Res Commun. 2010;394:1098–1104. doi: 10.1016/j.bbrc.2010.03.138. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, Deguchi T, Ito M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Di LG, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Garofalo M, Quintavalle C, Di LG, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008a;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008b;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nature Med. 2009a;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009b;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutova M, Najbauer J, Gevorgyan A, Metz MZ, Weng Y, Shih CC, Aboody KS. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS One. 2007;2:e243. doi: 10.1371/journal.pone.0000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagenson KK, Wu GS. The role of MAP kinases and MAP kinase phosphatase-1 in resistance to breast cancer treatment. Cancer Metastasis Rev. 2010;29:143–149. doi: 10.1007/s10555-010-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson AW, Haluska P. Resistance pathways relevant to insulin-like growth factor-1 receptor-targeted therapy. Curr Opin Investig Drugs. 2009;10:1032–1040. [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox S, Jiang WG, Obermeier K, Taylor K, Morgan L, Burmi R, Barrow D, Nicholson RI. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int J Cancer. 2006;118:290–301. doi: 10.1002/ijc.21355. [DOI] [PubMed] [Google Scholar]
- Hollier BG, Evans K, Mani SA. The epithelial-to-mesenchymal transition and cancer stem cells: a coalition against cancer therapies. J Mammary Gland Biol Neoplasia. 2009;14:29–43. doi: 10.1007/s10911-009-9110-3. [DOI] [PubMed] [Google Scholar]
- Hong SP, Wen J, Bang S, Park S, Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- Hopper-Borge EA, Nasto RE, Ratushny V, Weiner LM, Golemis EA, Astsaturov I. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert Opin Ther Targets. 2009;13:339–362. doi: 10.1517/14712590902735795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer. 2010;102:1276–1283. doi: 10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Casalini P, Piovan C, Di LG, Merlo A, Triulzi T, Menard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009a;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009b;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BH, Liu LZ. Role of mTOR in anticancer drug resistance: perspectives for improved drug treatment. Drug Resist Updat. 2008;11:63–76. doi: 10.1016/j.drup.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- Kajiyama H, Shibata K, Terauchi M, Yamashita M, Ino K, Nawa A, Kikkawa F. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int J Oncol. 2007;31:277–283. [PubMed] [Google Scholar]
- Kakarala M, Brenner DE, Korkaya H, Cheng C, Tazi K, Ginestier C, Liu S, Dontu G, Wicha MS. Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Choi HK, Cho KB, Kim HS, Kang KW. Involvement of Pin1 induction in epithelial-mesenchymal transition of tamoxifen-resistant breast cancer cells. Cancer Sci. 2009;100:1834–1841. doi: 10.1111/j.1349-7006.2009.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz JE, George SL, Marcucci G, Vij R, Powell BL, Allen SL, et al. P-glycoprotein inhibition using valdospar (PSC-833) does not improve outcomes for patients under age of 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia group B study 19808. Blood. 2010 doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny GE, Venkatesan N, Yang G, Dering J, Ginther C, Finn R, Rahmeh M, Fejzo MS, Toft D, Jiang SW, Slamon DJ, Podratz KC. Activity of lapatinib a novel HER2 and EGFR dual kinase inhibitor in human endometrial cancer cells. Br J Cancer. 2008;98:1076–1084. doi: 10.1038/sj.bjc.6604278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono SA, Marshall ME, Ware KE, Heasley LE. The fibroblast growth factor receptor signaling pathway as a mediator of intrinsic resistance to EGFR-specific tyrosine kinase inhibitors in non-small cell lung cancer. Drug Resist Updat. 2009;12:95–102. doi: 10.1016/j.drup.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Tabe Y, Zeng Z, Andreeff M. Therapeutic targeting of microenvironmental interactions in leukemia: mechanisms and approaches. Drug Resist Updat. 2009;12:103–113. doi: 10.1016/j.drup.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A, Ha D, Hsieh J, Rao PK, Schotte D, den Boer ML, Armstrong SA, Lodish HF. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- Lee TK, Poon RT, Wo JY, Ma S, Guan XY, Myers JN, Altevogt P, Yuen AP. Lupeol suppresses cisplatin-induced nuclear factor-kappaB activation in head and neck squamous cell carcinoma and inhibits local invasion and nodal metastasis in an orthotopic nude mouse model. Cancer Res. 2007;67:8800–8809. doi: 10.1158/0008-5472.CAN-07-0801. [DOI] [PubMed] [Google Scholar]
- Lemoli RM, Salvestrini V, Bianchi E, Bertolini F, Fogli M, Amabile M, et al. Molecular and functional analysis of the stem cell compartment of chronic myelogenous leukemia reveals the presence of a CD34- cell population with intrinsic resistance to imatinib. Blood. 2009;114:5191–5200. doi: 10.1182/blood-2008-08-176016. [DOI] [PubMed] [Google Scholar]
- Li Y, Kong D, Wang Z, Sarkar FH. Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res. 2010a;27:1027–1041. doi: 10.1007/s11095-010-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009a;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- Li Y, Vandenboom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009b;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vandenboom TG, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010b;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bai L, Chen W, Xu S. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin Ther Targets. 2010;14:45–55. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Qu L, Tao H. Cyclo-oxygenase 2 up-regulates the effect of multidrug resistance. Cell Biol Int. 2010a;34:21–25. doi: 10.1042/CBI20090129. [DOI] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:1–12. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Cheng W, Lai D, Huang Y, Guo L. Characterization of primary ovarian cancer cells in different culture systems. Oncol Rep. 2010b;23:1277–1284. doi: 10.3892/or_00000761. [DOI] [PubMed] [Google Scholar]
- Liu T, Xu F, Du X, Lai D, Liu T, Zhao Y, Huang Q, Jiang L, Huang W, Cheng W, Liu Z. Establishment and characterization of multi-drug resistant, prostate carcinoma- initiating stem-like cells from human prostate cancer cell lines 22RV1. Mol Cell Biochem. 2010c;340:265–273. doi: 10.1007/s11010-010-0426-5. [DOI] [PubMed] [Google Scholar]
- Lopez-Chavez A, Carter CA, Giaccone G. The role of KRAS mutations in resistance to EGFR inhibition in the treatment of cancer. Curr Opin Investig Drugs. 2009;10:1305–1314. [PubMed] [Google Scholar]
- LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat. 2008;11:32–50. doi: 10.1016/j.drup.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta K, Osipo C. Trastuzumab resistance: role for Notch signaling. Scientific World Journal. 2009;9:1438–1448. doi: 10.1100/tsw.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31:252–258. doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ong CW, Kim LG, Kong HH, Low LY, Iacopetta B, Soong R, Salto-Tellez M. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod Pathol. 2010;23:450–457. doi: 10.1038/modpathol.2009.181. [DOI] [PubMed] [Google Scholar]
- Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J, Costantino JP, Geyer CE, Jr, Wickerham DL, Wolmark N. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and A. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- Radisky DC, LaBarge MA. Epithelial-mesenchymal transition and the stem cell phenotype. Cell Stem Cell. 2008;2:511–512. doi: 10.1016/j.stem.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Rausch V, Liu L, Kallifaditis G, Baumann B, Mattern J, Gladkich J, et al. Synergistic activity of sorafenib and sulphorane abolishes pancreatic cancer stem cell characteristics. Cancer Res. 2010;70:5004–5013. doi: 10.1158/0008-5472.CAN-10-0066. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rho JK, Choi YJ, Lee JK, Ryoo BY, Na II, Yang SH, Kim CH, Lee JC. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer. 2009;63:219–226. doi: 10.1016/j.lungcan.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Riccioni R, Dupuis ML, Bernabei M, Petrucci E, Pasquini L, Mariani G, Cianfriglia M, Testa U. The cancer stem cell selective inhibitor salinomycin is a p-glycoprotein inhibitor. Blood Cells Mol Dis. 2010;45:86–92. doi: 10.1016/j.bcmd.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Rui W, Bing F, Hai-Zhu S, Wei D, Long-Bang C. Identification of microRNA profiles in docetaxel-resistant human non-small cell lung carcinoma cells (SPC-A1) J Cell Mol Med. 2010;14:206–214. doi: 10.1111/j.1582-4934.2009.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M, Emami S, Redeuilh G, Julien S, Prevost G, Zimber A, Ouelaa R, Bracke M, De WO, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11:123–151. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Saito Y, Uchida N, Tanaka S, Suzuki N, Tomizawa-Murasawa M, Sone A, Najima Y, Takagi S, Aoki Y, Wake A, Taniguchi S, Shultz LD, Ishikawa F. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat Biotechnol. 2010;28:275–280. doi: 10.1038/nbt.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13:57–66. doi: 10.1016/j.drup.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, Stanbridge EJ, Lee EY. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–3250. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14:3629–3637. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- Shi L, Zhang J, Pan T, Zhou J, Gong W, Liu N, Fu Z, You Y. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010a;1312:120–126. doi: 10.1016/j.brainres.2009.11.056. [DOI] [PubMed] [Google Scholar]
- Shi MF, Jiao J, Lu WG, Ye F, Ma D, Dong QG, Xie X. Identification of cancer stem cell-like cells from human epithelial ovarian carcinoma cell line. Cell Mol Life Sci. 2010b doi: 10.1007/s00018-010-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010 doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol JP, Neil JR, Schiemann BJ, Schiemann WP. The use of cystatin C to inhibit epithelial-mesenchymal transition and morphological transformation stimulated by transforming growth factor-beta. Breast Cancer Res. 2005;7:R844–R853. doi: 10.1186/bcr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:1–10. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styczynski J, Drewa T. Leukemic stem cells: from metabolic pathways and signaling to a new concept of drug resistance targeting. Acta Biochim Pol. 2007;54:717–726. [PubMed] [Google Scholar]
- Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Tang C, Ang BT, Pervaiz S. Cancer stem cell: target for anti-cancer therapy. FASEB J. 2007;21:3777–3785. doi: 10.1096/fj.07-8560rev. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, Iwata KK, Gibson N, Haley JD. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- To K, Fotovati A, Reipas KM, Law JH, Hu K, Wang J, et al. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer Res. 2010;70:2840–2851. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, Stassi G. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Tokar EJ, Qu W, Liu J, Liu W, Webber MM, Phang JM, Waalkes MP. Arsenic-specific stem cell selection during malignant transformation. J Natl Cancer Inst. 2010;102:638–649. doi: 10.1093/jnci/djq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole BP, Slomiany MG. Hyaluronan, CD44 and Emmprin: partners in cancer cell chemoresistance. Drug Resist Updat. 2008;11:110–121. doi: 10.1016/j.drup.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2010;21:140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A, Oliveras-Ferraros C, Barco SD, Martin-Castillo B, Menendez JA. The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-0924-x. [DOI] [PubMed] [Google Scholar]
- Vere White RW, Vinall RL, Tepper CG, Shi XB. MicroRNAs and their potential for translation in prostate cancer. Urol Oncol. 2009;27:307–311. doi: 10.1016/j.urolonc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter G, Saumet A, Moes M, Vallar L, Le BA, Laurini C, Sabbah M, Arar K, Theillet C, Lecellier CH, Friederich E. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene. 2010 doi: 10.1038/onc.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Wang B, Hsu SH, Majumder S, Kutay H, Huang W, Jacob ST, Ghoshal K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010a;29:1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang H, Huang YZ, Yan RH, Liu FJ, Zhang JN. Biologic characteristics of the side population of human small cell lung cancer cell line H446. Chin J Cancer. 2010b;29:254–260. doi: 10.5732/cjc.009.10330. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ahmad A, Li Y, Kong D, Azmi AS, Banerjee S, Sarkar FH. Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim Biophys Acta. 2010c;1806:122–130. doi: 10.1016/j.bbcan.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kong D, Li Y, Sarkar FH. PDGF-D signaling: a novel target in cancer therapy. Curr Drug Targets. 2009a;10:38–41. doi: 10.2174/138945009787122914. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Y, Banerjee S, Sarkar FH. Exploitation of the Notch signaling pathway as a novel target for cancer therapy. Anticancer Res. 2008;28:3621–3630. [PubMed] [Google Scholar]
- Wang Z, Li Y, Banerjee S, Sarkar FH. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009b;279:8–12. doi: 10.1016/j.canlet.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Kong D, Ahmad A, Banerjee S, Sarkar FH. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010d;292:141–148. doi: 10.1016/j.canlet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009c;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, Chen CZ, Cleary ML. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Robles AI, Herschkowitz JI, Hollingshead MG, Anver MR, Perou CM, Varticovski L. Molecular analysis reveals heterogeneity of mouse mammary tumors conditionally mutant for Brca1. Mol Cancer. 2008;7:1–12. doi: 10.1186/1476-4598-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- Yang AD, Fan F, Camp ER, van BG, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–4153. doi: 10.1158/1078-0432.CCR-06-0038. [DOI] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, Modrusan Z, Lin CY, O’Neill V, Amler LC. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010 doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- Zhang N, Li R, Tao KS, Cao DY, Ti ZY, Ding R, Cai L, Zhang FQ, Dou KF. Characterization of a stem-like population in hepatocellular carcinoma MHCC97 cells. Oncol Rep. 2010;23:827–831. [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–588. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


