Abstract
Regular bouts of physical activity may cause changes in gene expression that accumulate over time and ultimately affect phenotypes, such as body weight, blood lipid profile, and tumor development. Furthermore, acute activity may affect gene expression and phenotypes differently depending on whether the individual is regularly inactive or active. One-month old male Sprague-Dawley rats (n=72) were equally divided into SED (standard laboratory cage, n=24), PA (large activity box, n=24), and EX (exercise wheel inside standard cage, n=24) groups. At three months of age, half the animals from each group were sacrificed at rest and the other half following 30 minutes of physical activity. RNA was extracted from cardiac tissue, and microarray analysis was performed on 27,000 genes. Select gene results were validated using qPCR. No gene expression differences occurred when comparing all 3-month old groups at rest. A relatively small percentage of genes (1.9%) were differentially expressed (p<0.05) following acute swim activity in all groups but only 37 unique and identifiable genes reached or exceeded two fold differences in expression. The genes Atf3, Fos, Apold1, and Pxdn were expressed differently among SED, PA and EX following acute activity, with a clear separation of the magnitude in gene expression with SED > PA > EX. Differences in gene expression levels in young physically inactive and active animals following acute activity have different regulatory roles in gene networks that affect health-related phenotypes.
Keywords: acute exercise, gene expression, physical activity
Introduction
The interaction of genes and environment influences vital health and disease related phenotypes. Certain genes are regulated by either chronic physical activity or inactivity (Booth et al., 2008; Hamilton, Hamilton, & Zderic, 2007; Alessio et al., 2005; Booth, Chakravarthy, & Spangenburg, 2002; Kannel & Gordon, 1978). The up- or down-regulation of activity-sensitive genes may affect health-related phenotypes such as body weight, cholesterol, glucose, and triglycerides (Alessio et al, 2005), as a result of being regularly active or sedentary. In addition to the well-known effects of chronic activity, we speculate that a single bout of acute physical activity may affect a number of gene expressions depending on whether or not the individual is accustomed to regular physical activity. It was hypothesized that an acute bout of swimming would have different effects on gene expression depending on the animal's prior access to physical activity.
Regular exercise has been shown to enhance the health and function of an organism by influencing selective gene expressions that play a role in metabolic processes (Alessio 2006; Ji, Gomez-Cabrera, & Vina, 2006). If an animal is sedentary, acute activity could influence the level of expression of some genes differently compared with an active animal. These genes may then either regulate other genes, or directly influence the translation of proteins associated with health-related phenotypes. Although gene expression is only one of multiple regulatory factors that influence phenotypes, it is likely that acute physical activity causes cellular adaptations that influence transient changes in gene transcription. In a study by Hargreaves and Cameron-Smith (2002), genes encoding growth factors and signaling intermediates were found to be more responsive in animals that were physically inactive while genes that encode for metabolic enzymes and transporters were more affected in physically active animals. The significance of changes to gene transcription, translation, and ultimately specific protein levels is not fully understood and involves many complicated and integrated physiological steps.
Physical inactivity or activity may also influence gene expression by modifying chromatin and opening up core promotor regions of the DNA for the binding of regulatory proteins (Nestler and Hyman, 2002). Following an acute bout of physical activity, key regulatory proteins may come in contact with each other causing transcription factor complexes to form and phosphorylation to occur that affect how proteins bind to one another. These events, possibly stimulated by physical activity, are likely to activate or repress transcription of responsive target genes that may ultimately influence phenotypes associated with cardiovascular health and metabolic disease (Nestler and Hyman, 2002).
To date few studies have systematically investigated gene expressions and associated phenotypes in physically inactive and active animals before and after a single bout of acute exercise. One study reported that an hour following vigorous exercise, sedentary female rat muscle exhibited 52 significant changes in gene transcription compared with muscle collected from resting animals (McKenzie & Goldfarb, 2007). It is likely that multiple bouts of vigorous exercise may influence selective gene expressions that play a role in metabolic processes, ultimately accumulating over time, and affecting certain phenotypes. When performed regularly, physical activity can produce a hormetic effect, with beneficial adaptations occurring as a result of regular, tolerable stress (e.g. Alessio, 2006; Ji, Gomez-Cabrera, & Vina, 2006). It has also been theorized that the absence of vigorous exercise can either silence or unleash different genes due to long periods of very low physical stimulation (Booth et al., 2002). This could have important implications in the use of animal models for research, whereby the standard laboratory cage allows for little movement, forcing an animal into a sedentary lifestyle. As such, the sedentary model may have different physiological responses to treatment, and may not necessarily be representative of a regularly active population.
The purpose of this study was to investigate whether gene expressions change as a result of acute exercise in animals that are physically inactive and physically active. Gene expression levels were assessed before and after 30 minutes of swimming in three-month old male rats that were sedentary (SED), physically active for two months (PA), or voluntarily ran for two months on a exercise wheel (EX). Body weight, blood cholesterol and triglycerides were also compared in all animals.
Methods
Ethical approval
EX and SED animals were housed in pairs and the PA animals in groups of eight in climate-controlled rooms (24 ± 2 °C) with a 12-hour light/dark cycle. All animals had free access to water and food (LabDiet 5001 Rodent Chow, Purina, St Louis, MO). The health surveillance program included comprehensive serology: samples were sent quarterly from each rat colony to the Research Animal Diagnostic Laboratory at the University of Missouri (Columbia, MO). The study was conducted in accordance with ethical procedures and policies and was approved by the Miami University Institutional Animal Care and Use Committee.
Animals
Seventy-two four week old male Sprague-Dawley rats were randomly divided into three groups and given access to varying levels of physical activity or chronic exercise: 1) EX: voluntary access to wheel running exercise, 2) PA: a large activity box, and 3) SED: sedentary control with no access to physical activity outside of a standard cage.
EX and SED animals were housed in standard plastic cages (0.454 × 0.238 m), with the exception of the EX group having access every other day to voluntary wheel running access. The PA animals were housed in a large plastic box (1.2 × 0.6 m) that had ledges to encourage climbing and tubes to encourage exploration. Video recordings over 24 h were used to determine distance covered and intensity of exercise for all groups. The exercise wheels (Nalgene, Rochester, New York) recorded revolutions per day, which were translated into meters per day for analysis. All animals were handled and weighed weekly.
Rats were sacrificed after two months of activity treatment at age three months. Animals from each treatment group (EX, PA, SED) were randomly divided into two sets before sacrifice. One subset of animals (rest) from each treatment group was sacrificed at rest, and the other subset (swim) after vigorous swimming (approximately 30 min, water temperature = 35°C). All animals were naïve to swimming but appeared to tolerate the exercise, with no animals having to be rescued before 30 min. All animals were sacrificed by decapitation between 0800 and 1200 h. Whole blood was collected, chilled and centrifuged to recover serum. Heart, liver, kidneys, the soleus, and red and white muscle tissue from the quadriceps were removed immediately and flash frozen in liquid nitrogen. All serum, organs, and tissues were deep frozen (-80°C) for future assays and RNA extraction.
Blood cholesterol and triglycerides
Cholesterol was measured by an enzymatic method using cholesterol esterase, cholesterol oxidase and peroxidase (Wako Cholesterol CII, Wako Chemicals USA, Inc., Richmond, VA). Triglycerides were measured by an enzymatic method using glycerol kinase, glycerol-3-phosphate oxidase, and peroxidase (L-Type TG H, Wako Chemicals USA, Inc., Richmond, VA).
RNA extraction
The left ventricles (20-30mg) from 72 heart tissue samples (rest EX, n=12; rest PA, n= 12; rest SED, n=12; swim EX, n=12, swim PA, n=12; swim SED, n=12) were used for all RNA extractions and gene analysis. The RNA extraction was performed using Qiagen RNeasy (Valencia, CA) RNA extraction kits. The absorbance ratio (A260/A280) was determined using a ND-1000 UV/VIS spectrophotometer (Nanodrop Technologies, Inc., Montchanin DE) and was used to verify RNA purity. RNA integrity was assessed with an Agilent BioAnalyzer system by the Biomedical Genomics Core at the Research Institute at Nationwide Children's Hospital (Columbus, OH). Samples passing quality control were pooled within each treatment group to yield 4 gene chips per treatment group, with RNA from 3 animals on each chip, to ensure correct quantity of total RNA. Samples were labeled with the Affymetrix® Whole Transcript Labelling system and then hybridized to the Affymetrix GeneChip Rat Gene 1.0 ST Array (Santa Clara, CA). The GeneChip® Rat Gene 1.0 ST Array is the latest product in the family of Affymetrix expression arrays offering whole-transcript coverage. The design of the Rat Gene 1.0 ST Array is based primarily on a subset of GeneChip® Rat Exon 1.0 ST Array probes that map to well-supported exons of known genes. The array comprised of more than 700,000 unique 25-mer oligonucleotide features constituting more than 27,000 gene-level probe sets. Data was preprocessed using the RMA approach for background correction, normalization and probe set summarization using the Bioconductor affy package in R.
qPCR
Real Time RT-PCR gene expression analysis was performed at the SABiosciences laboratory with a RT2 qPCR Primer Assays kit (SABiosciences, Fredrick, MD). Glyceraldehyde-3-phospate dehydrogenase (Gapdh) was used as a control and the genes Atf3, Fos, Apold1, Pxdn, and Creb1 were analyzed. The first strand reaction for each sample was carried out with 1ug of RNA, and a dissociation (melting) curve was run for quality control.
Gene networks
Gene networks were generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). A gene network is a graphical representation of the molecular relationships between molecules. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least 1 reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. Microarray data from the current study has been overlaid onto these constructed networks to illustrate relationships between genes that were differentially expressed in response to acute exercise.
Statistical Analysis
Phenotype data are presented as means ± S.E.M. One-way ANOVAs were used to evaluate differences among the groups and a Bonferroni correction was applied for multiple comparisons. Significance Analysis of Microarrays (SAM) used the Bioconductor siggenes package to identify differentially expressed genes between rest and swim conditions for each treatment group. A two-class unpaired analysis with a false discovery rate (FDR) of 10% was used to maximize sensitivity and minimize the effect on accuracy. Analysis between conditions was used to determine the effect of acute exercise on gene expression. Comparisons were made between 3 mo swim EX vs 3 mo rest EX, 3 mo swim PA vs 3 mo rest PA, and 3 mo swim SED vs 3 mo rest SED. Statistical analysis of the QPCR delta-Ct values was performed using the Mann-Whitney Rank Sum test. Comparisons with a fold change ≥1.5 and a p <0.05 were considered significantly different.
Results
Phenotype data
All reported phenotype differences are among SED, PA, and EX groups, combining both rest and swim conditions, and are presented in Table 1. Weight differences were observed between all three groups. Body weight for the animals in the EX group was lower than for animals in the SED group (p<0.005) and the PA group (p=0.084). The differences between PA and SED were not found to be significant (p=0.793). Heart to body weight ratios differed between EX and both SED and PA (p < 0.05).
Table 1. Phenotype data comparisons among SED, PA, and EX groups.
| Body Weight (g) | Heart:Body Weight (×100) | Distance (m/day) | Cholesterol (mg·dl-1) | Triglycerides (mg·dl-1) | |
|---|---|---|---|---|---|
| SED | 408 ± 7 b | 0.34±0.007 | 130 ± 2b | 24 ± 1a,b | 152 ± 11 |
| PA | 397 ± 5 | 0.34 ±0.006 | 209 ± 3c | 21 ± 1a | 140 ± 16 |
| EX | 375 ± 8b | 0.37±0.007 | 11900 ± 2335b,c | 20 ± 1b | 139 ± 18 |
Values are means ± S.E.M.
SED≠PA,
SED≠EX,
PA≠EX;
p<0.05.
The EX group covered a greater distance per day than the PA and SED groups (p<0.001), but there were no differences between the PA and SED groups (p=1.000).
The SED group had higher blood cholesterol levels than PA and EX (p<0.050). Blood triglycerides followed the same trend where EX<PA<SED, however the differences did not reach statistical significance due to large variance.
Microarray
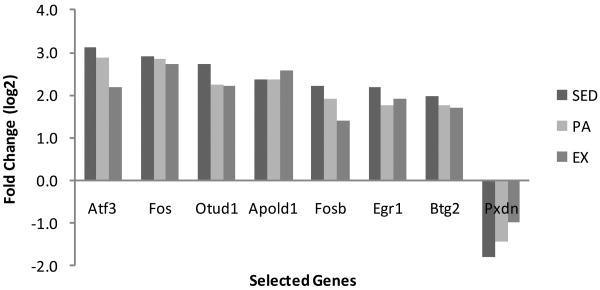
No gene expression differences occurred when comparing all 3-month old groups at rest. For each treatment, a relatively small number of genes (466) were up- or down-regulated (10% FDR) in the swim group compared to rest (Table 2) as determined by Significance Analyses of Microarrays (SAM). For the current study, SAM analysis was implemented using the Bioconductor siggenes package. A two-class unpaired analysis with a FDR of 10% was used to maximize sensitivity without significantly impacting accuracy. Less than 1% of the total 27,000 probe sets (113) expressed fold changes greater than 2. Only the genes that have been linked to expression of a known protein are listed in Tables 3 and 4, which brought the number of unique identifiable genes that were differentially expressed to 37. All the genes differentially expressed by at least two-fold and the respective fold change values for selected genes can be found in Tables 3 and 4 and Figure 1.
Table 2. Comparison of the number of significantly (10% FDR) up- and down-regulated genes in 3 month rats across all conditions.
| Comparison | Test | Control | Up-regulated | Down-regulated | Up-regulated at least 2 fold | Down-regulated at least 2 fold |
|---|---|---|---|---|---|---|
| 1 | Swim EX | Rest EX | 73 | 85 | 24 | 15 |
| 2 | Swim PA | Rest PA | 39 | 27 | 22 | 13 |
| 3 | Swim SED | Rest SED | 102 | 140 | 32 | 7 |
Table 3. Microarray results for exercise-induced gene expression changes, in alphabetical order for remaining identifiable genes with at least a two-fold change.
| Symbol | Gene Name | SED | PA | EX | |
|---|---|---|---|---|---|
| Abra | actin-binding Rho activating protein | 2.8 | 2.9 | 2.7 | |
| Adamts1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 2.6 | 2.3 | 2.2 | |
| Alb | albumin | 2.3 | |||
| Arl4a | ADP-ribosylation factor-like 4A | 2.0 | 2.1 | 2.2 | |
| Btg2 | B-cell translocation gene 2, anti-proliferative | 3.9 | 3.4 | 3.2 | |
| Car3 | carbonic anhydrase 3 | -2.6 | |||
| Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 | -2.0 | |||
| Cyr61 | cysteine-rich, angiogenic inducer, 61 | 4.9 | 3.6 | 3.8 | |
| Dusp1 | dual specificity phosphatase 1 | 2.0 | 2.3 | ||
| Dyrk3 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 | 2.5 | |||
| Egr1 | early growth response 1 | 4.5 | 3.4 | 3.8 | |
| Egr2 | early growth response 2 | 2.1 | 2.0 | ||
| Enc1 | ectodermal-neural cortex 1 | 2.0 | 2.0 | ||
| Errfi1 | ERBB receptor feedback inhibitor 1 | 3.5 | 3.0 | 3.0 | |
| Fosb | FBJ osteosarcoma oncogene B | 4.6 | 3.7 | 2.6 | |
| Gadd45g | growth arrest and DNA-damage-inducible, gamma | 2.4 | 2.4 | 2.4 | |
| Jun | Jun oncogene | 2.1 | 2.1 | ||
| Junb | jun B proto-oncogene | 2.5 | 2.2 | 2.5 | |
| LOC500300 | similar to hypothetical protein MGC6835 | 2.4 | 2.1 | 2.2 | |
| Myl7 | myosin, light chain 7, regulatory | -6.0 | |||
| Nr4a1 | nuclear receptor subfamily 4, group A, member 1 | 8.1 | 5.7 | 6.3 | |
| Nr4a2 | nuclear receptor subfamily 4, group A, member 2 | 7.4 | 6.0 | 4.4 | |
| Nr4a3 | nuclear receptor subfamily 4, group A, member 3 | 3.3 | 3.2 | ||
| Otud1 | OTU domain containing 1 | 6.7 | 4.8 | 4.7 | |
| Ptgs2 | prostaglandin-endoperoxide synthase 2 | 2.1 | |||
| Reg3b | regenerating islet-derived 3 beta | 2.0 | |||
| Rgs1 | regulator of G-protein signaling 1 | 2.1 | |||
| Rgs2 | regulator of G-protein signaling 2 | 2.2 | 2.0 | ||
| Serpine1 | serine (or cysteine) peptidase inhibitor, clade E, member 1 | 2.0 | 2.0 | ||
| Sgk1 | serum/glucocorticoid regulated kinase 1 | 2.0 | 2.1 | 2.0 | |
| Snf1lk | SNF1-like kinase | 2.8 | 2.7 | 2.6 | |
| Sox18 | SRY (sex determining region Y)-box 18 | -2.1 | |||
Fold changes in gene expression are for SWIM compared to REST within each group.
Table 4. Microarray and qPCR for exercise-induced gene expression changes, in alphabetical order, for select genes with at least a two-fold change.
| Symbol | Gene Name | SED | PA | EX | |||
|---|---|---|---|---|---|---|---|
| microarray | qPCR | microarray | qPCR | microarray | qPCR | ||
| Apold1 | apolipoprotein L domain containing 1 | 5.1 | 7.1 | 5.1 | 3.1 | 6.0 | 3.2 |
| Atf3 | activating transcription factor 3 | 8.7 | 15.0 | 7.3 | 5.1 | 4.5 | 3.7 |
| Creb1 | cAMP responsive element binding protein 1 | 0.0 | 1.5 | 0.0 | 1.0 | 0.0 | -1.1 |
| Fos | FBJ osteosarcoma oncogene | 7.5 | 50.0 | 7.2 | 8.0 | 6.6 | 8.1 |
| Pxdn | peroxidasin homolog (Drosophila) | -3.5 | 1.7 | -2.7 | -1.1 | -2.0 | 1.0 |
Fold changes in gene expression are for SWIM compared to REST within each group.
Figure 1. Microarray values of significantly differentially expressed genes following acute exercise in SED, PA, and EX experimental groups.
Fold changes for Creb1 did not differ following acute exercise. Reported as log2 transform. n=6, p<0.05
qPCR
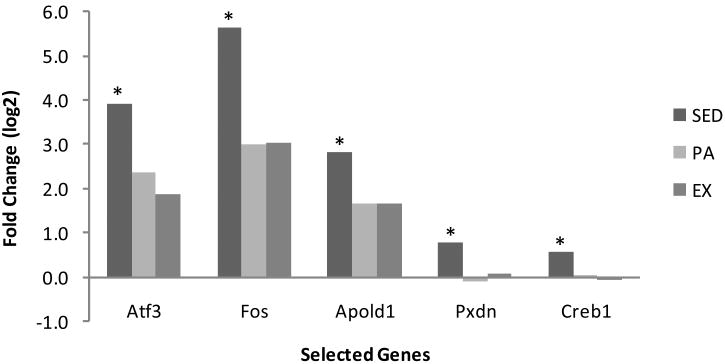
qPCR was performed on five genes in addition to the standard control Gapdh. These genes were selected for investigation based on relatively high fold changes in microarray, reported functions relating to health, and/or position within complex gene network systems (Figures 3a and 3b). The SED group was found to be significantly different from both the PA and EX groups (p<0.05). For the genes Atf3, Fos, and Apold1 the direction of the fold change was the same as microarray data, with an increase in gene expression in acutely exercised rats relative to control. The magnitude of change was similar to microarray data except for Fos in the SED group, which had a 7.5-fold increase in microarray but a 50-fold increase with qPCR. Differential expression of Pdxn as detected by microarray was not confirmed with qPCR. For Creb1, both microarray analysis and qPCR confirmed no change in gene expression levels in any of the three groups. Thus, qPCR confirmed microarray results at a level closely matching the 10% FDR used as a statistical cutoff. The qPCR results are reported with corresponding microarray data in Table 3 and Figures 1 and 2.
Figure 3.
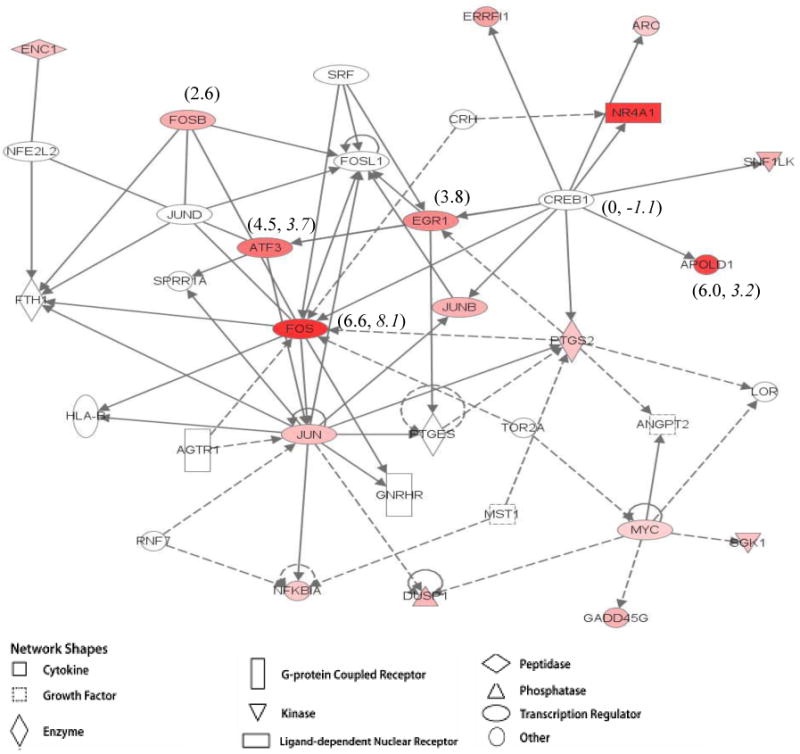
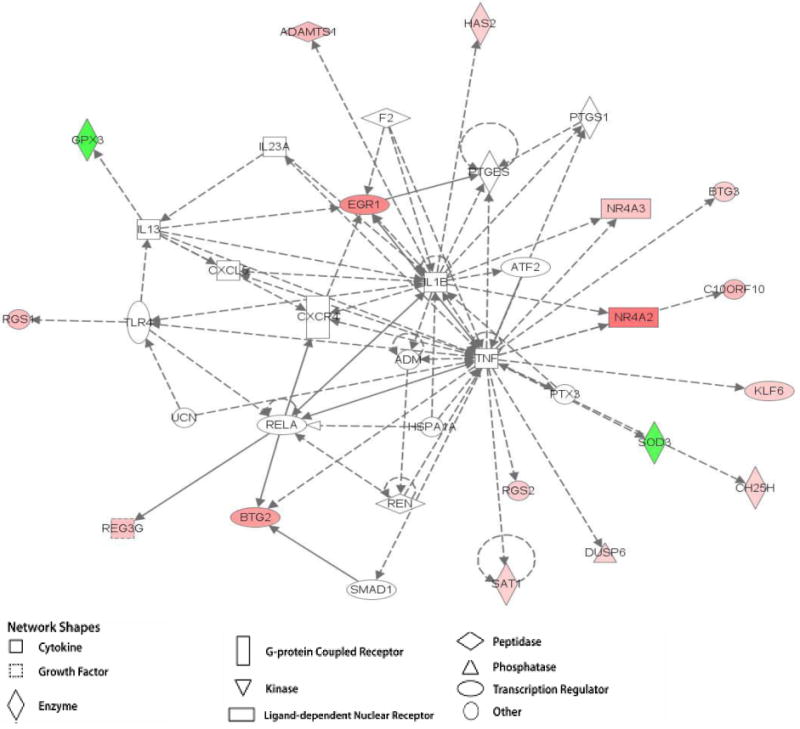
Figure 3a. Ingenuity pathway analysis of differentially expressed genes in 3 month-old male rats at rest and following acute exercise. Example of previously established gene maps overlaid with EX microarray data showing gene interaction and differential gene expression. Where indicated, values in parenthesis represent microarray fold changes followed by qPCR fold changes, when available. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product.
Figure 3b. Ingenuity pathway analysis of differentially expressed genes in 3 month-old male rats at rest and following acute exercise. Example of previously established gene maps overlaid with EX microarray data showing gene interaction and differential gene expression. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product.
Figure 2. qPCR values of significantly differentially expressed genes following acute exercise in SED, PA, and EX experimental groups.
Reported as log2 transform. n=6, * indicates significance p<0.05
Gene networks
Microarray data was overlaid on gene networks to show genes that were up- or down-regulated following acute exercise among the treatment groups in this study (Figures 3a and 3b). These pathways visually describe the relative magnitude and direction of gene expression changes specific to this study. The gene networks in Figures 3a and 3b include genes that were differentially expressed when comparing acute swim to rest, as well as those genes that did not differ. Often, a gene that was not differentially expressed, such as Creb1, was found to interact with several genes that were significantly up or down-regulated. This may suggest a change in function of this gene at the protein level. In other instances, several genes that were differentially expressed are all linked together in a common chain, such as seen with Fos, Atf3, Egr1, and Apold1.
Discussion
Even at a relatively young age of three months, the current results report differences in gene expressions and phenotypes including body weight, blood cholesterol, and a trend in triglycerides among the three treatment groups (Table 1). SED, with no access to physical activity outside a standard cage, had the highest cholesterol, and the PA group, with freedom to roam a large box, had cholesterol levels similar to EX. Triglyceride levels showed similar trends, although due to large variation, differences did not reach statistical significance. Thus, it seems that physical inactivity has a detrimental effect on some phenotypes, whereas relatively moderate to high levels of physical activity prevent them.
Less than 1.9% of the total rat genome was differentially expressed among the groups following swimming. All genes that had at least a two -fold change in expression and their specific fold change values for each group can be found in Tables 3 and 4. The majority of genes exhibited expression changes in the same direction across all three groups, implying that an acute bout of exercise elicited a similar gene response regardless of the animal's activity level. However, the trend of gene changes were consistent: SED > PA > EX, indicating that regular physical inactivity makes a large and potentially lasting impact on gene expressions, while regular physical activity attenuates the magnitude of the response of these genes to acute stress. This trend was observed in both microarray and qPCR, although the magnitude of the gene expression fold changes were typically higher in qPCR, providing compelling evidence that a sedentary condition may activate or suppress genes in such a way that contributes to unhealthy phenotypes, whereas being moderate to highly active may prevent some of these deleterious changes.
Changes in gene expression are not always associated with similar changes in protein expression. Bey and Hamilton (2003) reported a decreased skeletal muscle lipoprotein lipase (LPL) during physical inactivity that was independent of any change in LPL mRNA. Hamilton et al (2008) reported increased SOCS2 and SOCS3 mRNA but no difference in the protein expression. Gene networks are used to portray the interactions between genes in multiple related pathways and have been generated by studies of specific gene functions and causal connections (D'haeseleer, Liang, & Somogyi, 2000). Essentially, gene networks are a snapshot in time of gene expression levels when comparing rats sacrificed immediately following swimming with referent-control rats sacrificed at rest (Figures 3a and 3b). A network produced in the EX group gene array revealed Creb1 (cAMP-responsive element binding protein) as a strategically placed gene connected to pathways linking Fos, Atf3, Egr1, and Apold1. Creb1 plays is believed to play a prominent role in breast cancer (e.g. Sakamoto & Frank, 2009; Chhabra et al, 2007) and leptin signaling pathway associated with obesity related endometrial cancer (Catalano et al, 2009). While Creb1 was not differentially expressed, other genes linked in the pathway were up-regulated following acute exercise.
Atf3 (Activating transcription factor 3)
Atf3 (Activating transcription factor 3) expression responded differently to an acute bout of exercise in all three groups. Atf3 is considered an “adaptive response” gene, and its over-expression may suppress cell growth (Yin, Dewille, & Hai, 2008; Tamura et al, 2005; Fan et al., 2002). While increased expression of Atf3 is linked to tumor cell suppression, it may also cause the over-expression of genes known to enhance the growth of cancer cells. Acute swim-induced Atf3 expression changes were smallest in the EX group (4.5-fold), higher in the PA (7.3-fold) and highest (8.7-fold) in SED. It may be that expression of this gene is increased in all groups following a stressful episode of exercise in response to cell damage, with EX experiencing the least and SED experiencing the most exercise-induced cell damage.
Fos (FBJ murine osteosarcoma viral oncogene homolog)
Fos (FBJ murine osteosarcoma viral oncogene homolog) followed a similar pattern (e.g. SED>PA>EX) as Atf3 with a 7.5 fold increase in the SED and slightly less increase in the EX (6.6). The current study reported opposite results than Collins et al (2009), where Fos was up-regulated the most in their exercise trained group. The reason for the different results may be because in the Collins et al (2009) study, animals were sacrificed two hours following swimming- a time they thought to be peak for Fos induction compared with immediately following swimming in the present study. The fold-change trends were similar in both microarray and qPCR data, however, SED Fos qPCR results were extremely high (50-fold) and represented a lone outlier.
Apold1 (apolipoprotein)
Apold1 (apolipoprotein) regulates endothelial cell differentiation, activation, cell signaling, and may also be related to vascular function (Regard et al, 2004). One study associated the Apold gene cluster with triglyceride and high density lipoprotein (HDL) cholesterol levels (Lu et al, 2006), while another related it to HDL and atherosclerosis regulation (Plump, Scott, & Breslow, 1994). In the current study, Apold1 was over-expressed in all groups following swimming. Microarray fold changes were similar in all three groups while the qPCR trend was SED > PA > EX. This was the only gene of five measured where microarray and qPCR inconsistency was observed.
Pxdn (peroxidasin homolog)
Pxdn (peroxidasin homolog) is involved in peroxidase activity and is associated with oxidation-reduction and oxidative stress (Cheng et al, 2008). Microarray results indicated that Pxdn was down-regulated in all three groups (SED = -3.5-fold, PA=-2.7-fold, EX=-2-fold) but qPCR did not always show the same direction (SED = 1.7-fold, PA=1.1-fold, EX =-1.1-fold). It is possible that the whole transcript microarray used in this study may be capturing a change in differential splicing of the Pdxn transcript that may not be captured by qPCR analysis which typically targets a single 3′ region of the mRNA.
Results of this study are consistent with previous studies (e.g. Lambertucci et al., 2006; Bronikowski et al., 2002) that reported marked differences in a relatively small number of genes between untrained and exercised-trained groups of animals at rest. Four genes (Dyrk3, Alb, Ptgs2, Sox18) responded to acute exercise only in the SED group, three genes (Myl7, Car3, Arl4a) changed following acute exercise only in the PA group, and three other genes (Rgs1, Cyp1a1, Reg3b) changed following acute exercise only in the EX group. While different access to physical activity did not seem to affect most gene expressions considering the entire genome, healthier phenotypes were found in animals that regularly exercised over a period of 2 months. Results from the current study support previous work that transcriptional activation can occur during exercise and recovery from exercise (Kraniou et al., 2000). It is possible that phenotypic differences that endure and intensify into adulthood are a result of a relatively small number of modified gene expressions that occur relatively early in life in response to regular physical inactivity or activity.
In summary, 1.9% of all rat genes were differentially expressed (p<0.05) when comparing the three treatment groups (SED, PA, EX) before and after acute swimming. Microarray results indicated Atf3, Fos, Apold1, and Pxdn were expressed differently among SED, PA and EX groups following an acute bout of swimming, with a very clear separation of the magnitude in gene expression of SED > PA > EX. Favorable phenotypes including body weight, total blood cholesterol, and blood triglycerides were observed in animals that voluntarily ran on an exercise wheel compared with animals residing in a standard cage. Differences in inactivity and activity-induced gene expression levels in young animals may have important regulatory roles in gene networks and ultimately affect health-related phenotypes in adulthood.
Acknowledgments
Funding for this research was supported by the National Institute of Aging Grant 1R15AG029653-01A1.
References
- Alessio HM. Alessio HM, Hagerman AE. Oxidative stress in exercise and aging. London: Imperial College Press; 2006. Oxidative stress and the exercise continuum; pp. 58–84. [Google Scholar]
- Alessio HM, Hagerman AE, Nagy S, Philip B, Byrnes RN, Woodward JL, et al. Exercise improves biomarkers of health and stress in animals fed ad libitum. Physiology & Behavior. 2005;84(1):65–72. doi: 10.1016/j.physbeh.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Alessio HM, Schweitzer NB, Snedden AM, Callahan P, Hagerman AE. Revisiting influences on tumor development: Focusing on laboratory housing. Journal of the American Association for Laboratory Animal Science. 2009;48:1–5. [PMC free article] [PubMed] [Google Scholar]
- Bey L, Hamilton MT. Suppression of skeletal muscle lipoprotein lipase activity during physical inactivity: a molecular reason to maintain daily low-intensity activity. Journal of Physiology. 2003;551(2):673–682. doi: 10.1113/jphysiol.2003.045591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW, Chakravarthy MV, Spangenburg EE. Exercise and gene expression: Physiological regulation of the human genome through physical activity. Journal of Physiology. 2002;543(2):399–411. doi: 10.1113/jphysiol.2002.019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW, Laye MJ, Lees SJ, Rector RS, Thyfault JP. Reduced physical activity and risk of chronic disease: The biology behind the consequences. European Journal of Applied Physiology. 2008;102:381–390. doi: 10.1007/s00421-007-0606-5. [DOI] [PubMed] [Google Scholar]
- Bronikowski AM, Carter PA, Morgan TJ, Garland T, Ung N, Pugh TD. Lifelong voluntary exercise in the mouse prevents age-related alterations in gene expression in the heart. Physiological Genomics. 2002:1–36. doi: 10.1152/physiolgenomics.00082.2002. Articles in Press. [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Baldwin LA. Defining hormesis. Human and Experimental Toxicology. 2002;21:91–97. doi: 10.1191/0960327102ht217oa. [DOI] [PubMed] [Google Scholar]
- Catalano S, Giordano C, Rizza P, Gu G, Barone I, Bonofiglio D, et al. Evidence that leptin through STAT and CREB signaling enhances cyclin D1 expression and promotes human endometrial cancer proliferation. Journal of Cell Physiology. 2009;8(3):490–500. doi: 10.1002/jcp.21622. [DOI] [PubMed] [Google Scholar]
- Cheng G, Salerno JC, Cao Z, Pagano PJ, Lambeth JD. Identification and characterization of VPO1, a new animal heme-containing perosidase. Free Radical Biology & Medicine. 2008;45:1682–1694. doi: 10.1016/j.freeradbiomed.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra A, Fernando H, Watkins G, Mansel RE, Jiang WG. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncology Report. 2007;18(4):953–958. [PubMed] [Google Scholar]
- Collins A, Hill LE, Chandramohan Y, Whitcomb D, Droste SK, Reul JM. Exercise improves cognitive responses to psychological stress through enhancement of epigenetic mechanisms and gene expression in the dentate gyrus. Public Library of Science. 2009;4(1):4330. doi: 10.1371/journal.pone.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'haeseleer P, Liang S, Somogyi R. Genetic network inference: From co-expression clustering to reverse engineering. Bioinformatics. 2000;16(8):707–726. doi: 10.1093/bioinformatics/16.8.707. [DOI] [PubMed] [Google Scholar]
- Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene. 2002;21(49):7488–7496. doi: 10.1038/sj.onc.1205896. [DOI] [PubMed] [Google Scholar]
- Hamilton KL, Lin L, Wang Y, Knowlton AA. Effect of ovariectomy on cardiac gene expression: Inflammation and changes in SOCS genes expression. Physiological Genomics. 2008;32:254–263. doi: 10.1152/physiolgenomics.00039.2007. [DOI] [PubMed] [Google Scholar]
- Hamilton MT, Hamilton DG, Zderic TW. Role of low energy expenditure and sitting in obesity, metabolic syndrome, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:2655–2667. doi: 10.2337/db07-0882. [DOI] [PubMed] [Google Scholar]
- Hargreaves M, Cameron-Smith Exercise, diet, and skeletal muscle gene expression. Medicine and Science in Sports and Exercise. 2002;34:1505–1508. doi: 10.1097/00005768-200209000-00017. [DOI] [PubMed] [Google Scholar]
- Ji LL, Gomez-Cabrera MC, Vina J. Exercise and hormesis: Activation of cellular antioxidant signaling pathway. Annals of the New York Academy of Sciences. 2006;1067:425–435. doi: 10.1196/annals.1354.061. [DOI] [PubMed] [Google Scholar]
- Kannel WB, Gordon T. Evaluation of cardiovascular risk in the elderly: The Framingham study. Bulletin of the New York Academy of Medicine. 1978;54(6):573–591. [PMC free article] [PubMed] [Google Scholar]
- Kraniou Y, Cameron-smith D, Misso M, Collier G, Hargreaves M. Effects of exercise on -4 and glycogenin gene Expression in human skeletal muscle. Journal of Applied Physiology. 2000;88:794–796. doi: 10.1152/jappl.2000.88.2.794. [DOI] [PubMed] [Google Scholar]
- Lambertucci RH, Levada-Pires AC, Rossoni LV, Curi R, Pithon-Curi TC. Effects of aerobic exercise training on antioxidant enzyme activities and mRNA levels in soleus muscle from young and aged rats. Mechanisms of Ageing and Development. 2006;128(2007):267–275. doi: 10.1016/j.mad.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Lu Q, Liu S, Rifai N, Hunter D, Hu FB. Associations of the apolipoprotein A1/C3/A4/A5 gene cluster with triglyceride and HDL cholesterol levels in women with type 2 diabetes. Atherosclerosis. 2006;192(1):204–210. doi: 10.1016/j.atherosclerosis.2006.05.006. [DOI] [PubMed] [Google Scholar]
- McKenzie MJ, Goldfarb AH. Aerobic exercise bout effects on gene transcription in the rat soleus. Medicine and Science in Sport & Exercise. 2007;39(7):1515–1521. doi: 10.1249/mss.0b013e318074c256. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Regulation of gene expression. In: Davis KL, Charney D, Coyle JT, Nemeroff C, editors. Neuropsychopharmacology: The Fifth Generation of Progress. American College of Neuropsychopharmocology; 2002. pp. 217–228. [Google Scholar]
- Pilegaard H, Ordway GA, Saltin B, Neufer D. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. American journal of physiology endocrinology and metabolism. 2000;279:806–814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- Plump AS, Scott CJ, Brewlow JL. Human apolipoprotein A-1 gene expression increases high density lipoprotein and suppresses atherosclerosis in apolipoprotein E- deficient mouse. Proceedings of the National Academy of Sciences. 1994;91:9607–9611. doi: 10.1073/pnas.91.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard JB, Scheek S, Borbiey T, Lanahan AA, Schneider A, Demetriades AM, et al. Verge: A novel vascular early response gene. Journal of Neuroscience. 2004;24(16):4092–4103. doi: 10.1523/JNEUROSCI.4252-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: Implications for targeting transcription factors for cancer therapy. Clinical Cancer Research. 2009;15(8):2583–2587. doi: 10.1158/1078-0432.CCR-08-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Hua B, Adachi S, Guney I, Kawauchi J, Morioka M, et al. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. The EMBO Journal. 2005;24(14):2590–601. doi: 10.1038/sj.emboj.7600742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov VT, Volkl S, Friedrich J, Kunz-Schughart LA, Hehlgans T, Vermeulen L, et al. Role of CREB1 and NF{kappa}B-p65 in the down-regulation of renin gene expression by tumor necrosis factor {alpha} Journal of Biological Chemistry. 2005;280(26):24356–24362. doi: 10.1074/jbc.M502968200. [DOI] [PubMed] [Google Scholar]
- Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27(15):2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]