Abstract
The tissue tropism of Musca domestica salivary gland hypertrophy virus (MdSGHV) infecting adult house flies was examined by transmission electron microscopy (TEM) and quantitative real-time PCR. TEM demonstrated that characteristic MdSGHV-induced nuclear and cellular hypertrophy was restricted to the salivary glands. Both nucleocapsids and enveloped virions were present in salivary gland cells. In contrast, thin sections of midguts, ovaries, abdominal fat body, crops, air sacs and brains showed the presence of enveloped virions in vacuoles of tracheal cells associated with these tissues. However, no sites of viral morphogenesis were detected in the tracheal cells. Quantitative analysis of MdSGHV DNA and transcript titers revealed that viral DNA was present in all hemolymph and tissue samples collected from MdSGHV-infected flies. Average numbers of MdSGHV genome copies per 50 ng of DNA varied significantly between examined tissues and ranged from 3.83 × 108 (± 3.75 × 107) in salivary gland samples to 7.98 × 105 (± 2.91 × 105) in hemolymph samples. High levels of viral genome copies were detected in midgut, fat body and brain samples. Viral transcripts were present in all examined samples, and transcript abundance was also at the highest level in salivary glands and at the lowest level in hemolymph. However, over the range of different tissues that were analyzed, there was no correlation between estimated quantities of genome copies and viral transcripts. The function of viral transcripts in host tissues that do not show sites of viral morphogenesis remains to be elucidated.
Keywords: insect virus, dsDNA virus, Hytrosaviridae, house fly, histology, real-time PCR
1. Introduction
The Musca domestica salivary gland hypertrophy virus (MdSGHV) is an entomopathogenic dsDNA virus that infects adult house flies. Recently, this virus has been classified as a member of a newly proposed virus family, the Hytrosaviridae, which potentially contains several viruses found to infect and induce salivary gland enlargement in other dipteran species (Abd-Alla et al., 2009). The circular genome of the MdSGHV consists of 124, 279 bp and contains 108 putative open reading frames (ORFs), of which 101 have been validated to be transcriptionally active (Garcia-Maruniak et al., 2008; Salem et al., 2009). The MdSGHV replicates in the nuclei of salivary gland cells and induces characteristic symptoms of salivary gland hypertrophy (SGH) within a few days after infection (Lietze et al., 2007). Upon passage through the nuclear membrane, the nucleocapsids assemble an envelope in the cytoplasm and are transported to and released into the lumen of the salivary glands (Geden et al., 2008). During feeding, infected flies release high numbers of virions in salivary secretions onto the solid food substrate, which can result in horizontal transmission to healthy conspecifics that consume the contaminated substrate (Lietze et al., 2009). Typically, about 50% of orally challenged house flies develop SGH (Prompiboon et al., 2010). Potential barriers to per os infection can be circumvented by injecting the virus directly into the hemocoel of adult flies (Lietze et al., 2007). In addition to causing SGH, symptomatic MdSGHV infection suppresses vitellogenesis in infected females (Lietze et al., 2007; Lietze et al., 2009).
It is not known if the MdSGHV can infect and replicate in tissues other than salivary glands. Bioassays conducted on tissue homogenates dissected from infected flies have implicated the presence of infectious virus in non-salivary gland tissues. For example, Lietze et al. (2007) demonstrated that ovarian homogenates contained viral titers capable of inducing SGH when injected into healthy adult flies. Hence, the objective of this study was to define the tissue tropism of the MdSGHV by determining the spatial virus distribution and the presence of viral DNA and transcripts in non-salivary gland tissues. Specifically, we examined the salivary glands, midgut, ovaries, abdominal fat body, crop, air sacs, and brain of infected female flies by transmission electron microscopy (TEM) and quantitative real-time PCR (qPCR).
2. Materials and methods
2.1. Infection of adult flies
House flies were obtained from the “Orlando Normal” colony of insecticide-susceptible M. domestica maintained at the Center for Medical, Agricultural and Veterinary Medicine (CMAVE), United States Department of Agriculture-Agricultural Research Service (USDA-ARS) in Gainesville, Florida. For histological studies, newly eclosed adults were infected per os by feeding them 0.1-μl droplets of filter-sterilized viremic salivary gland homogenate in a 4% powdered milk solution as described in Lietze et al. (2009). Healthy control flies were fed with 4% powdered milk solution. To produce cohorts of synchronously infected house flies for qPCR assays, newly emerged females were injected with filter-sterilized viremic salivary gland homogenate as described in Lietze et al. (2007). This treatment guarantees symptomatic infection in 100% of the injected flies (Lietze et al., 2007). In a preliminary experiment, DNA samples from flies extracted at 1 h and 24 h post-injection did not contain PCR-detectable levels of MdSGHV DNA, whereas samples extracted at 72 h and 120 h post-injection were PCR-positive (V.-U. Lietze, unpublished data). Healthy control flies were mock-injected with saline. All treated flies were maintained in separate groups at constant conditions (26 °C, 12L: 12D photoperiod, 40% relative humidity) and provided with food and water ad libitum until used for sample preparation.
2.2. Sample preparation for microscopy
For TEM, per os infected and healthy house flies were cold anesthetized and dissected in fixative to collect salivary glands, crops, air sacs, ovaries, abdominal fat body tissue, midguts, and brains. Sample processing has been described elsewhere (Lietze et al., 2009). To compare the widths, lengths and cell numbers of salivary glands between healthy and MdSGHV-infected house flies, additional salivary gland pairs were fixed in 4% formaldehyde (in phosphate buffered saline [PBS]), permeabilized with Triton-X-100 (0.8%), treated with RNase A (10 μg/ml), equilibrated in PBS, and incubated with 600 nM 4′, 6- diamidino-2-phenylindole (DAPI, Invitrogen Molecular Probes, Carlsbad, CA). Stained glands were visualized, photographed, and measured using a combination of epifluorescent and phase contrast microscopy with calibrated Spot Advanced™ software. Stained nuclei were counted in separate gland sections and related to the entire lengths of dissected glands. Square root transformed width measurements and untransformed numbers of nuclei in healthy and infected salivary glands were analyzed by one-way ANOVA using SPSS software (SPSS for Windows, release 15.0.0; SPSS Inc., Chicago, IL), and Tukey’s HSD was used for pairwise comparison. Untransformed data are presented as mean widths or mean numbers ± SE.
2.3. Sample collection and preparation for nucleic acid extraction and cDNA synthesis
Seventy-two hours after injection, all flies were dissected in sterilized saline. In all cases, three replicate samples were collected from different cohorts of injected flies. For an initial set of samples, five viremic flies were dissected as follows: upon opening the ventral side of the abdomen with fine forceps, salivary glands were carefully removed and placed into 500 μl of Tri-Reagent (Fisher Scientific, Pittsburgh, PA). Head and thorax were discarded, and the remaining abdomen (including crop, air sacs, ovaries, fat body, and entire midgut) was collected into 1 ml of Tri-Reagent. Healthy fly samples consisted of five dissected abdomens including the salivary gland pairs. For a second set of samples, individual abdominal tissues, brains and hemolymph samples from viremic flies were collected and stored in 500 μl of Tri-Reagent per sample. Five flies per sample were used to dissect salivary gland pairs, midguts, ovary pairs, and abdominal fat body. Twenty flies per sample were used to dissect crops, air sacs, and brains. Hemolymph was collected from 50 flies per sample as described in Lietze et al. (2007).
Each sample was homogenized by adding approximately twenty zirconium beads (BioSpec Products, Bartlesville, OK) followed by vigorous shaking in a bead-homogenizer (FastPrep® Instrument, Qbiogene, Carlsbad, CA) for 30 s. DNA and RNA were then isolated following the manufacturer’s instructions. DNA samples were stored at −20 °C until used for qPCR. A maximum of 10 μg of total RNA per sample was treated with RQ1 RNase free DNase (Promega, Madison, WI) according to the manufacturer’s protocol. RNA was then re-isolated by adding a defined volume of Tri-Reagent and repeating the isolation protocol. DNase-treated RNA samples were quantified by spectrophotometer readings and stored as ethanol precipitates in 1-μg aliquots at −70°C. For quality control, 50 ng and 10 ng of DNase-treated RNA were subjected to one-step reverse-transcriptase PCR with and without reverse transcriptase as described in Salem et al. (2009) and to PCR using the 1 X Sensi Mix Plus SYBR® + fluorescein reaction mix (as described in section 2.5), respectively, with house fly- and MdSGHV-specific primers. One-microgram aliquots of the second set of DNA-free RNA samples were subjected to cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) according to the manufacturer’s protocol, and cDNA samples were stored at −20 °C until used for qPCR.
2.4. One-step reverse transcriptase PCR (RT-PCR)
The initial set of RNA samples (abdomens from healthy flies, salivary gland pairs from viremic flies, and corresponding abdomens without salivary gland pairs from viremic flies) was subjected to one-step RT-PCR to screen for the presence of MdSGHV transcripts of 10 selected open reading frames (ORFs). These ORFs (1, 10, 13, 29, 45, 47, 62, 84, 86, and 106) were selected after an initial screening of 62 viral ORFs in one replicate set of samples (data not shown), so that they would represent genes that are dispersed over the genome and that encode for both structural and non-structural proteins as annotated by Garcia-Maruniak et al. (2008) and Salem et al. (2009). The M. domestica ribosomal 28S gene (GenBank accession number DQ656974) served as a reference gene and internal positive control. Primer pairs produced amplicons ranging from 245 bp to 666 bp and primer sequences have been recently published (Salem et al., 2009). Using the Access RT-PCR System (Promega, Madison, WI), each 20-μl reaction contained 50 ng of DNase-treated total RNA, 5 pmol of each gene-specific forward and reverse primer, 2 units of avian myeloblastosis virus (AMV) reverse transcriptase, 1 X AMV/Tfl reaction buffer, 400 pmol of each dNTP, 1.25 mM MgSO4, and 2 units of Tfl DNA polymerase. The one-step RT-PCR program was 45 °C for 1 h, 70 °C for 15 min and 94 °C for 3 min, then 35 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 2 min, and an extension step at 72 °C for 7 min. Five microliters per reaction were electrophoresed on agarose gels and stained with ethidium bromide to visualize PCR products.
2.5. Quantitative real-time PCR (qPCR)
For qPCR analysis of cDNA and DNA samples, specific reverse (or forward) primers were designed that could be used with the respective forward (or reverse) primers (see section 2.4) to amplify shorter fragments (between 130 and 180 bp) of the M. domestica ribosomal 28S gene and of five selected MdSGHV ORFs 1, 10, 47, 86, and 106 (Table 1) representing three non-structural and two structural proteins, respectively (Salem et al., 2009). Samples obtained from healthy flies were included to verify the absence of viral transcripts and viral DNA. The qPCR was conducted using SensiMix Plus SYBR® + fluorescein (Quantace, Norwood, MA). Each 20-μl reaction was run in duplicate and contained 10 ng of reverse transcribed RNA (cDNA) or 50 ng of DNA as template, 10 pmol of each forward and reverse primer, and 1X SensiMix Plus SYBR® + fluorescein reaction mix. Reverse transcriptase qPCR utilized the M. domestica ribosomal 28S gene as an internal control for normalizing the threshold cycles (Ct), and the 2−ΔΔCt method was employed to determine and compare the relative abundance of MdSGHV transcripts in the different cDNA samples (Livak and Schmittgen, 2001; Pfaffl, 2001; Schmittgen and Livak, 2008). For absolute quantification of viral genome copies in the DNA samples, tenfold serial dilutions of purified genomic MdSGHV DNA representing 101 to 107 MdSGHV genome copies per reaction served as an external standard, and a previously published qPCR protocol was followed (Lietze et al., 2009).
Table 1.
Primers used for real-time PCR and evaluation of standard curves obtained with each primer pair
| ORF | Primer name | Primer sequence (5′ to 3′) | Amplicon sizea | qPCR standard curve resultsb |
||
|---|---|---|---|---|---|---|
| R2 | slope | PCR % efficiency | ||||
| 1 | MdvO1F1714 qMdvO1R1882 |
ATTTCCGCCACACCATACAT ATTGTTGGGCATCGAGTTGT |
169 bp (1714–1882) | 0.999 | −3.309 | 100.6 |
| 10 | MdvO10F133 qMdvO10R7964 |
CGCAATTGTTCCATGTATCG GAAGGAGGGCGGTATGGTA |
165 bp (8128–7964) | 1.000 | −3.299 | 101.0 |
| 47 | SGHVenvF1 qSGHVenvR1 |
ACCTACGGTCTGGATATTCGTG ATAAAGACCCATGCCTGTGC |
149 bp (49471–49619) | 0.999 | −3.242 | 103.4 |
| 86 | qMdvO88(86)F94757 MdvO88(86)R767 |
TGCCATCAAGTGTCTGGAAG ACAAAAATCTGTCGGGCATC |
180 bp (94757–94936) | 0.998 | −3.187 | 106.0 |
| 106 | MdvO108(106)F110 qMdvO108(106)R119549 |
CGGCAGCAACCTTATTCATT ACGACGACATCATTTGACGA |
130 bp (119420–119549) | 0.999 | −3.275 | 102.0 |
| Reference | Mdv28sFDQ656974 qMdv28sRDQ656974 |
GTTAAGCCCGATGAACCTGA TTGAATAAACTTTCGCCATCG |
158 bp | N/A | N/A | N/A |
Numbers in parentheses indicate the position of the amplicon in the MdSGHV genome.
N/A = not applicable.
Statistical analyses were conducted using the Statistical Analysis System (SAS) for Windows (SAS, 2004). Viral genome copy numbers were transformed by the square-root of the square-root to obtain normal distribution. Transformed genome copy numbers were subjected to two-way ANOVA using the general linear model (GLM) procedure of SAS, and means were separated using Tukey’s studentized range test (Tukey) (Cody and Smith, 2006; Köhler et al., 1992). Untransformed numbers were presented as means ± standard error (SE). The ΔCt values of viral transcripts (i.e., threshold cycle values normalized to the 28S reference gene) were also analyzed by GLM and Tukey. Correlation between relative abundance of viral genome copies and transcripts was examined using the CORR procedure of SAS (Cody and Smith, 2006; Köhler et al., 1992).
3. Results
3.1. Histology
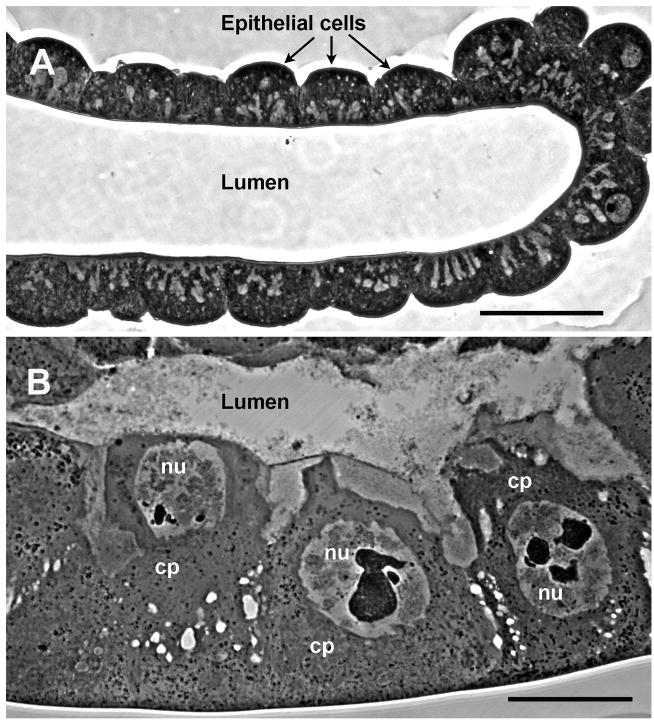
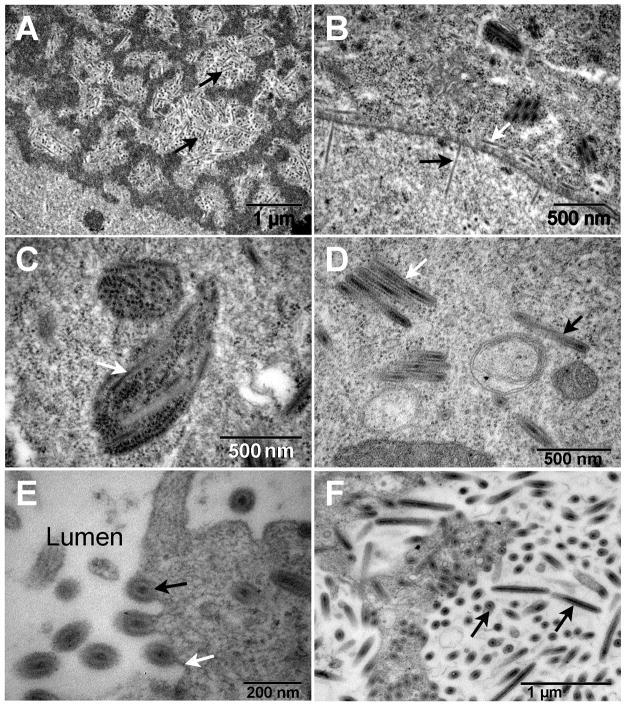
Infected flies displayed characteristic hypertrophied, blue-white, viremic salivary glands (Fig. 1). The salivary gland, comprised of giant cells with enlarged nuclei (Fig. 1B), was the only tissue that displayed detectable cytopathology. The width of the posterior portion significantly increased over time from an average 75.2 ± 1.5 μm in healthy glands to an average 148.3 ± 2.7 μm at 4 d post-infection (dpi) (F =183.44, df = (4, 576), P > 0.05), while gland length doubled from 14.0 mm to 28.8 mm. Enlargement was due to individual cell and nuclear hypertrophy and not due to cell proliferation (Fig. 1). Average numbers of nuclei in healthy and hypertrophied single glands were 4067 ± 304 and 4011 ± 188, respectively, and were not significantly different (F = 1.528, df = (1, 55), P > 0.05). Examination by TEM revealed virogenic stroma, sites of viral DNA packaging, and assembly of progeny nucleocapsids within the hypertrophied nuclei (Fig. 2A). Nucleocapsids were commonly aligned perpendicular to the nuclear membrane and exited the nucleus via nuclear pores (Fig. 2B). Acquisition of the inner viral envelope took place in the cytoplasm (Figs. 2C, D). Enveloped particles migrated to the gland lumen and gained an additional membrane as they budded out of the infected cell (Fig. 2E). The lumina of the glands were filled with enveloped rod-shaped MdSGHV measuring 500–600 nm by 65–70 nm (Fig. 2F).
Fig. 1.
Light micrographs of thick sections through (A) healthy and (B) MdSGHV-infected salivary glands. Images were taken at identical magnification with bars equal to 50 μm. The salivary gland supports viral replication along its entire length. The relative number of cells in healthy and infected glands remains constant; the enlarged state of the infected gland is due to extensive cellular and nuclear hypertrophy induced by viral replication. Cp, cytoplasm; nu, nucleus.
Fig. 2.
Transmission electron micrographs of hypertrophied salivary glands. (A) The enlarged nuclei were characterized by virogenic stroma within which the nucleocapsids (back arrows) were assembled and then (B) translocated to the nuclear membrane where they were observed to exit (black arrow) the nucleus via the nuclear pores and to be released (white arrow) into the region of the perinuclear cistern. (C) Virus particles (white arrow) appear to be associated with endoplasmic reticulum. (D) Both clusters (white arrow) and individual (black arrow) enveloped virions are found throughout the cytoplasm. (E) The enveloped virions (black arrow) migrate preferentially to the cell membrane (white arrow) and are released into the gland lumen via a budding process. (F) As the infection progresses the lumen becomes filled with enveloped virions (black arrows).
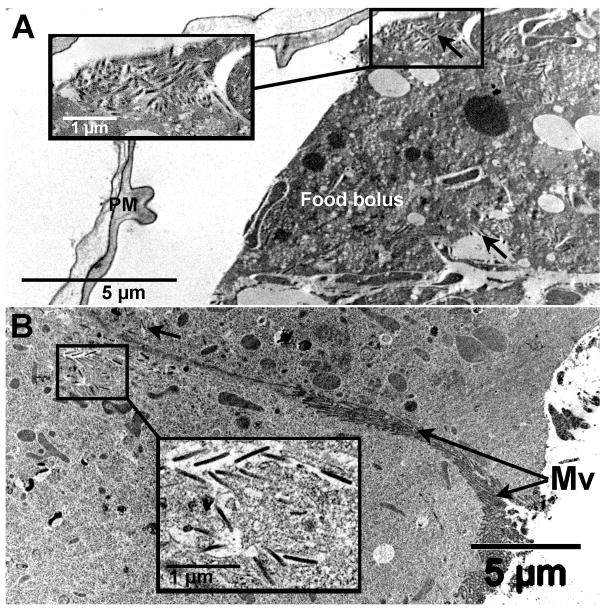
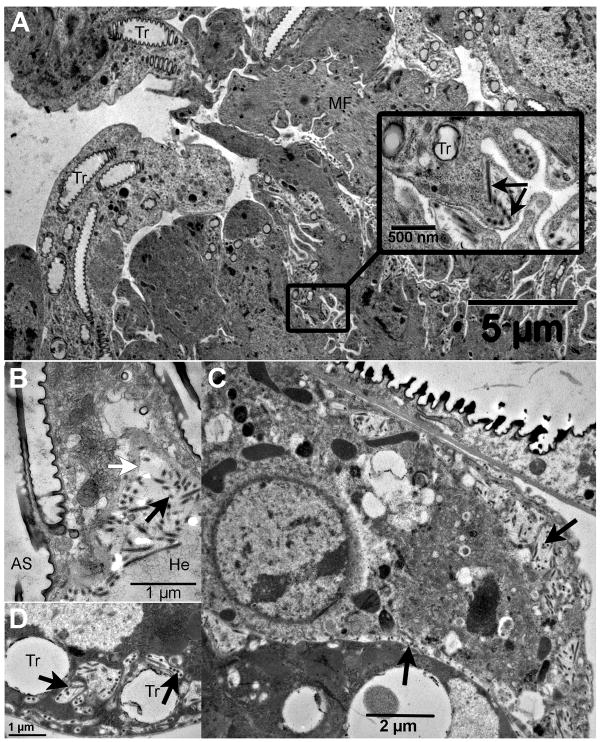
High densities of enveloped virus were detected in the food bolus of the midgut, confined within a well-developed multi-laminate peritrophic matrix (Fig. 3A). Midgut columnar cells contained enveloped virions in vacuoles throughout the cytoplasm (Fig. 3B) and in aggregates located in the basement membrane region along the hemocoel face (not shown in figure). The MdSGHV was not observed to replicate in the nuclei of midgut cells suggesting that virus associated with vacuoles had either bypassed the peritrophic matrix barrier to gain ingress into the midgut epithelium or originated from the virus associated with basement membrane. There was no evidence of MdSGHV replication in the nuclei of the fat body, ovarian, crop, brain, or tracheal tissues. However, all sections of these tissues demonstrated the presence of enveloped virions along the basement membrane and in the cytoplasm of the tracheal cells that were associated with these insect tissues. For example, the external layer of the ovaries (Fig. 4A) contained numerous enveloped virions along the cell margins of the sheath layer encompassing the multiple ovarioles. No virus was detected within the follicular epithelia, internal nurse cells and developing oocytes. Numerous enveloped virions were depicted in sections of the abdominal air sac and often found aggregated along the hemolymph face of the basement membrane (Fig. 4B). As observed with the ovaries, enveloped virions were frequently seen in tracheal cells associated with the brain, located in intracellular vacuoles at the cell periphery and in intercellular regions (Fig. 4C, D).
Fig. 3.
Transmission electron micrographs of the MdSGHV-infected house fly midgut. (A) The food bolus enclosed by a well-developed peritrophic matrix contains numerous enveloped virions (see insert and black arrows). (B) Virions (see insert and black arrow) were detected in vesicles in the cytoplasm of the midgut columnar cells, note microvilli. Mv, microvilli; PM, peritrophic matrix.
Fig. 4.
Electron micrographs of the MdSGHV (A) in tracheoles associated with the ovaries (see insert), (B) in the abdominal air sacs, and (C, D) the air sac in the head tagma that is closely opposed to the brain tissue. Black arrows in all figures indicate virions. White arrow in (B) indicates the basement membrane at the hemolymph face. AS, air sac; He, hemolymph; MF, muscle fibers; Tr, tracheoles.
3.2. MdSGHV DNA titers
The standard curves obtained with purified MdSGHV DNA showed low inter-ORF variation (average variation coefficient = 6.38%, minimum 3.60%, maximum 9.58%), and each standard curve had a very good fit over a range of seven orders of magnitude (101 to 107 genome copies) with correlation coefficients (R2 values) ranging from 0.998 to 1.000 (average 0.999), slopes ranging from −3.187 to −3.309 (average −3.262), and PCR efficiencies ranging from 100.6% to 106.0% (average 102.6%) (Table 1). These results confirmed that all of the five selected genes (ORFs) were equally suitable to be used for qPCR (Stratagene, 2004). Viral DNA was not present in samples from healthy flies (data not shown), whereas in all samples collected from infected flies MdSGHV DNA was detected with all five primer pairs. All quantitative data obtained by qPCR were within the linear range of the standard curve. Statistical analysis of estimated genome copy numbers showed no difference between the five ORFs examined (F = 1.74, df = 4, P = 0.1495) but significant differences between the tissues (F = 84.26, df = 7, P < 0.0001). Average numbers of MdSGHV genome copies per 50 ng of DNA ranged from 3.83 × 108 (± 3.75 × 107) in salivary gland samples to 7.98 × 105 (± 2.91 × 105) in hemolymph samples (Table 2). Midgut and fat body samples contained lower numbers (an average 2.03 × 108 and 1.63 × 108, respectively) than salivary gland samples but higher numbers than all other samples. Average genome copy numbers in brain samples (5.14 × 107) were significantly higher than numbers in air sac samples (3.10 × 107) and ovary samples (2.51 × 107), which in turn were significantly higher than average numbers estimated in crop samples (1.49 × 106) and hemolymph samples.
Table 2.
Mean (±SE) MdSGHV genome copy numbers (times 103) detected by real-time PCR in 50 ng of DNA isolated from different tissues of house flies at 72 h post-infectiona
| Tissue | ORF 1 | ORF 10 | ORF 47 | ORF 86 | ORF 106 | all ORFsb |
|---|---|---|---|---|---|---|
| Salivary glands | 319,667 (27,449) | 246,667 (14,310) | 368,667 (24,633) | 616,333 (84,294) | 362,667 (39,976) | 382,800 (37,454)a |
| Midgut | 172,967 (61,687) | 141,800 (48,258) | 205,867 (72,526) | 307,400 (125,758) | 186,133 (70,234) | 202,833 (33,832)b |
| Fat body | 147,867 (38,875) | 113,600 (26,801) | 159,667 (45,322) | 247,000 (65,126) | 148,133 (38,498) | 163,253 (20,692)b |
| Brain | 40,100 (2,318) | 39,967 (786) | 56,467 (273) | 73,000 (2,196) | 47,667 (1,719) | 51,440 (3,364)c |
| Air sacs | no data | 23,484 (16,090) | 32,878 (23,169) | 41,131 (29,899) | 26,346 (18,921) | 30,960 (9,862)d |
| Ovaries | 24,567 (884) | 20,033 (1,033) | 25,867 (899) | 32,167 (1,517) | 22,733 (581) | 25,073 (1,151)d |
| Crop | 877 (503) | 1,601 (1,114) | 2,213 (1,093) | 997 (796) | 1,778 (911) | 1,493 (369)e |
| Hemolymph | 1,980 (n/a) | 620 (620) | 709 (701) | 771 (770) | 696 (692) | 798 (291)e |
Means are based on three replicate samples per tissue. Between five and 50 flies were used to obtain each pooled sample (see methods).
Mean copy numbers followed by different letters are significantly different (P < 0.05, SNK)
3.3. Quality control of RNA preparations
Prior to analyzing the presence and abundance of MdSGHV transcripts by one-step RT-PCR and qPCR, all RNA samples were subjected to quality control reactions. All RNA samples were free of contaminating DNA as determined by one-step RT-PCR with and without the addition of reverse transcriptase and by PCR. One-step RT-PCR only produced amplicons of the M. domestica 28S gene when reverse transcriptase was added to the reaction, whereas no product was detected in reactions without the enzyme (data not shown). During PCR (using reaction mixtures and cycling conditions identical to the qPCR protocol), none of the RNA samples produced amplicons, confirming that preparations were free of contaminating DNA (data not shown). In addition, residual reverse transcriptase activity of the DNA polymerase (present in the Sensi Mix Plus SYBR® + fluorescein reaction mix), which may potentially produce false positives during qPCR (Martel et al., 2002), could be excluded under the conditions used to determine transcript titers.
3.4. One-step reverse transcriptase PCR
Initial analysis of MdSGHV transcript abundance by one-step RT-PCR showed that viral transcripts of all ten examined ORFs were present in the salivary glands of viremic flies as well as in the corresponding abdomen samples of viremic flies from which the salivary glands had been removed (Table 3). Samples obtained from healthy flies did not contain MdSGHV transcripts. Band intensities of RT-PCR products on ethidium bromide stained agarose gels were visually related to the intensity of the M. domestica reference gene. Subsequent comparison of band intensities between corresponding replicate samples of salivary glands and abdomens from virus-infected flies indicated lower quantities of most transcripts in the three replicate abdomen samples (Table 3), with the following exceptions: transcripts of ORFs 13 and 84 were more or equally abundant in one replicate abdomen sample than in the corresponding salivary gland sample; transcripts of ORFs 29, 45, and 106 were more or equally abundant in two replicate abdominal samples; and transcript of ORF 86 was equally abundant in all three corresponding abdomen and salivary gland samples.
Table 3.
Intensity of bandsa detected on ethidium bromide stained agarose gels after one-step reverse transcriptase PCR with 50 ng of DNase-treated RNA isolated from different samples at 72 h post-infection
| ORF | Viremic salivary glands (SG) | Viremic abdomen w/o SG | Healthy abdomen with SG | ||||||
|---|---|---|---|---|---|---|---|---|---|
| sample 1 | sample 2 | sample 3 | sample 1 | sample 2 | sample 3 | sample 1 | sample 2 | sample 3 | |
| 1 | +++ | +++ | +++++ | − | + | ++++ | − | − | − |
| 10 | ++++ | ++++ | +++++ | ++ | +++ | ++++ | − | − | − |
| 13 | ++++ | ++ | +++++ | − | +++b | +++ | − | − | − |
| 29 | +++++ | + | ++++ | +++ | ++b | ++++c | − | − | − |
| 45 | +++ | ++++ | ++++ | ++++b | +++ | ++++c | − | − | − |
| 47 | +++++ | +++++ | +++++ | + | +++ | ++++ | − | − | − |
| 62 | ++++ | ++++ | ++++ | +++ | +++ | +++ | − | − | − |
| 84 | +++++ | +++ | +++ | +++++c | ++ | ++ | − | − | − |
| 86 | ++++ | ++++ | ++++ | ++++c | ++++c | ++++c | − | − | − |
| 106 | +++ | +++ | +++ | +++c | ++ | +++c | − | − | − |
| Reference | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
Visually compared and related to the intensity of the reference gene band.
Band intensity was higher than in the corresponding salivary gland sample.
Band intensity was equal to the corresponding salivary gland sample.
3.5. MdSGHV transcript titers
As shown in Table 4, normalized Ct values obtained for each ORF and sample type ranged from 4.13 to 17.33. Transcript abundance varied between the five ORFs (F = 44.80, df = 4, P < 0.0001) and between the tissues (F = 38.89, df = 7, P < 0.0001) examined. The highest transcript abundance was detected for ORF 86, followed by transcripts of ORFs 10 and 47, which in turn were significantly more abundant than transcripts of ORFs 1 and 106 (Tukey, P < 0.05). When the transcript abundance of each ORF was compared among tissues (by relating transcript abundance in each sample type to the salivary gland samples using the 2−ΔΔCt method), a similar distribution pattern was found for each ORF examined (Table 5). For all ORFs examined, transcript abundance was highest in the salivary gland samples and lowest in the hemolymph samples. High levels of MdSGHV transcription were also found in air sac, fat body and brain samples. For example, transcript levels of ORF 1 were reduced 4-, 7- and 10-fold, respectively, compared with the levels of ORF 1 transcripts in the salivary gland samples (Table 5). In addition, ORF 1 transcript abundance was 15-fold lower in the crop samples, 77-fold lower in the midgut samples, and 112-fold lower in the ovary samples. The data in Table 5 show that this pattern was similar for the transcripts of ORFs 10, 47, 86, and 106.
Table 4.
Normalizeda threshold cycle values of five MdSGHV ORFs in different tissues of female house flies at 72 h post-infection
| Tissue | ORF1 | ORF10 | ORF47 | ORF86 | ORF106 |
|---|---|---|---|---|---|
| Salivary glands | 9.16 | 7.25 | 6.94 | 4.13 | 10.57 |
| Midgut | 15.43 | 13.03 | 12.49 | 10.11 | 15.30 |
| Ovaries | 15.97 | 13.96 | 13.65 | 10.79 | 15.94 |
| Fat body | 11.89 | 11.79 | 11.39 | 8.31 | 12.54 |
| Crop | 13.11 | 11.48 | 10.99 | 8.98 | 13.92 |
| Air sacs | 11.06 | 10.65 | 9.92 | 7.36 | 11.62 |
| Brain | 12.43 | 10.76 | 11.82 | 9.04 | 12.80 |
| Hemolymph | 16.60 | 14.90 | 14.07 | 11.53 | 17.33 |
Threshold cycle values were obtained by real-time PCR and normalized to the Musca domestica ribosomal 28S reference gene as described in Pfaffl (2001). Each reaction contained 10 ng of reverse transcribed RNA.
Table 5.
Relative reduced level of transcriptiona of five MdSGHV ORFs detected in different tissues of female house flies at 72 h post-infection, presented as x-fold difference compared with salivary glands for each ORF
| Tissue | ORF1 | ORF10 | ORF47 | ORF86 | ORF106 |
|---|---|---|---|---|---|
| Salivary glands | 1 | 1 | 1 | 1 | 1 |
| Midgut | −77 | −55 | −47 | −63 | −27 |
| Ovaries | −112 | −105 | −105 | −102 | −42 |
| Fat body | −7 | −23 | −22 | −18 | −4 |
| Crop | −15 | −19 | −17 | −29 | −10 |
| Air sacs | −4 | −11 | −8 | −9 | −2 |
| Brain | −10 | −11 | −29 | −30 | −5 |
| Hemolymph | −174 | −201 | −140 | −169 | −109 |
For each ORF normalized threshold cycle values obtained from Table 4 were related to the threshold cycle in salivary gland samples by using the 2−ΔΔCt method as described in Pfaffl (2001).
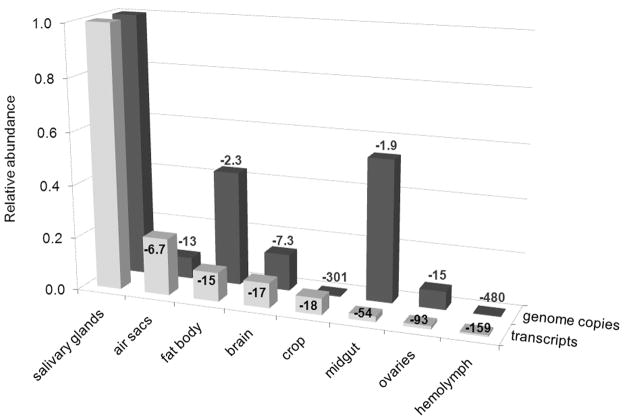
A comparison of the relative abundance of MdSGHV genome copy numbers and transcripts (averaged for all ORFs examined) showed that the level of genome copy numbers did not always correspond with the abundance of transcripts (Fig. 5). For instance, midgut samples contained high levels of viral DNA (reduced 1.9-fold compared with salivary gland samples) but low levels of transcripts (reduced 54-fold compared with salivary gland samples) whereas air sac and crop samples contained significantly lower levels of viral DNA (reduced 13-fold and 301- fold, respectively, compared with salivary gland samples) but higher levels of transcripts (reduced 6.7-fold and 18-fold, respectively, compared with salivary gland samples). There was no correlation between the relative abundance of viral genome copies and viral transcripts in these samples (Pearson correlation coefficient = −0.00919, P = 0.9588).
Fig. 5.
Average relative abundance of MdSGHV transcripts and genome copy numbers in different tissues of female house flies at 72 h post-infection. Abundance was estimated by real-time PCR and expressed relative to the levels in salivary glands. Numbers indicate the x-fold reduction of transcripts and genome copy numbers in each tissue sample relative to the abundance in salivary glands. For more details, refer to the methods section.
4. Discussion
MdSGHV infects adult house flies and causes salivary gland hypertrophy due to viral replication in the nuclei of salivary gland cells (Geden et al., 2008). Infected salivary gland cells, containing high levels of enveloped virus in the cytoplasm, continuously shed large numbers of virus into the gland lumen to be released in the salivary secretions (Lietze et al., 2009). The gland hypertrophy is caused by a massive increase in the size of existing cells and is not due to cell proliferation (or hyperplasia) as reported previously for the Merodon equestris SGHV (Amargier et al., 1979) and the Glossina pallidipes SGHV (Jaenson, 1978). Furthermore, the MdSGHV is non-lytic; infected cells persist throughout the adult housefly lifespan serving as foci for virus production. The mechanisms underlying the cytopathology exhibited by infected salivary gland cells is unknown, unfortunately, the vast majority of the 108 MdSGHV genes have no known homologues (Salem et al., 2009). One ORF (ORF 78) identified as a homologue of inhibitor of apoptosis (iap) contains a single baculoviral inhibition of apoptosis protein repeat domain (BIR) and may be involved in preventing the onset of the programmed cell death.
This study demonstrated the presence of numerous MdSGHV virions in asymptomatic tissues such as the midgut, ovaries, abdominal fat body, crop, air sacs and brain of the infected insect. When these samples were examined by TEM, no definitive sites of viral morphogenesis were found in any tissue other than the salivary glands. However, enveloped virions were detected in cytoplasmic vacuoles, in zones adjacent to the basal lamina, and in tracheole cells associated with all tissues. These observations were confirmed by qPCR. Considerable numbers of MdSGHV genome copies were found in all examined tissues and in the hemolymph of 72-h-infected flies. Non-enveloped nucleocapsids commonly observed in the cytoplasm and nuclei of infected salivary glands were not detected in these other tissues.
The source of virions detected in the various tissues is unclear. Preliminary assays demonstrated that the viruses injected into these flies were undetected by PCR in samples collected at 1 h and 24 h post-injection, i.e., the virus load injected was below the detection threshold. The enveloped virions produced in the salivary gland cells have been shown to be directionally translocated to the gland lumen (Geden et al., 2008). This observation, combined with the low virus titer in the hemolymph, suggests that the virions present in the various tissues are not derived from budding through the basal lamina of the salivary gland. An alternative source of the enveloped virus may be the food bolus. However, the peritrophic matrix in this insect is fully formed at the time of food ingestion and has been speculated to be an effective barrier against per os infection in 1-d-old flies (Prompiboon et al., 2010). All sites harboring enveloped virus in these non-gland tissues were in proximity to the tracheal system which terminates in intracellular tracheoles on or in individual cells of other tissues. Potentially, the MdSGHV is undergoing replication in the tracheal cells without displaying the cytopathology observed in the salivary gland. Additionally, the MdSGHV could mimic the baculovirus (Engelhard et al., 1994; Washburn et al., 2003) and use the tracheal system as a conduit providing a mechanism to breach basal laminal barriers. With the exception of hemolymph, all tissues were penetrated by tracheoles. Significantly, the salivary gland is laden with tracheae that originate from the well-developed abdominal air sacs. The notion that MdSGHV is capable of using the tracheal system as a conduit might explain how the orally ingested virus could breach the basal lamina surrounding midgut cells and enter the salivary gland cells. Likewise, such a conduit would provide for the dissemination of enveloped virus produced in the salivary gland to other insect tissues.
The presence of virus particles and high titers of viral DNA in all examined tissues suggests that MdSGHV may be undergoing transcriptional and translational events in non-salivary gland tissues. Although histological examination did not reveal any replication sites, RT-PCR was conducted to elucidate potential transcriptional activity. The initial screen of MdSGHV transcripts showed that all of the tested genes were transcribed not only in salivary gland tissue but also in the corresponding abdomen samples from which the salivary glands had been removed. These results lead to the detailed examination of abdominal tissues, hemolymph and also brain samples. Brain samples were included because of the impact of infection on vitellogenesis and oogenesis (Lietze et al., 2007), processes known to be controlled by neurosecretions (Adams et al., 1997). Transcripts of five selected ORFs were present in all tissue samples that were obtained from 72-h-infected flies and examined by qPCR. While the transcript abundance of these ORFs differed among the tissues, their relative frequency patterns were similar to that observed in the salivary glands (Table 6). Significantly, transcripts of the non-structural DNA polymerase (ORF 1) gene and the structural ORF 86 gene, both considered in other viruses to represent late gene products, were detected in all tissues suggesting that the entire viral genome is undergoing low levels of transcription. Detection of viral RNA in non-salivary gland tissues of house flies does not imply that replication and morphogenesis occur. The examined ORFs may be transcribed constitutively, i.e., their transcription may not require assistance from early expressed genes as it is the case in baculoviruses (Miller, 1997). While the MdSGHV replicates in the permissive salivary gland cells, non-salivary gland tissues may be considered non-permissive for viral replication. Semi- or non-permissiveness of certain cells or tissues to viral replication may be regulated by viral and/or host factors (Miller and Lu, 1997; Morris and Miller, 1992, 1993). The virus can enter these cells and undergo partial transcription, but no replication occurs. For instance, in a study to test the permissiveness of different lepidopteran, coleopteran, dipteran, and homopteran cell lines to Autographa californica multiple nucleopolyhedrosis virus replication, transcription of the red fluorescent protein gene driven by the heat shock protein 70 promoter was detected in cell lines that did not support viral replication (McIntosh et al., 2005).
Table 6.
Relative reduced level of transcriptiona of five MdSGHV ORFs detected in different tissues of female house flies at 72 h post-infection, presented as x-fold difference compared with ORF86 for each tissue
| Tissue | ORF1 | ORF10 | ORF47 | ORF86 | ORF106 |
|---|---|---|---|---|---|
| Salivary glands | −33 | −9 | −7 | 1 | −87 |
| Midgut | −40 | −8 | −5 | 1 | −36 |
| Ovaries | −36 | −9 | −7 | 1 | −36 |
| Fat body | −12 | −11 | −8 | 1 | −19 |
| Crop | −18 | −6 | −4 | 1 | −31 |
| Air sacs | −13 | −10 | −6 | 1 | −19 |
| Brain | −10 | −3 | −7 | 1 | −14 |
| Hemolymph | −34 | −10 | −6 | 1 | −56 |
For each tissue normalized threshold cycle values obtained from Table 4 were related to the threshold cycle of ORF86 by using the 2−ΔΔCt method as described in Pfaffl (2001).
In the non-salivary gland tissues, high numbers of MdSGHV genome copies were not necessarily accompanied by high transcriptional activity (Fig. 5). For instance, relative to the level in the salivary glands, the highest numbers of viral genome copies were detected in the midgut, but the relative abundance of transcripts in the midgut was much lower than in four of the other tissue samples. This could be explained by the horizontal route of oral MdSGHV transmission within groups of house flies (Lietze et al., 2009). Infected flies deposit high numbers of virus particles onto the shared food substrate, which are subsequently ingested by flies and are found in high numbers in the alimentary tract. In contrast, air sac and crop samples contained relatively low numbers of viral genome copies but relatively high levels of viral transcripts. It should be noted that the air sac is a modified trachea and that the crop, a diverticulum of the insect foregut, is comprised of a thin monolayer of tracheated epithelial cells. Whether transcription is occurring in only the tracheoles and/or in the associated cells is not clear. Significantly, the transcriptional profiles for both hypertrophied salivary glands and tissues lacking hypertrophy are similar suggesting that a viral transcription is not blocked in these tissues.
In summary, pathological symptoms of MdSGHV infection such as the characteristic nuclear and cellular hypertrophy are only seen in salivary glands. All other examined tissues of infected flies were found to contain enveloped virions, considerable numbers of MdSGHV genome copies, and viral transcripts. Although complete viral replication (morphogenesis) was found to be restricted to salivary gland cells of the infected host, the omnipresent transcriptional activity in all other tissues may play a significant role in modulating the female sterility induced by MdSGHV infection.
Acknowledgments
The authors would like to thank Chris Geden and Melissa Doyle (USDA-ARS CMAVE, Gainesville, Florida) for generously providing and rearing house flies, and Karen Kelley (UF-ICBR EM, Gainesville, Florida) for assistance during electron microscopy. Financial support was provided in part by USDA/NRI grant 2007-35302-18127 and National Institute of Health (NIAID R21 A1073501-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abd-Alla A, Vlak JM, Bergoin M, Maruniak JE, Parker A, Burand J, Jehle JA, Boucias D. Hytrosaviridae: a proposal for classification and nomenclature of a new insect virus family. Arch Virol. 2009;154(6):909–918. doi: 10.1007/s00705-009-0398-5. [DOI] [PubMed] [Google Scholar]
- Adams TS, Gerst JW, Masler EP. Regulation of ovarian ecdysteroid production in the housefly, Musca domestica. Arch Insect Biochem Physiol. 1997;35(1–2):135–148. [Google Scholar]
- Amargier A, Lyon JP, Vago C, Meynadier G, Veyrunes JC. Mise en evidence et purification d’un virus dans la proliferation monstreuse glandulaire d’insectes. Etude sur Merodon equestris (Diptera, Syrphidae) Note Comp Rend Seanc Acad Sci Ser D Sci Natur. 1979;289:481–484. [PubMed] [Google Scholar]
- Cody RP, Smith JK. Applied statistics and the SAS programming language. Pearson Prentice Hall; Upper Saddle River: 2006. [Google Scholar]
- Engelhard EK, Kam-Morgan LNW, Washburn JO, Volkman LE. The insect tracheal system: a conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc Natl Acad Sci USA. 1994;91:3224–3227. doi: 10.1073/pnas.91.8.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maruniak A, Maruniak JE, Farmerie W, Boucias DG. Sequence analysis of a non-classified, non-occluded DNA virus that causes salivary gland hypertrophy of Musca domestica, MdSGHV. Virology. 2008;377:184–196. doi: 10.1016/j.virol.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geden CJ, Lietze VU, Boucias D. Seasonal prevalence and transmission of salivary gland hypertrophy virus of house flies (Diptera: Muscidae) J Med Entomol. 2008;45(1):42–51. doi: 10.1603/0022-2585(2008)45[42:spatos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jaenson TGT. Virus-like rods associated with salivary gland hyperplasia in tsetse, Glossina pallidipes. Trans R Soc Trop Med Hyg. 1978;72(3):234–238. doi: 10.1016/0035-9203(78)90200-6. [DOI] [PubMed] [Google Scholar]
- Köhler W, Schachtel G, Voleske P. Einführung in die Biometrie für Biologen und Agrarwissenschaftler. Springer; Berlin: 1992. Biostatistik; p. 255. [Google Scholar]
- Lietze VU, Geden CJ, Blackburn P, Boucias DG. Effects of salivary gland hypertrophy virus on the reproductive behavior of the house fly, Musca domestica. Appl Environ Microbiol. 2007;73(21):6811–6818. doi: 10.1128/AEM.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietze VU, Sims KR, Salem TZ, Geden CJ, Boucias DG. Transmission of MdSGHV among adult house flies, Musca domestica (Diptera: Muscidae), occurs via salivary secretions and excreta. J Invertebr Pathol. 2009;101:49–55. doi: 10.1016/j.jip.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta Ct) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martel FZ, Gründemann D, Schömig E. A simple method for elimination of false positive results in RT-PCR. J Biochem Mol Biol. 2002;35(2):248–250. doi: 10.5483/bmbrep.2002.35.2.248. [DOI] [PubMed] [Google Scholar]
- McIntosh AH, Grasela JJ, Popham HJR. AcMNPV in permissive, semipermissive, and nonpermissive cell lines from Arthropoda. In Vitro Cell Dev Biol-Anim. 2005;41(8–9):298–304. doi: 10.1290/0412083R.1. [DOI] [PubMed] [Google Scholar]
- Miller LK. The baculoviruses. Plenum Press; New York: 1997. [Google Scholar]
- Miller LK, Lu A. The molecular basis of baculovirus host range. In: Miller LK, editor. The baculoviruses. Plenum Press; New York: 1997. pp. 217–235. [Google Scholar]
- Morris TD, Miller LK. Promoter influence on baculovirus-mediated gene expression in permissive and nonpermissive insect cell lines. J Virol. 1992;66(12):7397–7405. doi: 10.1128/jvi.66.12.7397-7405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TD, Miller LK. Characterization of productive and non-productive AcMNPV infection in selected insect cell lines. Virology. 1993;197(1):339–348. doi: 10.1006/viro.1993.1595. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for the relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompiboon P, Lietze VU, Denton JSS, Geden CJ, Steenberg T, Boucias DG. Musca domestica salivary gland hypertrophy virus: a globally distributed insect virus that infects and sterilizes female houseflies. Appl Environ Microbiol. 2010;76(4):994–998. doi: 10.1128/AEM.02424-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem TZ, Garcia-Maruniak A, Lietze VU, Maruniak JE, Boucias DG. Analysis of transcripts from predicted open reading frames of the Musca domestica salivary gland hypertrophy virus. J Gen Virol. 2009;90(5):1270–1280. doi: 10.1099/vir.0.009613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS. User’s guide, version 9.1. SAS Institute; Cary, NC: 2004. [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Stratagene. Methods and application guide. Stratagene; La Colla, CA: 2004. Introduction to quantitative PCR. [Google Scholar]
- Washburn JO, Trudeau D, Wong JF, Volkman LE. Early pathogenesis of Autographa californica multiple nucleopolyhedrovirus and Helicoverpa zea single nucleopolyhedrovirus in Heliothis virescens: a comparison of the ‘M’ and ‘S’ strategies for establishing fatal infection. J Gen Virol. 2003;84:343–351. doi: 10.1099/vir.0.18701-0. [DOI] [PubMed] [Google Scholar]