Abstract
A large body of evidence indicates that the risk of developing chronic diabetic complications is under the control of genetic factors. Previous studies using a candidate gene approach have uncovered a number of genetic loci that may shape this risk, such as the VEGF gene for retinopathy, the ELMO1 gene for nephropathy, and the ADIPOQ gene for coronary artery disease. Recently, a new window has opened on identifying these genes through genome-wide association studies. Such systematic approach has already led to the identification of a major locus for coronary artery disease on 9p21 as well three potential genes for nephropathy on 7p, 11p, and 13q. Further insights are expected from a broader application of this strategy. It is anticipated that the identification of these genes will provide novel insights on the etiology of diabetic complications, with crucial implications for the development of new drugs to prevent the adverse effects of diabetes.
Keywords: Diabetic nephropathy, Diabetic retinopathy, Coronary artery disease, Atherosclerosis, Polymorphisms, Candidate genes, Genome-wide association studies
Introduction
Long-term complications affecting the eyes, the kidneys, and the cardiovascular system are the principal cause of morbidity and mortality among diabetic subjects (1). Because of the high prevalence of diabetes, these complications are significant public health problems, being responsible for a large proportion of blindness, renal replacement therapy, and cardiovascular interventions in the US. Large clinical trials have demonstrated that normalization of glycemia can greatly reduce the incidence of diabetic complications (2;3). However, in clinical practice, normalizing blood glucose is not a trivial task and almost 50% of diabetic subjects fail to reach the recommended target of an HbA1c lower than 7% (4). There is a clear need for additional interventions decreasing the impact of high glucose among those subjects who do not manage to reach normoglycemia.
A critical requirement for the design of such new therapies is a thorough knowledge of the mechanisms linking the diabetic milieu to tissue damage. One of the approaches that are being used to expand our knowledge in this field is to search the genome for variants that are associated with an increased risk of complications among diabetic subjects, with the expectation that the location and function of these variants will point to genes and hence molecular pathways that are involved in the etiology of these conditions. Such genetic approach may also generate algorithms for early identification of diabetic individuals at high risk of complications who may benefit from especially aggressive prevention programs. Below is a review of the findings obtained through this approach, with a special emphasis on the most recent developments.
Heritability of diabetic complications
The role of genetic factors in shaping susceptibility to diabetic complications has been known for many years. Family studies have clearly indicated that susceptibility to diabetic nephropathy has a familial basis, with diabetic siblings of diabetic probands with kidney disease having a three-fold increase in the risk of diabetic nephropathy as compared to siblings of normoalbuminuric subjects (5). About 30% of the variance of urinary albumin excretion rate, considered as a quantitative trait, has been found to be explained by familial factors (6). As far as cardiovascular complications are concerned, 40–50% of the variance in coronary calcium and carotid intima-media thickness – two indices of the extent of atherosclerosis – can be attributed to familial factors among individuals with diabetes (7;8). Interestingly, this figure is only modestly affected by adjustment for known cardiovascular risk factors, indicating that this effect involves as yet unidentified atherogenic pathways. Heritability estimates ranging from 25 and 50% have been reported for diabetic retinopathy (9;10).
Candidate genes studies
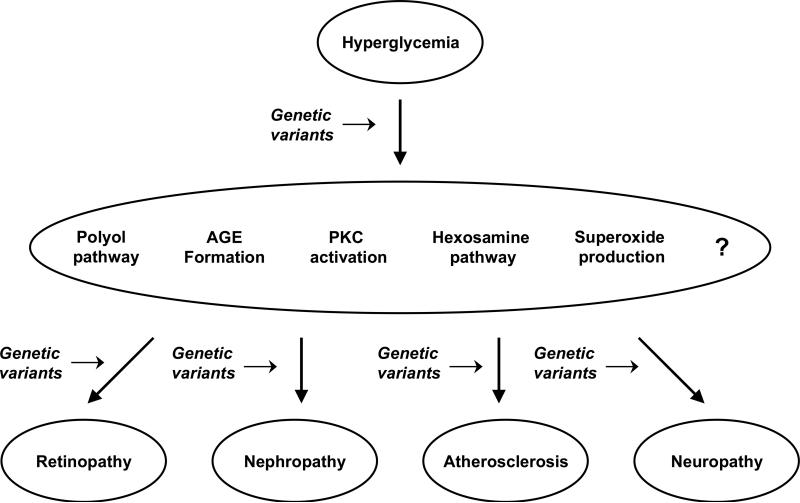
Until recently, our limited knowledge of the human genome and the low-throughput of the genotyping technology constrained the search for genetic determinants of diabetic complications to a small number of candidate genes selected on the basis of their postulated role in cellular pathways linking glucose to tissue damage. As schematically shown in Figure 1, the candidate genes that have been studied can be subdivided in two categories: i. genes placed in pathways that mediate the production of cellular toxins in response to excess glucose (e.g., reactive oxygen species, sorbitol, advanced glycation-end products), and ii. genes placed in pathways involved in the tissue-specific organ damage induced by these toxins.
Figure 1.
Schematic representation of the sites at which genetic variants may act in modulating the risk of diabetic complications.
While candidate gene studies have been valuable, it is difficult to draw firm conclusions from these efforts. Studies have often been small and have yielded p values that are only nominally significant and cannot withstand an adjustment for multiple comparisons. In some cases, limited or no attempts were made to replicate findings. In other cases, multiple reports were published with conflicting findings resulting either from false positives due to the marginal p values, or from false negatives due to the lack of statistical power or the fact that different phenotypes were investigated in different studies (e.g., clinically significant CAD vs. coronary artery calcium). Nonetheless, despite these problems, interesting association signals have emerged, some of which are reviewed below.
Candidate genes involved in glucose-induced pathways
The most robust finding in this category concerns the gene coding for aldose reductase (AKR1B1) – an enzyme that catalyzes the reduction of carbonyl-containing compounds to their respective alcohols. Aldose-reductase is a key regulator of the polyol-pathways through which excess glucose is converted to sorbitol and fructose in insulin-independent tissues. Accumulation of these metabolites, especially sorbitol, has been shown to induce complex biochemical changes contributing to the development of diabetic complications (11). An (AC)n microsatellite polymorphism, located in the 5' flanking region of AKR1B1, was found to be associated with diabetic retinopathy in a study from Hong Kong (12) and with diabetic nephropathy in a study from the UK (13). While subsequent attempts to replicate these initial findings yielded conflicting results, a recent meta-analysis indicates that this polymorphism may indeed influence the risk of diabetic retinopathy, with a summary odd ratio of about 2.0 and a p value of 2×10−4 (14). Also, a novel prospective study of 1,074 type 2 diabetic patients from Hong-Kong has confirmed the association between this locus and incident kidney disease (CKD3) as well as cardiovascular events (15). Altogether, these findings reaffirm the prominent role played by the polyol pathway in mediating the deleterious effect of hyperglycemia and call for a revamping of research on using aldose reductase inhibitors to prevent diabetic complications.
Candidate genes modulating tissue-specific alterations
The VEGF gene stands out from all the other candidates that have been investigated for contributions to diabetic retinopathy. VEGF is a growth factor that specifically acts on endothelial cells, increasing vascular permeability and inducing angiogenesis (16). High VEGF levels can be found in the vitreous of individual with proliferative diabetic retinopathy and intraocular injection of anti-VEGF drugs slows retinal vessels proliferation (17). Several polymorphisms in the promoter of the gene coding for VEGF (VEGFA) were initially investigated in small cross-sectional studies for association with retinopathy with conflicting results. However, convincing findings have recently come from a study of the DCCT/EDIC cohort, which differed from previous efforts for its prospective nature, the relatively large sample size, and the use of a large number of tag SNPs capturing most of the common variation at the VEGF locus (18). While individual SNPs showed fairly modest associations, a global multi-SNP test yielded a p value of 6 × of 10−5 for association with time to severe, non-proliferative retinopathy. The association was confirmed by family studies demonstrating increased risk allele transmission to severe retinopathy cases. Thus, in addition to be the principal mediator of new vessel proliferation, VEGF may also be involved in the vascular alterations that precede the proliferative stage of diabetic retinopathy.
The genetics of diabetic nephropathy has also been a very active area of research. Several review papers have been published providing a detailed description of the many candidate genes that have been implicated (see for instance ref. (19). One gene that is worth mentioning here is ELMO1 (engulfment and cell motility 1). This molecule, which is induced by high glucose, promotes increased expression of transcription growth factor β, collagen type 1, fibronectin, and integrin-linked kinase expression (20). Genetic variants at the ELMO1 locus have been found to be associated with diabetic nephropathy in Japanese, African-American, and, very recently, in Caucasians from the GoKinD study (21–23). Interestingly, the variants associated with kidney disease are different in the three races, suggesting the presence of allelic heterogeneity probably resulting from the diverse ancestral genetic backgrounds of the different racial groups. If these findings are confirmed and supported by further functional studies, ELMO1 could be an attractive target for the development of new reno-protective drugs for diabetic patients.
Tong et al. identified a SNP in the promoter of the gene coding for the potent erythropoietic and angiogenic factor EPO that was associated with a combined proliferative retinopathy (PDR)/end-stage renal disease (ESRD) phenotype in multiple datasets (24). The risk allele was also associated with increased EPO protein levels in the vitreous and was shown to enhance gene expression in in vitro studies (24). Such functional data lend support to the genetic findings. However, the design of the study, based on the examination of a combined eye/kidney phenotype, make these results difficult to interpret. Most of the subjects who develop nephropathy have retinopathy but only a fraction of the subjects who develop retinopathy have significant nephropathy. In the absence of data about the association between EPO and retinopathy not accompanied by nephropathy, one cannot determine whether the association is with retinopathy, nephropathy, or both complications.
The most interesting results concerning candidate genes for cardiovascular complications have come from studies of the adiponectin axis. Adiponectin is a cytokine exclusively produced by adipocytes that has insulin-sensitizing effects (25). Adiponectin also has direct anti-atherogenic actions by inhibiting monocyte adhesion to the endothelium, smooth muscle cell proliferation, and foam cell formation in the arterial wall (26). In a meta-analysis of four different studies, an intronic SNP in the adiponectin gene (rs1501299) was significantly associated with a two-fold increase in the risk of coronary artery disease (CAD) among diabetic subjects (27). Such association appears to be mediated by an effect of this SNP, or other variants in linkage disequilibrium with it, on adiponectin levels. Variability in the receptors mediating adiponectin action also seems to play a role. In studies from Boston and Italy, three SNPs tagging the 3' half of ADIPOR1 - one of the adiponectin receptors - were found to be associated with CAD among individuals with type 2 diabetes, with allelic odds ratios in the 1.3–1.4 range. This effect appears to be related to lower ADIPOR1 mRNA levels in carriers of the risk genotypes, possibly blunting the antiatherogenic effects of adiponectin on the vascular wall (28). An association with CAD in type 2 diabetes has been also described for SNPs in another adiponectin receptor (CDH13), but these findings have not been confirmed in other populations (29). These results suggest that interventions aimed at enhancing adiponectin actions may be worth pursuing.
Genome-wide studies
Candidate genes studies are useful, but by focusing on genes already implicated in diabetic complications, they are inherently geared towards confirming disease pathways rather than discovering new ones. Initial attempts to extend the study to the entire genome and overcome these constraints were based on linkage studies in families. This approach, however, did not have sufficient power to identify genetic effects of the magnitude expected for multifactorial disorders such as diabetic complications. Also, linkage studies can only identify the rough location of a disease gene and require extensive positional cloning efforts to pinpoint the causal variants responsible for linkage.
Two recent developments have allowed the genetic field to move beyond linkage and conduct sensitive genome-wide searches for variants contributing to complex traits. One of these developments is the genome-wide characterization of linkage disequilibrium – the phenomenon by which adjacent polymorphisms are correlated with each other because of their cosegregation from one generation to the other. Using these data, publicly available from the HapMap database, a set of single nucleotide polymorphisms (SNPs) can be selected that most efficiently capture (or “tag”) the 10 million or so common polymorphisms in the human genome (30;31). The other advancement is the development of technologies based on microarrays through which several hundred thousand tag SNPs can be typed in a single assay (32). Combined together, these two elements have made genome wide association (GWA) studies of multifactorial disorders possible.
The GWA approach is not without problems. First, in its current implementation, it is limited to the analysis of common genetic variants. Second, it requires very large sample sizes because of the stringent significant thresholds that must be used to account for the extremely large number of tests that are conducted. This requirement can be addressed by conducting meta-analyses of GWAS results from different studies, but this increases the risk of genetic and phenotypic heterogeneity, potentially “diluting” association signals. Despite these limitations, however, the GWAS approach offers unprecedented opportunities for and unbiased screen of the entire genome for genes affecting susceptibility to diabetic complications. This approach has been thus far pursued for diabetic nephropathy. Also, several GWAS for coronary artery disease have been conducted in the general population, the findings of which are relevant to the cardiovascular complications of diabetes.
GWAS of diabetic nephropathy
The GWAS for kidney complications stemmed from the Genetics of Kidneys in Diabetes (GoKinD) initiative, under which a large collection of type 1 diabetic individuals with and without diabetic nephropathy was assembled. The scan, including ~360,000 single nucleotide polymorphisms (SNPs) from the Affymetrix 500K array, was conducted in 820 nephropathy cases, one third with proteinuria and two thirds with end-stage renal disease (ESRD), and 885 controls without kidney complications despite a duration of diabetes of 15 years or longer (33). Replication of the strongest association signals was sought in the DCCT/EDIC cohort.
None of the SNPs in the array achieved significance according to an alpha error level that took into account the number of SNPs that were analyzed (α=0.05/360,000=1.4 × 10−7). However, 11 SNPs placed in four distinct chromosomal regions had p values < 1 × 10−5. The strongest association (OR=1.45, p=5 × 10−7) was found with a SNP placed on chromosome 9q near the 5' end of FRMD3 coding for the exrin, radixin, moesin (FERM) domain-containing protein 3 (Table 1). The other three loci displaying association were found near CHN2 (β-chimerin) and CPVL (serin carboxypeptidase vitellogenic-like) on chromosome 7p (OR=1.39, p=5 × 10−6), in an intron of CARS (cysteynil-tRNA synthetase) on 11p (OR=1.36, p=3 × 10−6), and in an intergenic region at 13q (OR=1.41, p=2 × 10−6) (Table 1). These associations were not affected by duration of diabetes or end-stage renal disease, excluding the occurrence of survival bias. The association at 9q and 11p were replicated in the EDIC/DCCT cohort with nominal significance (HR=1.33, p=0.02) and 11p (HR=1.32, p=0.01), whereas the SNP at 13q was found to be associated with nephropathy among type 2 diabetic subjects in a recent meta-analysis of four studies from Japan (34) (Table 1).
Table 1.
Loci associated with diabetic nephropathy in a GWAS of the GoKind collection and their association with diabetic nephropathy in other studies.
| GoKind GWAS (Pezzolesi et al.)† |
DCCT/EDIC (Pezzolesi et al.)‡ |
JAPAN (Maeda et al.)∥ |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Representative SNP | Chr | Mb | Nearest gene | Risk allele | OR | p value | HR | p value | OR | p value |
| rs39075 | 7p | 29.2 | CPVL/CHN2 | G | 1.43 | 6.5 × 10−7 | 0.85 | NS | 1.07 | 0.19 |
| rs1888747 | 9q | 85.3 | FRMD3 | G | 1.45 | 6.3 × 10−7 | 1.33§ | 0.02 | 0.96 | 0.51 |
| rs451041 | 11p | 3.0 | CARS | A | 1.36 | 3.1 × 10−6 | 1.32 | 0.01 | 1.06 | 0.30 |
| rs1411766 | 13q | 109.1 | IRS2/MYO16 | A | 1.41 | 1.8 × 10−6 | 1.17 | 0.11 | 1.26 | 0.004 |
820 cases/885 controls with T1D,
132 events/1304 subjects with T1D,
1535 cases/1915 controls with T2D from four different studies.
Data refer to rs1888746, which is in complete LD (r2=1.00) with rs1888747.
In considering these findings, several factors should be taken into account. First, the GoKinD collection is heavily weighted with cases of ESRD. Thus, there was little power to detect genetic effects that specifically act on the transition from normoalbuminuria to proteinuria rather than on that from proteinuria to ESRD. Second, the individuals with ESRD in the collection were prevalent cases who had survived for many years on hemodialysis or with a kidney transplant. Under these circumstances, a disease allele that not only increase susceptibility to nephropathy but also increases mortality could have gone undetected. Third, because of the stringent significance threshold, one cannot exclude the presence of other genetic effects that did not emerge as significant due to the limited power. Fourth, although replication of some of these association signals in the EDIC/DCCT cohort is encouraging, the significance of those findings does not completely exclude the possibility of false-positives.
If we accept these results as genuine, they offer some potentially interesting leads for the identification of novel pathways involved in the etiology of diabetic nephropathy. The FRMD3 gene on 7p encodes a structural protein of the 4.1O family, which is involved in the maintenance of cell shape and form in a variety of cell types, including the mouse nephron (35). CARS on 11p has been implicated in the pathogenesis of cystinosis – a recessive disorder caused by accumulation of free cystine in renal tubules resulting to a deficit in lysosomal cystine transport (36). CPVL on 7p is a carboxypeptidase that is highly expressed in the kidney and specifically in the proximal tubule. Other carboxypeptidases, such as ACE and bradikinin, have been implicated in the etiology of diabetic nephropathy because of their regulatory role in kidney hemodynamics (37). Thus, these initial GWAS findings could open new research avenues to expand our knowledge of the pathogenesis of kidney damage in diabetes.
GWAS of coronary artery disease
There have been no published reports of GWAS for coronary artery disease that were specifically conducted in the diabetic population. However, several GWAS for coronary artery disease performed in the general population have generated results that may also be relevant to diabetic subjects. All of these studies used a two-stage strategy consisting of a GWA screen in an initial cohort of unrelated cases and controls followed by replication of the most significant findings in additional case-control sets (38–44) A total of 12 loci have been identified by these studies as being associated with CAD or MI in the general population with genome-wide significance (Table 2). Most of these variants are associated with a relative small increase in cardiovascular risk, with allelic odds ratios that do not exceed 1.30 and are mostly below 1.20. Also, as observed with other complex disorders, most of the CAD-associated SNPs variants are placed in non-coding regions, suggesting an effect on regulatory elements and gene expression rather than on the amino acid sequence. Another interesting feature is that most of the genes that appear to be involved were not previously implicated in the etiology of atherosclerosis. Notable exceptions are LDLR, which codes for the low density lipoprotein (LDL) receptor, PCSK9, which codes for a serine protease that is mutated in Mendelian forms of hypercholesterolemia (45), and the SLC22A3-LPAL2-LPA cluster, which includes the gene for the atherogenic lipoprotein Lp(a).
Table 2.
Genetic associations with CAD/MI identified by GWA studies in the general population.
| Representative SNP | Closest Gene(s) | Chromosome | Position (Mb) | Risk allele | OR | 95% CI | Reference |
|---|---|---|---|---|---|---|---|
| rs11206510 | PCSK9 | 1 | 55.3 | T | 1.15 | 1,10–1.21 | (40) |
| rs646776 | CELSR2-PSRC1-SORT1 | 1 | 109.6 | T | 1.19 | 1.13–1.26 | (40;42;44) |
| rs17465637 | MIA3 | 1 | 220.8 | C | 1.14 | 1.10–1.19 | (40;42;44) |
| rs6725887 | WDR12 | 2 | 203.5 | C | 1.17 | 1.11–1.23 | (40) |
| rs9818870 | MRAS | 3 | 139.6 | T | 1.15 | 1.11–1.19 | (38) |
| rs12526453 | PHACTR1 | 6 | 13.0 | C | 1.12 | 1.08–1.17 | (40) |
| Haplotype | SLC22A3- LPAL2- LPA | 6 | 160.7 | CCTC | 1.82 | 1.57–2.12 | (43) |
| Haplotype | SLC22A3- LPAL2- LPA | 6 | 160.7 | CTTG | 1.20 | 1.13–1.28 | (43) |
| rs4977574 | CDKN2A/CKN2B | 9 | 22.1 | G | 1.29 | 1.25–1.34 | (39-42;44) |
| rs1746048 | CXCL12 | 10 | 44.1 | C | 1.17 | 1.11–1.24 | (40;42;44) |
| rs2259816 | HNF1A-C12orf43 | 12 | 119.9 | T | 1.08 | 1.05–1.11 | (38) |
| rs1122608 | LDLR | 19 | 11.0 | G | 1.15 | 1.10–1.20 | (40) |
| rs9982601 | MRPS6-SLC5A3-KCNE2 | 21 | 34.5 | T | 1.20 | 1.14–1.27 | (40) |
The strongest and most extensively replicated of these associations is placed on chromosome 9p21 (39;41). While this locus was discovered in the general population, its predisposing effect on coronary artery disease can also be observed in the presence of diabetes. In fact, data from the Joslin Heart Study (JHS) suggest that this locus may have a larger effect on CAD risk among individuals with type 2 diabetes than in the general population (46). This study compared cases with type 2 diabetes and angiographic evidence of CAD with controls having a negative cardiovascular history of CAD and a normal exercise treadmill test despite 5 years or more of type 2 diabetes. The odds ratios were 1.45 (0.94–2.22) for heterozygotes and 2.37 (1.52–3.70) for homozygotes, consistent with an additive mode of inheritance. These effect estimates ere larger than those reported in the general population, suggesting the possibility of an interaction with some aspect of diabetes such as hyperglycemia. Consistent with this hypothesis, the odds of CAD were almost four times higher in risk allele homozygotes who were also in poor glycemic control as compared to subjects who were not homozygous for the risk allele and were in good glycemic control (OR=3.77, 2.13–6.7). By contrast, subjects with only one risk factor (either the homozygote genotype or poor glycemic control) had only a little increase in cardiovascular risk. Such interaction between homozygous genotype and glycemic control was significant and was replicated with respect to cardiovascular mortality in a separate prospective study of 475 type 2 diabetic subjects from the Joslin Clinic. The 10-year cardiovascular mortality was 36% among risk allele homozygotes with a history of poor glycemic control as compared to only 15–20% of all other subjects (46).
The synergism between 9p21 locus and poor glycemic control may be instrumental in clinical practice to improve estimation of cardiovascular risk among diabetic subjects. Studies of other complex disorders have shown that common genetic variants with small effects do not significantly improve predictive algorithms (47). However, the magnitude of the joint effect of poor glycemic control and 9p21 locus is such to suggest some clinical benefit, even if the cost-effectiveness of using this combined predictors remains be determined. The implications for our understanding of the biology of atherogenesis in diabetes are not as straightforward since the CAD-associated SNPs reside in a 60 Kb linkage disequilibrium block. (41). This block is devoid of protein-coding genes but includes the most 3' exons of a non-coding gene known as ANRIL (a.k.a. CDKN2BAS) (48;49). The ANRIL gene is transcribed as two alternatively spliced transcripts: a long one, including the 3' exons placed in the CAD-associated block, and a short one missing those exons. Jarinova et al. recently reported that the risk allele is associated with a decrease in the long-to-short variant ratio (49). While the meaning of this finding is still unclear, the altered balance of ANRIL splice variants might affect the expression of other genes through mechanisms such as RNA interference or chromatin remodeling. Prime candidates as ANRIL targets are the adjacent cell-cycle genes CDKN2A and CDKN2B involved in the control cell proliferation, cell aging, and apoptosis (50). Indeed, deletion of the 9p21 CAD-associated region determines a marked decrease in the expression of CDKN2A and CDKN2B along with a doubling of the proliferative capacity of cultured smooth muscle cells (51). Interestingly, hyperglycemia and other components of the diabetic milieu are also known to induce vascular smooth muscle cell proliferation (52). Thus, the two atherogenic factors – 9p21 variant and excess glucose – may enhance each other by “double hitting” the same target. Altogether, these findings point to the pathways regulating cell cycle as potential targets for the development of new drugs to prevent atherosclerosis. They also suggest that these drugs could be especially effective in individuals with poorly controlled diabetes.
Preliminary data from the Joslin, the Nurses Health Study, and the Health Professional Follow-up Study suggest that several other of the 12 CAD genes identified in the general population may also influence the risk of CAD in the diabetic population (53). The strongest of these non-9p21 effects is observed on chromosome 6p24, in an intron of the PHACTR1 (phosphatase and actin regulator 1) gene (54).
Conclusions
The past two decades have seen a rapid expansion of the research on the genetic determinants of diabetic complications. These efforts, although often underpowered and haphazard, have led to the identification of a few credible associations pointing to cellular pathways playing a prominent role in linking the diabetic milieu to tissue damage. Are these findings all we can expect from the genetics of diabetic complications? The answer is no. First, the GWAS approach has not yet been exploited to its full extent. The only GWAS study that has been thus far published on a diabetic complication (i.e., diabetic nephropathy) was not sufficiently powered to detect genetic effects of the small magnitude observed with other complex disorders. For cardiovascular complications, we have thus far relied on the GWAS findings in the general population. However, the interaction between 9p21 locus and poor glycemic control suggest that other genetic effects might be found by conducting a GWAS focused on diabetic subjects. No GWAS data are available to date for diabetic retinopathy. Second, all the studies conducted thus far, including the GWAS, have been based on the assumption that susceptibility to diabetic complications results from common polymorphisms (i.e., with minor allele frequencies >5%). While there are theoretical grounds for such an assumption (55), the experience with other complex disorders suggest that less frequent sequence differences might play a role as well (56). Thus, as a complete catalog of these variants is compiled by initiatives such as the 1000 Genomes Project (www.1000genomes.org), it is likely that additional genetic effects will be identified. The same is true for structural variants, such as copy-number variants, insertions, deletions, and duplications, which at this point remain almost completely unexplored (57). With this additional information on hand, we will be in the position of reconstructing the genetic architecture of diabetic complications. This will allow us to tell which genetic factors are shared and which ones are specific to each complication and to build prediction model to identify high-risk individuals. Perhaps more importantly, this knowledge will provide us with unprecedented insights on the pathogenesis of diabetic complications, with crucial implications for the development of new drugs specifically aimed at contrasting the deleterious effects of diabetes.
ACKNOWLEDGMENTS
Results of the author's work described in this article were supported by NIH grants HL73168, HL71981, and DK36836
References
- 1.Krolewski AS, Warram JH. Epidemiology of late complications of diabetes: A basis for the development and evaluation of preventive program. In: Kahn CR, Weir GC, King GL, Jacobson AM, Moses AC, Smith RJ, editors. Joslin's Diabetes Mellitus. Lippincott, Williams & Wilkins; New York: 2005. [Google Scholar]
- 2.The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. The Diabetes Control and Complications Trial Research Group. N Engl J Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 3.Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet. 1998;352:837–853. [PubMed] [Google Scholar]
- 4.Hoerger TJ, Segel JE, Gregg EW, Saaddine JB. Is glycemic control improving in U.S. adults? Diabetes Care. 2008;31:81–86. doi: 10.2337/dc07-1572. [DOI] [PubMed] [Google Scholar]
- 5.Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia. 1996;39:940–945. doi: 10.1007/BF00403913. [DOI] [PubMed] [Google Scholar]
- 6.Fogarty DG, Rich SS, Hanna L, et al. Urinary albumin excretion in families with type 2 diabetes is heritable and genetically correlated to blood pressure. Kidney Int. 2000;57:250–257. doi: 10.1046/j.1523-1755.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- 7.Lange LA, Bowden DW, Langefeld CD, et al. Heritability of carotid artery intima-medial thickness in type 2 diabetes. Stroke. 2002;33:1876–1881. doi: 10.1161/01.str.0000019909.71547.aa. [DOI] [PubMed] [Google Scholar]
- 8.Wagenknecht LE, Bowden DW, Carr JJ, et al. Familial aggregation of coronary artery calcium in families with type 2 diabetes. Diabetes. 2001;50:861–866. doi: 10.2337/diabetes.50.4.861. [DOI] [PubMed] [Google Scholar]
- 9.Arar NH, Freedman BI, Adler SG, et al. Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Invest Ophthalmol Vis Sci. 2008;49:3839–3845. doi: 10.1167/iovs.07-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hietala K, Forsblom C, Summanen P, Groop PH. Heritability of proliferative diabetic retinopathy. Diabetes. 2008;57:2176–2180. doi: 10.2337/db07-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene DA, Lattimer SA, Sima AA. Sorbitol, phosphoinositides, and sodium-potassium-ATPase in the pathogenesis of diabetic complications. N Engl J Med. 1987;316:599–606. doi: 10.1056/NEJM198703053161007. [DOI] [PubMed] [Google Scholar]
- 12.Ko BC, Lam KS, Wat NM, Chung SS. An (A-C)n dinucleotide repeat polymorphic marker at the 5' end of the aldose reductase gene is associated with early-onset diabetic retinopathy in NIDDM patients. Diabetes. 1995;44:727–732. doi: 10.2337/diabetes.44.7.727. [DOI] [PubMed] [Google Scholar]
- 13.Heesom AE, Hibberd ML, Millward A, Demaine AG. Polymorphism in the 5'-end of the aldose reductase gene is strongly associated with the development of diabetic nephropathy in type I diabetes. Diabetes. 1997;46:287–291. doi: 10.2337/diab.46.2.287. [DOI] [PubMed] [Google Scholar]
- 14.Abhary S, Hewitt AW, Burdon KP, Craig JE. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009;58:2137–2147. doi: 10.2337/db09-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First attempt to summarize the findings on the role of aldose reductase gene in shaping susceptibility to diabetic diabetic retinopathy.
- 15.So WY, Wang Y, Ng MC, et al. Aldose reductase genotypes and cardiorenal complications: an 8-year prospective analysis of 1,074 type 2 diabetic patients. Diabetes Care. 2008;31:2148–2153. doi: 10.2337/dc08-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung DW, Cachianes G, Kuang WJ, et al. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 17.Jardeleza MS, Miller JW. Review of anti-VEGF therapy in proliferative diabetic retinopathy. Semin Ophthalmol. 2009;24:87–92. doi: 10.1080/08820530902800330. [DOI] [PubMed] [Google Scholar]
- 18.Al Kateb H, Mirea L, Xie X, et al. Multiple variants in vascular endothelial growth factor (VEGFA) are risk factors for time to severe retinopathy in type 1 diabetes: the DCCT/EDIC genetics study. Diabetes. 2007;56:2161–2168. doi: 10.2337/db07-0376. [DOI] [PubMed] [Google Scholar]; • Systematic study of genetic variability in VEGF as a determinant of the risk of diabetic retinopathy.
- 19.Ng DP, Krolewski AS. Molecular genetic approaches for studying the etiology of diabetic nephropathy. Curr Mol Med. 2005;5:509–525. doi: 10.2174/1566524054553504. [DOI] [PubMed] [Google Scholar]
- 20.Shimazaki A, Tanaka Y, Shinosaki T, et al. ELMO1 increases expression of extracellular matrix proteins and inhibits cell adhesion to ECMs. Kidney Int. 2006;70:1769–1776. doi: 10.1038/sj.ki.5001939. [DOI] [PubMed] [Google Scholar]
- 21.Shimazaki A, Kawamura Y, Kanazawa A, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 22.Pezzolesi MG, Katavetin P, Kure M, et al. Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Extension of the findings of association between variants at the ELMO1 genes and diabetic nephropathy to Europen Whites. The paper shows how information about candidate genes can be extracted from GWA studies.
- 23.Leak TS, Perlegas PS, Smith SG, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet. 2009;73:152–159. doi: 10.1111/j.1469-1809.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study extends the finding od association between variants at the ELMO1 genes and diabetic nephropathy to African-Americans.
- 24.Tong Z, Yang Z, Patel S, et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A. 2008;105:6998–7003. doi: 10.1073/pnas.0800454105. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study shows that a variant affecting expression of the angiogenic factor EPO may underlie the simulatneous occurrence of diabetic proliferative retinopathy and end-stage renal disease.
- 25.Scherer PE, Williams S, Fogliano M, et al. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 26.Kadowaki T, Yamauchi T. Adiponectin and adiponectin receptors. Endocr Rev. 2005;26:439–451. doi: 10.1210/er.2005-0005. [DOI] [PubMed] [Google Scholar]
- 27.Qi L, Doria A, Manson JE, et al. Adiponectin genetic variability, plasma adiponectin, and cardiovascular risk in patients with type 2 diabetes. Diabetes. 2006;55:1512–1516. doi: 10.2337/db05-1520. [DOI] [PubMed] [Google Scholar]
- 28.Soccio T, Zhang YY, Bacci S, et al. Common haplotypes at the adiponectin receptor 1 (ADIPOR1) locus are associated with increased risk of coronary artery disease in type 2 diabetes. Diabetes. 2006;55:2763–2770. doi: 10.2337/db06-0613. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YY, Boonyasrisawat W, Xu R, et al. Polymorphisms at the adiponectin aeceptor T-Cadherin (CDH13) locus are associated with increased cardiovascular risk in type 2 diabetes. Diabetes. 2007;56:A303. [Google Scholar]
- 30.Daly MJ, Rioux JD, Schaffner SF, et al. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 31.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sapolsky RJ, Hsie L, Berno A, et al. High-throughput polymorphism screening and genotyping with high-density oligonucleotide arrays. Genet Anal. 1999;14:187–192. doi: 10.1016/s1050-3862(98)00026-6. [DOI] [PubMed] [Google Scholar]
- 33.Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Only GWAS that has been performed to date for diabetic nephropathy.
- 34.Maeda S, Araki SI, Babazono T, et al. Diabetes. 2010. Replication study for the association between 4 loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First attempt to replicate GWAS findings for diabetic nephropathy in Asians. It provides some evidence of replication for two of the association signals.
- 35.Hoover KB, Bryant PJ. The genetics of the protein 4.1 family: organizers of the membrane and cytoskeleton. Curr Opin Cell Biol. 2000;12:229–234. doi: 10.1016/s0955-0674(99)00080-0. [DOI] [PubMed] [Google Scholar]
- 36.Town M, Jean G, Cherqui S, et al. A novel gene encoding an integral membrane protein is mutated in nephropathic cystinosis. Nat Genet. 1998;18:319–324. doi: 10.1038/ng0498-319. [DOI] [PubMed] [Google Scholar]
- 37.Riad A, Zhuo JL, Schultheiss HP, Tschope C. The role of the renal kallikrein-kinin system in diabetic nephropathy. Curr Opin Nephrol Hypertens. 2007;16:22–26. doi: 10.1097/MNH.0b013e328011a20c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]; • One of the most recent GWAS for coronary artery disease in the general population.
- 39.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]; •• One of the first two reports of the association between chromosome 9p21 locus and coronary artery disease.
- 40.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Largest GWAS effort to date to identify common genetic variants predisposing to early-onset myocardial infarction in the general population.
- 41.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• One of the first two reports of the association between chromosome 9p21 and coronary artery disease.
- 42.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tregouet DA, Konig IR, Erdmann J, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]; • One of the most recent GWAS for coronary artery disease in the general population.
- 44.Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Landmark report describing the coordinated effort to map genes for several common disorders through the GWAS approach. Useful to understand how GWAS are designed and analyzed.
- 45.Abifadel M, Varret M, Rabes JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 46.Doria A, Wojcik J, Xu R, et al. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. JAMA. 2008;300:2389–2397. doi: 10.1001/jama.2008.649. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper shows that the 9p21 locus has an especially powerful effect on cardiovascular risk among diabetic subjects because of an interaction with the diabetic milieu.
- 47.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 49.Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]; • This paper shows that the 9p21 CVD-associated haplotype is associated with different transcripts of the non-coding gene ANRIL.
- 50.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 51.Visel A, Zhu Y, May D, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First experimental demonstration that the 9p21 region where the variants predisposing to coronary artery disease are located is involved in the regulation of the CDKN2A/2B genes and controls cell proliferation.
- 52.Natarajan R, Gonzales N, Xu L, Nadler JL. Vascular smooth muscle cells exhibit increased growth in response to elevated glucose. Biochem Biophys Res Commun. 1992;187:552–560. doi: 10.1016/s0006-291x(05)81529-3. [DOI] [PubMed] [Google Scholar]
- 53.Qi L, Parast L, Powers C, et al. A genetic risk score to improve the prediction of coronary artery disease in type 2 diabetes. Diabetes. 2010;59:A217. [Google Scholar]
- 54.Novelli V, Powers C, Gervino EV, et al. Common genetic variants at the PHACTR1 locus are major determinants of coronary artery disease among individuals with type 2 diabetes. Diabetes. 2010;59:A219. [Google Scholar]
- 55.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 56.Fearnhead NS, Wilding JL, Winney B, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci U S A. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]