Abstract
Lys63-linked TAK1 polyubiquitination plays an essential role in the regulation of TAK1 activation. TRAF6-mediated Lys63-linked polyubiquitilation of TAK1 has been shown to be required for TGF-β-induced TAK1 activation. However, it remains unclear which lysine residue on TAK1 is TRAF6-mediated TAK1 polyubiquitination acceptor site in TGF-β-signaling pathway. Here we report that lysine 158 on TAK1 is required for TGF-β-induced TRAF6-mediated TAK1 polyubiquitination and TAK1-mediated IKK, JNK and p38 activation. Notably, in contrast to TAK1 wild-type and K34R mutant, TAK1 K158R mutant co-overexpression with TAB1 failed to induce Lys63-linked TAK1 polyubiquitination. TRAF6-induced K63-linked TAK1 polyubiquitination was blocked by TAK1 K158R mutation, but not by K34R mutation. Furthermore, TGF-β-induced TAK1 polyubiqutination was inhibited by TAK1 K158R mutation, but not by K34R mutation in HeLa cells. Reconstitution of TAK1-deficient mouse embryo fibroblast cells with TAK1 wild-type, K158R mutant, or K34R mutant reveals that TAK1 lysine 158 residue is required for TGF-β-induced IKK, p38 and JNK activation.
Keywords: TGF-β, TAK1, polyubiquitination, Lysine 158
1. Introduction
Transforming growth factor-β (TGF-β) acts as a double-edged sword to either suppress tumor formation or facilitate tumor initiation and progression through activating distinct intracellular signaling pathways [1, 2]. In the classical signaling pathway, a complex of type I (TbRI) and type II (TbRII) transmembrane receptors convey the signal from TGF-β to its intracellular mediators [3]. TbRII phosphorylates TβRI in its juxtamembrane-localized GS-domain, leading to the activation of the TβRI kinase domain. The activated TβRI then phosphorylates Smad proteins, leading to their accumulation in the nucleus to regulate target gene expression. In the Smad-independent pathway, ligand-bound TGF-β receptors also activate JNK-AP-1 and IKK-NF-kB pathways [4].
TAK1, a member of the MAPK kinase kinase family, was originally found to function in the TGF-β-mediated MAPK activation [5]. TAB1 is a TAK1-interacting protein and induces TAK1 kinase activity through promoting autophosphorylation of key serine/threonine sites of the kinase activation loop [6]. TAB2, TAB3, and TAB4 activate TAK1 through binding to polyubiquitinated proteins and promoting a larger complex formation during TNF α and IL-1β-induced TAK1 activation [7, 8].
The physiological significance of TAK1 in TGF-β signaling was revealed in recent studies of mouse embryonic fibroblasts (MEFs) in which the TAK1 gene was inactivated through homologous recombination [9, 10]. It was demonstrated that TAK1 is absolutely required for the TGF-β-induced JNK and NF-κB activation [9].
Currently, growing evidence suggests that protein phosphorylation and ubiquitination play an essential role in the regulation of TAK1 activation. Recently, two groups reported that TRAF6 was specifically required for the Smad-independent activation of JNK and p38, and its carboxyl TRAF homology domain physically interacted with TGF-β receptors [11, 12]. TGF-β-induced TRAF6-mediated K63-linked polyubiquitylation of TAK1 Lys 34 correlates with TAK1 activation [12]. However, we found that Lys63 linked polyubiquitination of TAK1 at lysine 158 is required for TNFα and IL1β induced IKK-NF-kB, JNK and P38 acitivation [13]. Therefore, the molecular mechanism of TAK1 polyubiquitination induced by different stimuli needs to be further defined.
In this report, we have further investigated the molecular mechanism of TAK1 activation induced by TGF-β. We found that TAK1 lysine 158, but not lysine 34, is required for TGF-β-induced TAK1 polyubiquitination and TGF-β-induced TRAF6-mediated Smad-independent IKK, JNK and p38 activation.
2. Materials and methods
2.1 Cell culture and transfection
HeLa, HEK-293T and TAK1 deficient MEF cells were maintained in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 µg/ml streptomycin at 37 °C in 5% CO2. HeLa, HEK-293T and TAK1 deficient MEF cells were transfected with mammalian expression plasmids using Fugene HD (Roche Applied Science), Fugene 6 (Roche Applied Science) and Lipofectamine 2000 reagents (Invitrogen), respectively.
2.2 Expression vectors
Full-length human TAK1 expression constructs were generated with a C-terminal V5-His tag in pcDNA3.1 vector. TAK1 Lys 158 and Lys 34 mutations were created as described previously [13]. Retroviral TAK1-K34R mutant construct was generated using pBabe-puro vector. Mammalian expression vectors for TAB1, TAB2, TAB3, TRAF2/6 and IKKβ were constructed as described previously [13]. The NF-κB and AP-1-dependent firefly luciferase reporter and pCMV promoter-dependent Renilla luciferase reporter plasmids were purchased from Clontech.
2.3 Antibodies and reagents
The following antibodies and reagents were used: TAK1, phospho-TAK1, phospho-JNK, JNK, phospho-p38, p38, phospho-IKKα/β and secondary antibodies conjugated to horseradish peroxidase (Cell Signaling); HA, TAB1 and Protein A-agarose (Santa Cruz Biotechnology); β-actin (Sigma); Recombinant TGF-β (R & D Systems).
2.4 Establishment of TAK1 stable cell lines
For retroviral infection, the pBabe-TAK1 wide type, or pBabe-TAK1-K158R, or pBabe-TAK1-K34R or pBabe empty vector were co-transfected with retrovirus packing vector Pegpam 3e and PLC-ECO in HEK-293T cells to obtain retroviral supernatants. Viral supernatants were collected after 48 and 72 h. MEF-TAK1 knockout cells were incubated with retroviral supernatant in the presence of 4 µg/ml polybrene (Sigma). Stable cell lines were established after 10 days of puromycin (2 µg/ml) selection. HeLa cells were transfected with Flag-tagged TAK1 wild type, or K158R mutant, or K34R mutant expression vectors using Fugene HD reagent. Stable cell lines were established after 14 days of G418 (2000 µg/ml) selection.
2.5 Immunoprecipitation and Immunoblotting
To prepare total cell lysates, cells were washed 3 times with ice-cold PBS on ice, then were lysed by adding lysis buffer without EDTA (25 mM HEPES (pH 7.7), 135 mM NaCl, 1% Triton X-100, 25 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 0.5 mM phenylmethylsulfonylfluoride, 1 mM dithiothreitol, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM Benzamidine, 20 mM disodium p-nitrophenylphosphate, 1 mM phenylmethylsulfonyl fluoride, phosphatase inhibitor cocktail A and B). After centrifugation, cell lysates were incubated with the Nickel beads with rotation for 3 h at 4°C. The precipitates were washed four times in cold washing buffer without EDTA (20 mM HEPES (pH 7.7), 50 mM NaCl, 2.5 mM MgCl2, and 0.05% Triton X-100), then the beads were resuspended in Laemmli sample buffer and boiled for 5 min. The immunoprecipitates or the whole cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The membranes were probed with appropriate antibodies. The IgG horseradish peroxidase-conjugated antibodies were used as the secondary antibodies. The proteins were detected by using the ECL-Plus Western blotting detection system.
2.6 Luciferase reporter assays
HEK-293T and different MEF cell lines were seeded at a concentration of 3 × 105 cells per well and cultured overnight in 6-well plates. NF-κB-dependent firefly luciferase reporter and effector plasmids were co-transfected along with the Renilla luciferase plasmid in HEK-293T cells and MEF cell lines. Forty-eight h after transfection, cells were harvested in lysis buffer (Promega), and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity was calculated by dividing the firefly luciferase activity by the Renilla luciferase activity. Data represent three independent experiments performed in triplicate.
3. Results
3.1 Co-overexpression of TAK1/TAB1 or TRAF6/TAK1 induces K63-linked TAK1 polyubiquitination at Lys 158, but not at Lys 34
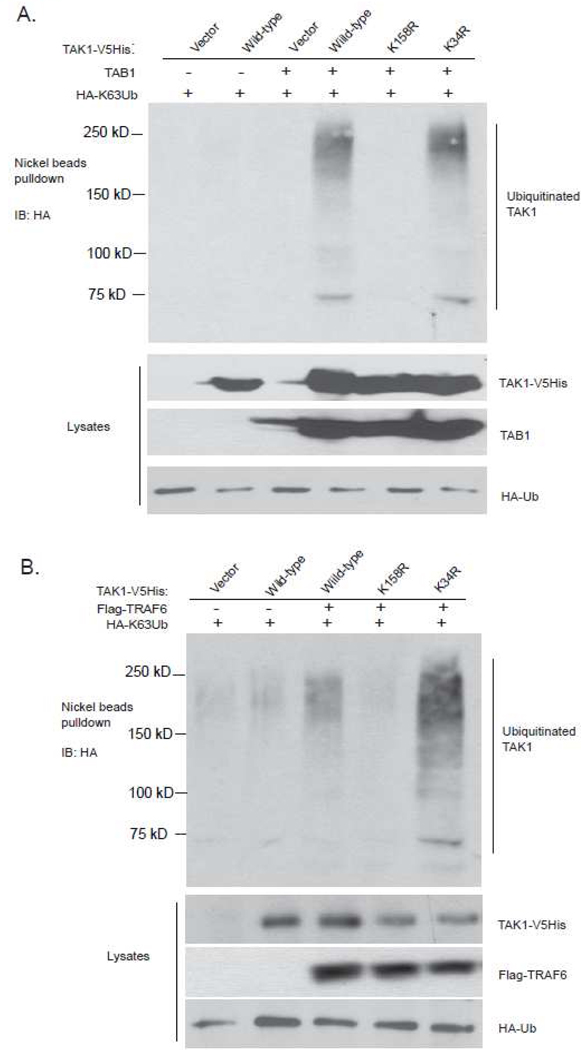
Overexpression of TAK1 with its regulatory subunit TAB1 leads to TAK1 K63-linked polyubiquitination and activation in the cells [13]. Our previous report suggests that TAK1 lysine 158 is required for TAK1-TAB1 co-overexpression-induced TAK1 polyubiquitination and TAK1-mediated IKK-NF-kB, JNK and p38 activations [13]. Interestingly, Sorrentino et al. reported that TAK1 lysine 34 is required for TGF-β-induced TAK1-mediated p38 activation [12]. To confirm which lysine residue is required for K63-linked TAK1 polyubiquitination and activation, the expression vectors encoding the C-terminal V5His-tagged TAK1 wild-type, K158R, K34R mutants were co-transfected with TAB1 into HEK-293T cells along with HA-Ub mutant containing one lysine only at position 63 (K63-only). Then The cell lysates from the transfected cells were boiled in the presence of 1% SDS and immunoprecipitated with Nickel beads, followed by SDS-PAGE and immunoblotting with anti-HA antibodies. In this assay, we found only TAK1 K158R mutant failed to induce TAK1 polyubiquitination compared to TAK1 wild-type and K34R mutant (Figure 1A).
Figure 1.
Lys63-linked TAK1 polyubiquitination acceptor site is Lys-158 residue, but not Lys-34 residue
a. Co-overexpression of TAK1/TAB1-induces TAK1 polyubiquitination at Lys-158, but not at Lys-34. Expression vectors encoding HA-ubiquitin (K63-only) and TAB1 were co-transfected into HEK-293T cells with control vector and expression vectors encoding TAK1-V5His-wild-type, -K34R and -K158R mutants, respectively. The cell lysates were heated in the presence of 1% SDS and diluted with lysis buffer in order to disrupt non-covalent protein-protein interactions. TAK1-V5His proteins in the transfected cells were precipitated with Nickel beads and immunoblotted with anti-HA antibodies to detect the presence of ubiquitinated TAK1-V5His.
b. TRAF6 mediates Lys63-linked TAK1 polyubiquitination at Lys-158 residue, but not at Lys-34 residue. Expression vectors encoding HA- ubiquitin (K63-only) and TRAF6 were co-transfected into HEK-293T cells with control vector and expression vectors encoding TAK1-V5His-wild-type, -K34R and -K158R, respectively. TAK1-V5His proteins in the transfected cells were precipitated with Nickel beads and immunoblotted with anti-HA antibodies to detect the presence of ubiquitinated TAK1-V5His.
TRAF6 is the E3 ligases that mediate K63-linked TAK1 polyubiquitination [13, 14]. To determine whether TAK1 lysine 34 or lysine 158 is required for TRAF6 mediate K63-linked TAK1 polyubiquitination, we co-transfected HA-Ub-K63-only and Flag-TRAF6 along with TAK1-V5His wild-type, K158R and K34R mutant into HEK-293T cells, respectively. As shown in Fig. 1B, TAK1 K158R mutant blocked TRAF6-induced K63-linked polyubiquitination of TAK1, whereas TAK1 K34R mutant failed to do so. This result suggests that TAK1 lysine 158 is required for TRAF6-mediated K63-linked TAK1 polyubiquitination in the cells.
3.2 TAK1 polyubiquitination at Lys-158, but not Lys-34, is required for TAK1/TAB1 co-overexpression-induced IKK-NF-κB and JNK-AP-1 activation
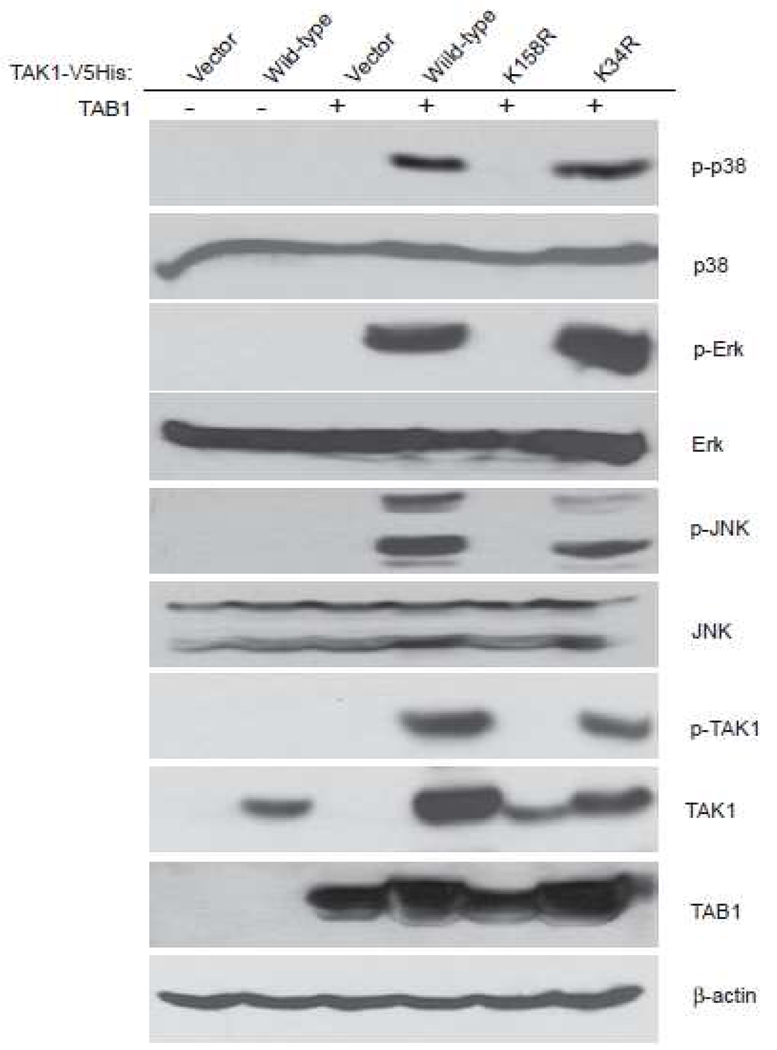
Protein ubiquitination plays an essential role in the tight regulation of the NF-κB activation [15]. K63-linked TAK1 polyubiquitination is critical for TAK1 activation [7, 16]. To clarify which lysine (Lysine 158 or Lysine 34) in TAK1 is important for TAK1 activation, we transfected TAB1 into 293T cells along with vector, TAK1 wild-type, K158R or K34R mutant, respectively. Overexpression of TAK1 wild-type and K34R mutant, but not K158R mutant, induced JNK and p38 activation (Figure 2A).
Figure 2.
TAK1 Lys-158 residue is required for TAK1/TAB1 Co-overexpression -induced p38 and JNK activation. TAB1 expression vector was co-transfected into 293T cells with empty vector or expression vectors encoding TAK1-wild-type, -K34R and -K158R mutants, respectively. Cell lysates were immunoblotted with antibodies indicated.
Consistent with these results, an AP-1-dependent luciferase reporter analysis showed that TAK1 K158R mutation, but not K34R mutation blocked TAK1/TAB1 co-overexpression-induced AP-1-dependent luciferase reporter gene activity [13]. This result indicates that TAK1 lysine 158 residue, but not lysine 34 residue, is required for TRAF6-mediated K63-linked TAK1 polyubiquitination and TAK1-mediated JNK and p38 activation.
3.3 Lysine 158, but not lysine 34, is required for TGF-β induces K63-linked polyubiquitination of TAK1
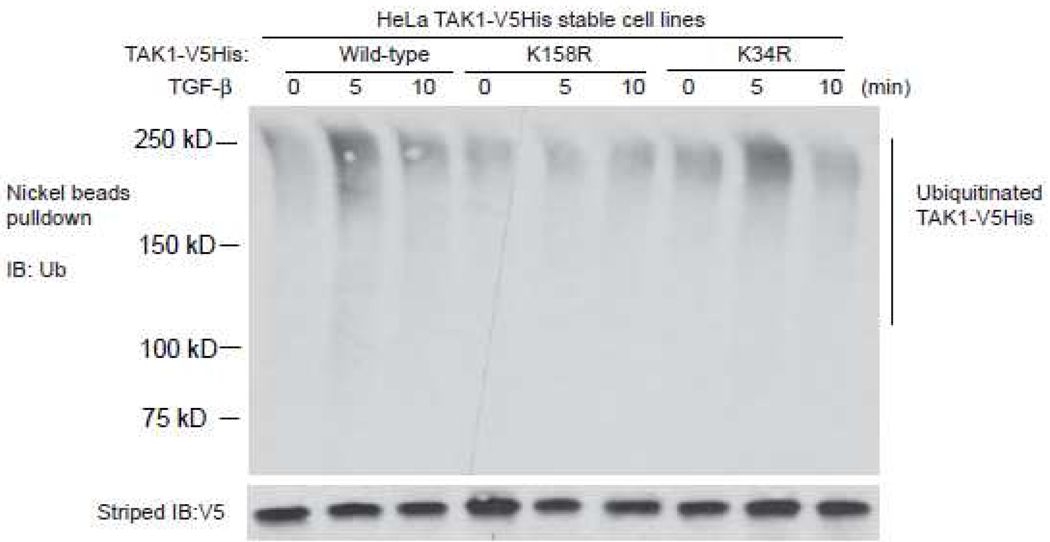
To determine whether Lysine 158 or Lysine 34 is required for TGF-β-induced TAK1 polyubiquitination, we establish three HeLa stable cell lines stably expressing TAK1 wild-type, K158R or K34R mutant with a His tag at the C-terminal. These cells were treated with TGF-β at different time points and then His-tagged TAK1 proteins in the cell lysates were pulled down by Nickel beads and immunoblotted with anti-ubiquitin antibodies to detect the polyubiquitination of TAK1. In this assay, we found that TGF-β induced polyubiqutination of TAK1 wild-type and K34R mutant, but failed to induce polyubiquitination of TAK1 K158R mutant. This result suggests that TAK1 lysine 158 is required for TGF-β-induced TAK1 polyubiquitination in the cells.
3.4 TAK1 lysine 158 residue is required for TGF-β-induced IKK, JNK and p38 activation
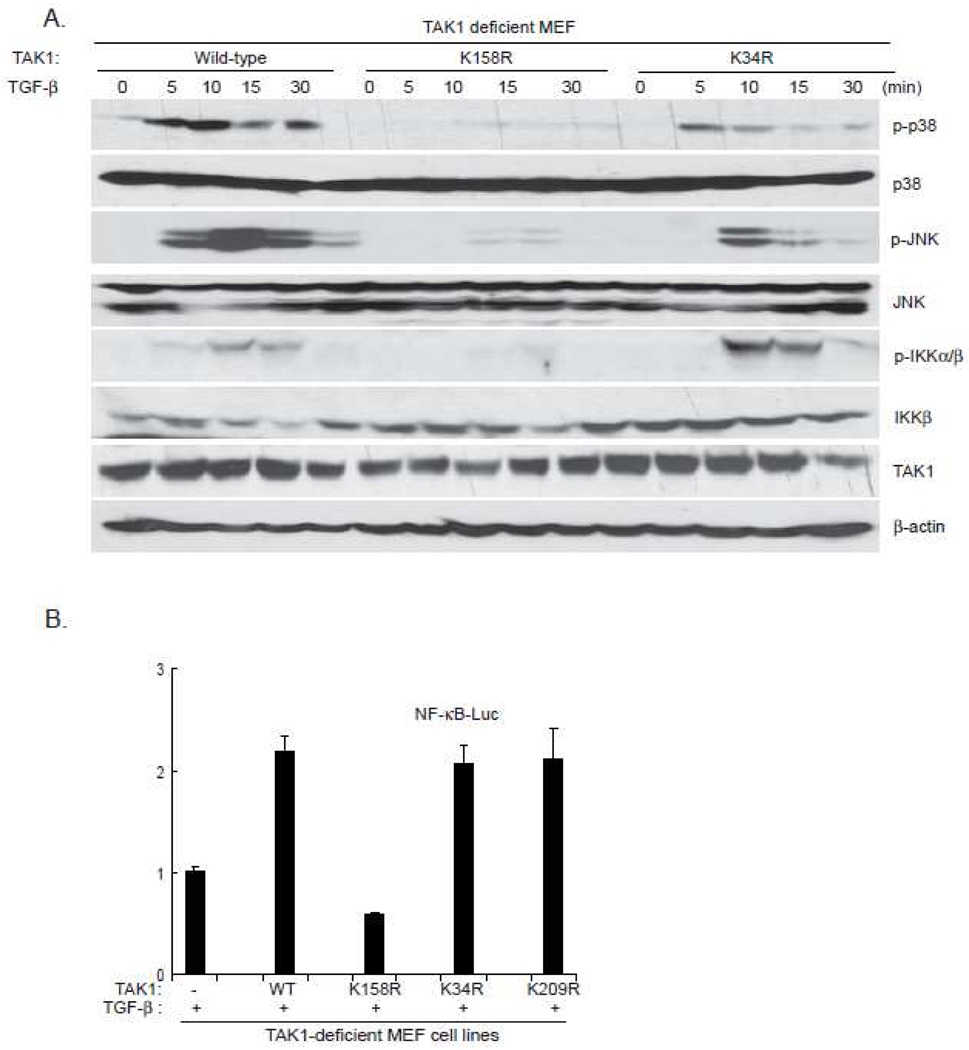
TAK1 mediates TGF-β-induced NF-κB activation via IKK phosphorylation and activation [17]. TAK1 also mediates TGF-β-induced p38 and JNK activation [4]. Therefore, we further analyzed the effect of TAK K158R and TAK1 K34R mutation on the TGF-β-induced IKK phosphorylation, P38 and JNK activation. The TAK1 wild-type, or K158R, or K34R mutant expression vectors were stably introduced back into the TAK1-deficient MEF cells by a retroviral transduction system. Then TAK1 wild-type, K158R mutant and K34R mutant reconstituted MEF cells were treated with TGF-β at different time points and then the cell lysates were immunoblotted with the indicated antibodies to examine TGF-β-induced JNK, p38, IKK phosphorylation. In this assay, TGF-β induced JNK and p38 phosphorylation were completely blocked in the TAK1 K158R mutant reconstituted MEF cells, whereas TGF-β-induced-normal JNK and p38 phosphorylation in the TAK1 wild-type or K34R mutant reconstituted cells (Figure 4). Similarly, TGF-β-induced IKK phosphorylation was impaired in the MEF cells with TAK1 K158R mutant compared to the cells with TAK1 wild-type or K34R mutant (Figure 4A). These results suggest that TAK1 lysine 158 residue is required for TGF-β-mediated IKK, p38 and JNK phosphorylation and activation.
Figure 4.
TAK1 Lys-158 but not Lys-34 residue is required for TGF-β-induced optimal p38, JNK and IKK activation
a. Expression of the TAK1-K158R mutant, but not -K34R, inhibits TGF-β-induced p38, JNK and IKK phosphorylation. TAK1-deficient MEF cells were transduced with the retrovirus encoding TAK1-wild-type, -K158R or -K34R mutant and subsequently selected with puromycin (3 µg/ml) to establish the TAK1-deficient MEF cell lines with the stable expression of either TAK1 wild-type, -K158R or -K34R mutant. The reconstituted TAK1-deficient MEF cells were untreated or treated with TGF-β(10 ng/ml) for the time points indicated and subsequently harvested.
b. Expression of the TAK1 K158R mutant inhibits TGF-β-induced NF-κB reporter activities. NF-κB reporter and 20 ng of Renilla-Luc plasmids were cotransfected into TAK1-deficient MEF control, TAK1 wild-type, K34R, K209R and K158R reconstituted cells for 48 h. Cells were then either untreated or treated with TGF-β (1 ng/ml) for 6 h. The relative luciferase activity was measured and normalized with the Renilla activity. Error bars indicate ± standard deviation in triplicate experiments.
Consistent with the above results, TGF-β induced a much higher NF-κB luciferase activities in MEF cells expressing the TAK1 wild-type, TAK1 K34R or TAK1 K209R, compared to the TAK1-deficient and K158R mutant cells (Figure 4B). Taken together, these results indicate that polyubiquitination of TAK1 at Lys158 site is required for TGF-β-induced optimal IKK/NF-κB and JNK/AP-1 activation.
4. Discussion
TAK1 plays important roles in immunological and developmental contexts across many species. In mammalian cells, TAK1 is essential for cells to respond to a variety of stimuli. TAK1 has emerged as a key regulator of signal transduction cascades leading to the activation of the transcription factors NF-κB and AP-1. Stimulation of cells with cytokines and microbial pathogens results in the activation of TAK1, which subsequently activates the IKK and MAP kinase, culminating in the activation of NF-kB and AP1, respectively [18, 19]. Recent studies have shown that K63-linked TAK1 polyubiquitination is required for TAK1 activation and its downstream signaling transduction. Sorrentino et al. showed that TGF-β-induced TRAF6-mediated K63-linked polyubiquitylation at TAK1 Lys 34 correlated with TAK1 activation [12]. Interestingly, we found that TAK1 lysine 158 was required for TNFα and IL-1β-induced TAK1 polyubiquitination and activation [13]. Our in vitro and in vivo analyses suggest that TAK1 lysine 158 is TRAF6-mediated TAK1 K63-linked polyubiqutination acceptor site in IL-1β-mediated signal transduction pathway. To rule out the possibility that different stimuli may induce TRAF6-mediated TAK1 K63-linked polyubiquitination at different sites, we have examined the effect of TAK1 K158R and K34R mutation on TGF-β-induced TAK1 polyubiquitination and IKK, JNK as well as p38 activation. Our results rule out the role of TAK1 lysine 34 residue in TGF-β-induced TAK1 polyubiquitination and IKK, JNK as well as p38 activation. TRAF family proteins are ubiquitin E3 ligases that promote TNFR signailing events [20]. TRAF6 is one of the most distinct members of its family, being uniquely able to transducer signals by all Toll/interleukin 1 receptors (TIRs) as well as a subclass of TNFR that includes CD40, RANK and NGFR [21]. Recently, TRAF6 has been shown to be a TAK1 E3 ligase [11, 12]. Interestingly, we found that TRAF6 failed to induce K63-linked polyubiquitination of TAK1 K158R mutant, but successfully induced K63-linked polyubiquitination of TAK1 wild-type and K34R mutant (Figure 1B), suggesting that TAK1 lysine 158 but not lysine 34 plays an essential role in TRAF6-mediated K63-linked TAK1 polyubiquitination and activation.
During preparation of this manuscript, Yamazaki et al. reported that TRAF6-mediated K63-linked TAK1 polyubiquitination at lysine 209 residue is required for IL-1β-induced TAK1-mediated IKK/NF-κB activation [14]. However, in our assay, we found that TAK1 lysine 209 residue is not essential for TRAF6-mediated K63-linked TAK1 polyubiquitination as well as TGF-β-induced IKK, JNK and p38 phosphorylation and activation (Figure 4B and data not shown).
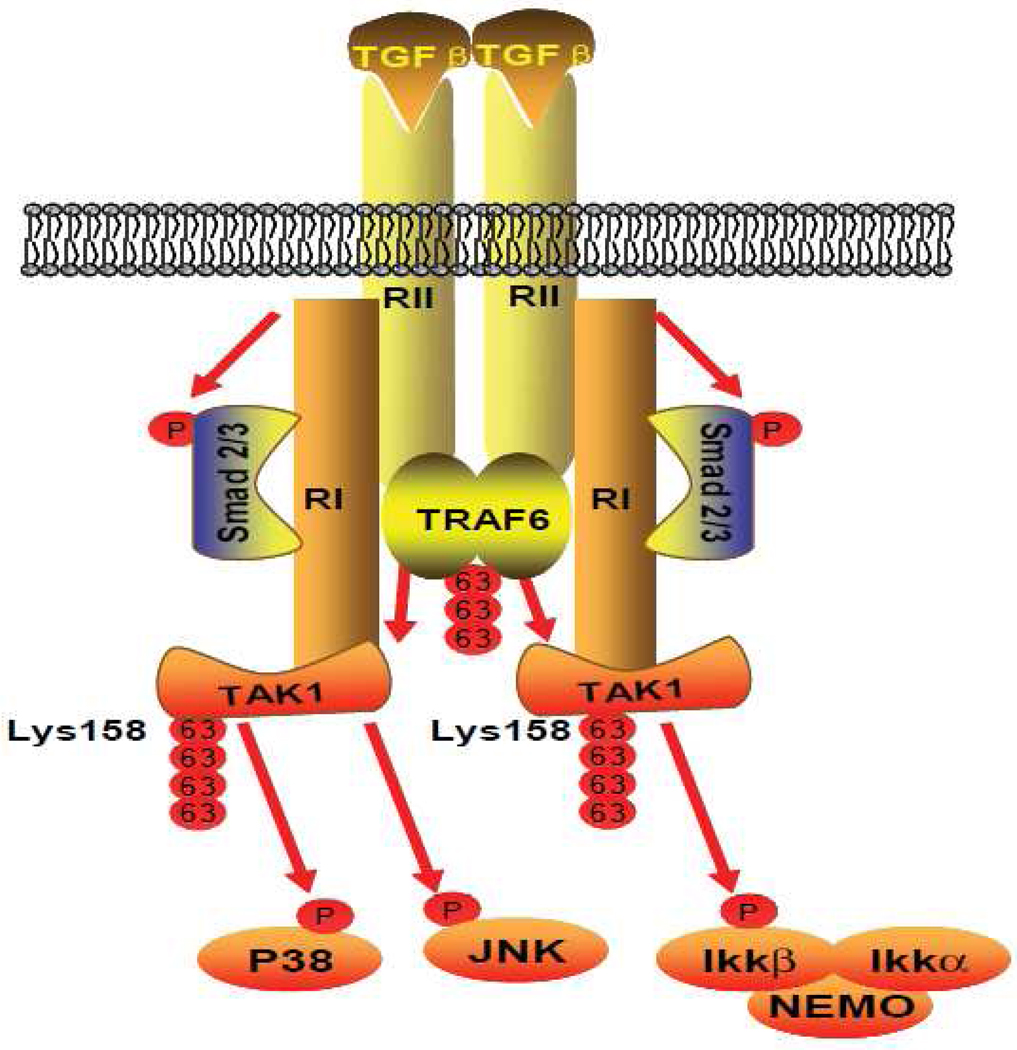
In view of the data presented here and in previous reports, we propose a working model (Figure 5) in which, upon TGF-β binding to their cognate receptors, the receptor-mediated signaling events induce TRAF6-mediated TAK1 polyubiquitination at lysine 158 within the kinase domain which in turn trigger the activation of TAK1-mediated IKK, JNK and p38 activation.
Figure 5.
Working Model: TGF-β binding to their cognate receptors, the receptor-mediated signaling events induce TRAF6-mediated TAK1 polyubiquitination at the Lys-158 within the kinase domain which in turn trigger the activation of TAK1-mediated p38, JNK and IKK activation.
Figure 3.
TGF-β induces TAK1 polyubiquitination at Lys-158 residue, but not at Lys-34 residue.
HeLa cells with stable expression of TAK1-V5His wild-type, -K34R and -K158R mutants were treated with TGF-β (10 ng/ml) for the time points indicated and subsequently lysed. The cell lysates were heated in the presence of 1% SDS and diluted with lysis buffer in order to disrupt non-covalent protein-protein interactions. Then, TAK1-V5His proteins in the cell lysates were precipitated with Nickel beads and immunoblotted with anti-ubiquitin antibodies to detect the presence of ubiquitinated TAK1-V5His.
Acknowledgments
This work was supported in part by the grants from the NIH/NCI 1R21CA106513-01A2 (to J.Y.), the American Cancer Society grant RSG-06-070-01-TBE (to J.Y.), the Virginia & L E Simmons Family Foundation Collaborative Research Fund (to J.Y.), 5P30CA125123-03 P30 Cancer Center Development Fund (to J.Y.) and a pilot fund from Dan L. Duncan Cancer Center (to J.Y.).
Abbreviations user are
- TGF-β
Transforming growth factor beta
- TAK1
TGF-β-activated kinase 1
- NF-κB
nuclear factor-κB
- IKK
IκB kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S, Deng C, Lu SL, Wang XJ. J Clin Invest. 2009;119:3408. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nawshad A, Lagamba D, Polad A, Hay ED. Cells Tissues Organs. 2005;179:11. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- 3.Heldin CH, Miyazono K, ten Dijke P. Nature. 1997;390:465. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 4.Derynck R, Zhang YE. Nature. 2003;425:577. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Science. 1995;270:2008. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 6.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. Science. 1996;272:1179. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. Nature. 2001;412:346. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 8.Prickett TD, Ninomiya-Tsuji J, Broglie P, Muratore-Schroeder TL, Shabanowitz J, Hunt DF, Brautigan DL. J Biol Chem. 2008;283:19245. doi: 10.1074/jbc.M800943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S. Genes Dev. 2005;19:2668. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jadrich JL, O'Connor MB, Coucouvanis E. Development. 2006;133:1529. doi: 10.1242/dev.02333. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. Mol Cell. 2008;31:918. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH, Landstrom M. Nat Cell Biol. 2008;10:1199. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 13.Fan YH, Yu Y, Shi Y, Sun W, Xie M, Ge N, Mao RF, Chang A, Xu G, Schneider MD, Zhang H, Fu S, Qin J, Yang J. J Biol Chem. 2010;285:5347. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki K, Gohda J, Kanayama A, Miyamoto Y, Sakurai H, Yamamoto M, Akira S, Hayashi H, Su B, Inoue J. Sci Signal. 2009;2:66. doi: 10.1126/scisignal.2000387. [DOI] [PubMed] [Google Scholar]
- 15.Skaug B, Jiang X, Chen ZJ. Annu Rev Biochem. 2009;78:769. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 16.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya-Tsuji J, Matsumoto K. Mol Cell. 2000;5:649. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 17.Delhase M, Hayakawa M, Chen Y, Karin M. Science. 1999;284:309. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 18.Adhikari A, Xu M, Chen ZJ. Oncogene. 2007;26:3214. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 19.Chen ZJ, Bhoj V, Seth RB. Cell Death Differ. 2006;13:687. doi: 10.1038/sj.cdd.4401869. [DOI] [PubMed] [Google Scholar]
- 20.Chung JY, Park YC, Ye H, Wu HJ. Cell Sci. 2002;115:679. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 21.Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J. J Biol Chem. 2005;280:35625. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]