Abstract
DNA methylation may restrict the activity of gene transfer vectors due to inadvertent silencing. In P19 embryonic carcinoma cells in vitro, we found that transgene expression regulated by the SFFV LTR and EF1α promoter declined rapidly within 16 days, but for A2UCOE derived from the human HNRPA2B1-CBX3 housekeeping gene locus, remained completely stable. Silencing correlated with extensive epigenetic methylation of CpG sites, whereas the A2UCOE was almost completely resistant. Linking of the A2UCOE upstream of the SFFV LTR protected this element from both DNA methylation and silencing. Analysis of engrafted hematopoietic cells in vivo transduced with the same vectors revealed a similar pattern. The A2UCOE displayed little or no methylation in either primary or secondary graft recipients, and gene expression profiles were highly conserved between the two groups. These studies provide convincing evidence that DNA methylation plays a direct role in regulating self-inactivating (SIN) lentiviral transgene expression, and that the stability of expression from the A2UCOE is, at least in part, due to methylation resistance. The A2UCOE therefore has considerable utility for gene therapy applications where reliable and sustained gene expression is desirable.
Introduction
Research in recent years has demonstrated that retroviral vector–mediated gene transfer into hematopoietic stem cells (HSCs) is a useful and promising tool for the treatment of inherited hematological disorders.1,2,3,4,5,6,7,8 Despite the recent successes and developments in this area, there remain two major potential problems that could compromise further clinical application of integrating vector-based gene transfer. First, there is a safety concern due to enhancer-mediated insertional mutagenesis, which has been observed in clinical trials using gammaretroviral vectors.6,9,10 This risk may be partially reduced, but not abolished, by the use of self-inactivating (SIN) configurations in which alternative regulatory elements are incorporated.11,12 Second, evidence has accumulated that transgene expression in both gammaretroviral and lentiviral vectors (LVs) may be subject to epigenetic (DNA methylation, histone modification) silencing both in vitro and in vivo.13,14,15,16,17,18,19,20,21 Efforts have been made to tackle this problem by flanking transgenes with chromatin insulators, including the chicken β-globin locus control region HS4 element (cHS4). However, introduction of this element has, at best, conferred only partial protection in a cell-dependent manner.22,23 In addition, the inclusion of the cHS4 insulator within LVs can result in a significant reduction in production efficiency, thereby further compromising its utility for gene therapy applications.24,25
We have previously assessed the use of a novel enhancer-less ubiquitous chromatin opening element (UCOE), derived from the human HNRPA2B1-CBX3 locus (A2UCOE),26,27 to regulate transgene expression within a SIN-LV context.27 We have shown that, unlike viral promoters, the A2UCOE gives rise to populations of cells that express transgenes at highly reproducible and stable levels in a variety of different cell lines and more importantly in vivo following ex vivo gene transfer to mouse bone marrow HSCs. We further tested the efficacy of the A2UCOE within a therapeutic context by employing it to control expression of the common cytokine receptor gamma chain gene (IL2RG), and demonstrated complete rescue of the SCID-X1 phenotype in a mouse model of this disease.28
Our previous studies suggested that efficient and stable expression of transgenes from the A2UCOE may be due to its resistance to DNA methylation–mediated silencing. Here, we have investigated the expression and DNA methylation status of the A2UCOE, EF1α, and SFFV regulatory elements within a SIN-LV vector system by functional analysis in mouse embryonic carcinoma P19 cells in vitro, and in vivo following ex vivo gene transfer to mouse HSCs. Our data show for the first time that, unlike the SFFV and EF1α elements, the stability of transgene expression from the A2UCOE within SIN-LVs is associated with a near total resistance to DNA methylation, thereby reinforcing its potential as an excellent regulatory element for gene therapy applications.
Results
A2UCOE confers stable gene expression in P19 cells
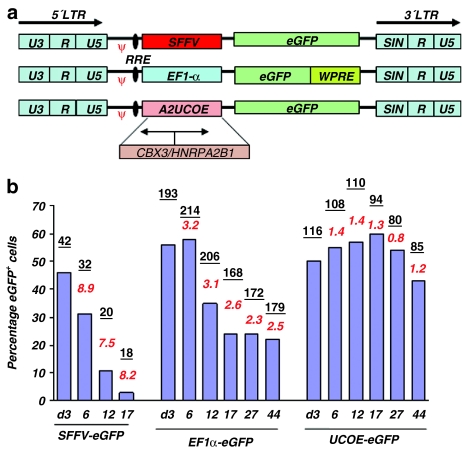
P19 cells are pluripotent embryonic carcinoma stem cells that are capable of differentiating into an array of cell types including neuronal, glial, cardiac, and skeletal muscle.29,30 A strong tendency toward repression of transgene expression in embryonic carcinoma stem cells has been described.31,32 This makes P19 cells a sensitive system in which to challenge and test the stability of expression from regulatory elements of interest. We therefore chose this tissue culture cell model system to compare the function of the commonly used viral SFFV LTR, the short variant of the human housekeeping gene EF1α promoter, and the A2UCOE (Figure 1a). SIN-LVs incorporating these elements were used to transduce P19 cells at a multiplicity of infection of 5 for the EF1α- and A2UCOE-containing vectors, and at a multiplicity of infection of 10 for the SFFV vector to give a similar proportion of eGFP+ cells at the start of the experiment when assessed by flow cytometry (Figure 1b; Supplementary Figure S1). Transgene eGFP expression was monitored every 3–5 days after transduction, for up to 44 days. Although a similar initial transduction efficiency was obtained with all vectors (46–56% eGFP+ cells), expression from the SFFV LTR rapidly declined from 46 to 2% positive cells within 17 days. This was associated with a low level of mean fluorescent intensity from the outset after transduction, which further declined over time. A relatively rapid decline in expression was also observed with the EF1α vector, with eGFP+ cell numbers falling from an initial level of 56 to 22% by day 17 after transduction, but which remained relatively stable at this level thereafter. In contrast, the proportion of eGFP+ cells from the A2UCOE vector remained completely stable over the 44-day period of culture. Parallel experiments with P19 cells transduced at varying virus concentration showed a similar eGFP expression profile over time for all three vectors testifying to the reproducibility of these observations regardless of LV load and copy number (Supplementary Figure S2).
Figure 1.
A2UCOE gives rise to stable eGFP expression in P19 cells compared to SFFV and EF1α regulatory elements. (a) Schematic of the lentiviral transfer vectors used in the study. The transcription direction of CBX3-HNRPA2B1 within A2UCOE is indicated. LTR, long-terminal repeat; RRE, rev-response element; ψ, packaging signal. (b) P19 cells were transduced with vectors shown in a, at multiplicity of infection (MOI) of 5 for EF1α and A2UCOE, and MOI 10 for SFFV. eGFP expression was assessed by flow cytometry at different time points after transduction up to 44 days, as indicated. The underlined numbers shown above each bar represent mean fluorescence intensity (MFI). Italicized numbers denote vector copy number per cell determined by Q-PCR. Note: Unlike SFFV and EF1α, eGFP expression driven from A2UCOE is stable throughout the culture period. This is achieved with a low vector copy number of one per cell compared to the high vector copy numbers present for EF1α and SFFV.
We next ascertained whether reduced eGFP expression was due to the loss of vector-positive cells from the culture. Quantitative PCR was performed on genomic DNA to determine average vector copy number (VCN) at each time point of the period of culture after transduction. Our data show that VCN per cell remained at a similar level for all vectors at all time points (Figure 1b, italicized numbers), suggesting that loss of eGFP expression in cells transduced with the SFFV and EF1α LVs was not due to loss of vector copies but from promoter silencing. This is despite the fact that cells transduced with both the SFFV and EF1α constructs contained multiple copies of the LVs with an average VCN ranging from 7.5 to 8.9 and 2.5 to 3.2, respectively, during the course of the experiment (Figure 1b, italicized numbers). This suggests that multiple copies of the EF1α and especially the SFFV LVs within the same cell are subject to silencing. However, the same effect was also observed with an average VCN of <1 per cell, indicating that the observed silencing is independent of this parameter (Supplementary Figure S2a–c), and implying that the majority of copies of the A2UCOE LV are actively and stably transcribed regardless of the level of VCN and the variable site of vector integration.
A2UCOE is not subject to DNA methylation in P19 cells
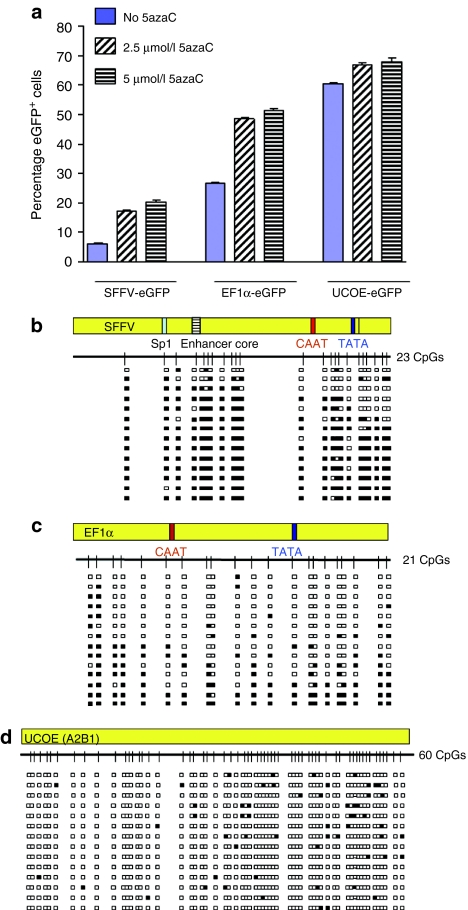
As a first indicator as to whether DNA methylation is associated with the reduction in eGFP expression from the SFFV and EF1α LVs, we treated transduced P19 cells with the DNA methylation inhibitor 5-azacytidine (5azaC).33 Cells transduced with the SFFV SIN-LV and grown in the presence of 5-azacytidine at day 8 after transduction showed a dose-dependent increase of two- to threefold in eGFP+ cell numbers following 5 days of incubation (Figure 2a). A similar effect (over a onefold increase in eGFP+ cells) was observed with cells transduced with the EF1α vector. In contrast, a much smaller change (<10% increase in eGFP+ cells) was observed with A2UCOE LV transduced cells (Figure 2a). Interestingly, we observed no effect on eGFP+ cell numbers from the SFFV and EF1α LVs following 5-azacytidine treatment when this was applied at 20 days after transduction (data not shown). This suggests that other epigenetic effects such as histone deacetylation following initial DNA methylation are also involved in the transgene silencing process, which leads to a nonpermissive chromatin state. Overall, these data suggest that DNA methylation is, at least in part, responsible for the decline in eGFP expression from the SFFV and EF1α promoters in P19 cells.
Figure 2.
A2UCOE is not subject to DNA methylation in P19 cells. (a) Abrogation of SFFV-eGFP and EF1α-eGFP silencing in P19 cells by addition of the DNA methylation inhibitor 5-azacytidine (5azaC). 5azaC was added to cultures 2.5 and 5 µmol/l on day 8 after transduction with eGFP expression assessed 5 days later. The mean and standard deviation of triplicate experiments for each vector are shown. Solid bars: no 5azaC. Hatched bars: 2.5 µmol/l 5azaC. Horizontal cross line bars: 5 µmol/l 5azaC. Note: percentage of eGFP+ cells transduced with the SFFV and EF1α vectors remained at a high level when treated with 5azaC compared to untreated cells; 5azaC treatment had a much smaller effect on eGFP expression from the A2UCOE. The P values of 5azaC treated to untreated cells: SFFV-eGFP 2.5 µmol/l: 1.9992 × 10−5, 5 µmol/l: 7.65488 × 10−6; EF1α-eGFP 2.5 µmol/l: 9.95594 × 10−7, 5 µmol/l: 5.95237 × 10−7; A2UCOE-eGFP 2.5 µmol/l: 0.000299, 5 µmol/l: 0.000143967. (b–d) DNA methylation analysis of SFFV (b), EF1α (c), and A2UCOE (d) proviral elements. Genomic DNA from cells transduced with lentiviruses at day 17 after transduction was isolated and subjected to methylation analysis by bisulfite conversion and sequencing. Methylation status of randomly selected PCR clones generated after conversion is shown. White boxes: unmethylated CpG sites. Black boxes: methylated CpG sites. Sp1: Sp1 transcription factor binding site. Enhancer core: SFFV enhancer core element; CAAT, TATA: CAAT and TATA box elements. Note: only scattered methylated CpG sites are found within the A2UCOE sequence compared to extensive methylation of CpG dinucleotides in EF1α and SFFV.
Based on these observations, we assessed provirus DNA methylation in P19 cells at day 17 after transduction, a point at which a high level of transgene silencing has occurred (Figure 1b). Genomic DNA was treated with sodium bisulfite to convert unmethylated cytosine residues to thymine and then subjected to a nested PCR using primers to specifically amplify full-length SFFV LTR and EF1α promoter fragments, and the HNRPA2B1 region of the A2UCOE. PCR-generated products were cloned and a random number selected for sequencing (Figure 2b–d). When the same CpG methylation pattern was obtained with different clones, it was assumed to be a repeat of PCR product from the same vector integration event and was therefore discounted from further analysis. We found <5% of repeated clones in the overall sequence analysis. This suggests that the PCR-generated library of products for each vector was not saturated and therefore, in the vast majority of cases, represented products from unique integration sites. Figure 2b shows the sequence analysis of cells transduced with the SFFV-eGFP vector. Out of 14 sequences analyzed, 11 (78%) displayed extensive (70–100%) methylation of CpG sites at the SFFV promoter and enhancer region, which is consistent with the substantial loss (95% reduction) of eGFP transgene expression (Figure 1b). Of note, the CpG dinucleotides in the vicinity of the Sp1/enhancer core at the 5′ end of the SFFV LTR region were almost completely methylated compared to sequences at the 3′ end of this element. DNA methylation of the EF1α promoter (Figure 2c) was less prominent compared to that of SFFV. Nevertheless, 6 of 14 (42%) vector copies analyzed exhibited 50–100% methylated CpG residues, which again is consistent with the 46% reduction in eGFP+ cell numbers at this culture time point (Figure 1b). To confirm that apparent DNA methylation of SFFV and EF1a regulatory sequences was not a reflection of the relatively higher VCN compared to A2UCOE, similar analysis was performed on cells with VCN <1(Supplementary Figure S3), which showed a very similar pattern and intensity of DNA methylation comparable with that observed at higher copy numbers (Figure 2b,c). It was also apparent here that most methylation on SFFV occurred at an early stage (day 4) after transduction (Supplementary Figure S3a).
Unlike and in marked contrast to the SFFV and EF1α vectors, the HNRPA2B1 promoter region of the A2UCOE-eGFP SIN-LV displayed only very low level and widely scattered methylation of CpG sites (Figure 2d). In addition, analysis of an 827 base-pair (bp) length of the CBX3 region of this element containing 86 CpG dinucleotides again showed only low numbers of scattered methylated CpG sites (Supplementary Figure S4a). Overall, these data provide clear evidence that stable eGFP transgene expression from the A2UCOE is due, at least in part, to its resistance to DNA methylation, whereas DNA methylation of the SFFV and EF1α promoters is a major contributory factor to their loss of function in P19 cells.
A2UCOE negates DNA methylation and silencing of the SFFV promoter in P19 cells
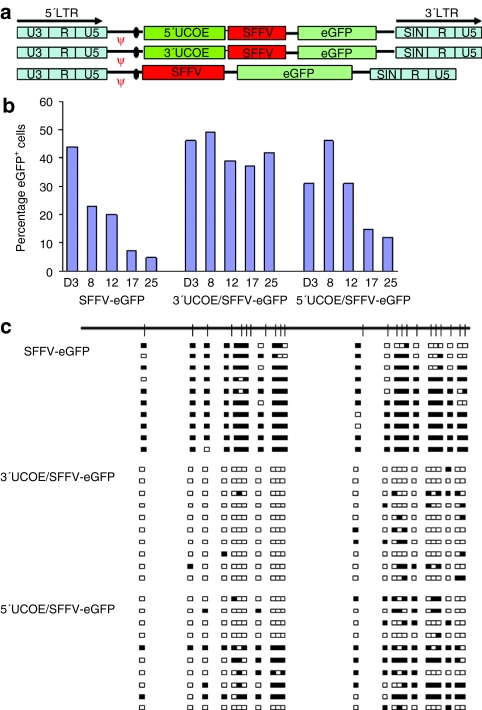
It has previously been shown that silencing of human cytomegalovirus promoter–driven transgenes in stably transfected tissue culture cells can be negated by linking the A2UCOE upstream of this element.27 We therefore assessed whether the A2UCOE is also able to confer stability of expression on the SFFV promoter within a SIN-LV context. A core 1.2 kb A2UCOE subfragment extending from a BsmBI site within the first intron of CBX3 to the exon I TthIII I site of HNRPA2B1 and which therefore still contains the dual divergently transcribed promoters of this element was linked in both orientation upstream of the SFFV promoter to give rise to the 5′UCOE/SFFV-eGFP (CBX3-A2B1), and 3′UCOE/SFFV-eGFP (A2B1-CBX3) vectors (Figure 3a). Viruses produced from these LVs were used to transduce P19 cells and expression monitored over time as before. Unlike the SFFV LTR in isolation, the 3′UCOE/SFFV-eGFP vector gave rise to stable expression throughout the culture period (Figure 3b). Interestingly, the 5′UCOE/SFFV-eGFP LV only displayed a partial protection to silencing, showing a gradual decline in eGFP expression over time. These data therefore agree with those obtained with the slightly larger 1.5 kb core A2UCOE previously employed and that extends a further 300 bp and into the first intron of HNRPA2B1 (ref. 26) and were confirmed when this fragment was similarly linked to the SFFV LTR (Supplementary Figure S5a,b). We then addressed whether stable eGFP expression in P19 cells from SFFV conferred by linkage to the A2UCOE was due to negation of DNA methylation on SFFV. DNA methylation status was assessed at day 17 after transduction by bisulfite conversion followed by DNA sequencing as before (Figure 2). In marked contrast to the extensive DNA methylation observed with the SFFV-eGFP vector, the presence of the A2UCOE in the 3′UCOE/SFFV-eGFP LV almost completely negated the appearance of this epigenetic mark (Figure 3c), which correlates well with the stability of eGFP expression (Figure 3b). By comparison, placement of the A2UCOE in the reverse orientation in 5′UCOE/SFFV-eGFP conferred only limited protection against DNA methylation of the SFFV promoter (Figure 3c), which again is also consistent with the gradual decline in eGFP expression from this configuration (Figure 3b). A similar DNA methylation pattern was observed with the larger 1.5 kb A2UCOE (Supplementary Figure S5c). Overall, our data clearly demonstrate that the DNA methylation–free, transcriptionally permissive environment generated by the A2UCOE can extend through a linked promoter leading to enhanced stability of transgene expression. To understand the basis behind the orientation-dependence for protection of the SFFV LTR against methylation, the linked A2UCOE sequences were also analyzed. Interestingly, in the protective configuration (3′UCOE/SFFV-eGFP), little methylation was noted, whereas in the alternative configuration, a cluster of methylated CpGs were located close to the adjacent SFFV LTR (Supplementary Figure S6b). This suggests that the context of the A2UCOE element is important for determination of methylation of both this element and any linked sequences, and that methylation of a small region of the A2UCOE adjacent to other sequences is sufficient to abrogate protection.
Figure 3.
Negation of silencing and DNA methylation by incorporating the A2UCOE upstream of SFFV. (a) Schematic of chimeric lentiviral vector constructs. A 1.2 kb A2UCOE element in both orientations, 5′UCOE (CBX3-A2B1) and 3′UCOE (A2B1-CBX3) is linked immediately upstream of the SFFV LTR in the SFFV-eGFP vector (Figure 1a). (b) Percentage of eGFP+ P19 cells transduced with lentiviruses made from the constructs shown in a. P19 cells were transduced with lentiviruses at multiplicity of infection of 40, and eGFP expression was assessed by flow cytometry at different time points up to 25 days, as indicated. Note: placing A2UCOE in a 3′–5′ orientation upstream of SFFV gives rise to stable eGFP expression from SFFV. (c) DNA methylation analysis on the SFFV regulatory element from P19 cells transduced with lentiviruses as in b. Genomic DNA was isolated from cells transduced with lentiviruses at day 17, and subjected to methylation analysis of the SFFV LTR. Upper panel: SFFV-eGFP. Middle panel: 3′A2UCOE/SFFV-eGFP. Lower panel: 5′A2UCOE/SFFV-eGFP. Note: placing 3′A2UCOE upstream of SFFV results in virtually complete protection of SFFV sequences from DNA methylation compared to partial protection in the 5′A2UCOE-SFFV construct.
A2UCOE is resistant to DNA methylation in vivo
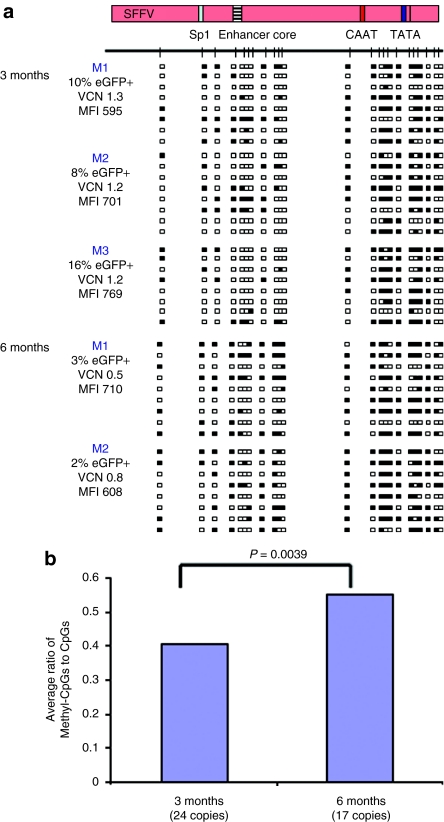
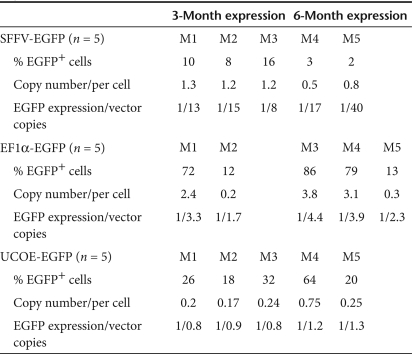
In order to further assess the role of DNA methylation in regulating transgene expression in vivo, we performed ex vivo gene transfer of murine HSC with the same SFFV, EF1α, and A2UCOE LVs. Vector DNA methylation status was determined by bisulfite conversion and sequencing as before (Figure 2) within whole bone marrow samples recovered at 3 and 6 months following transplantation. The SFFV promoter region was found to be extensively methylated at both 3 months (n = 3) and 6 months (n = 2) after transplantation (Figure 4a). However, in contrast to the findings in P19 cells (Figure 2b), the majority of the CpG sites around the Sp1/enhancer core region of this element were unmethylated, compared to high-level methylation observed in the vicinity of the proximal promoter CAAT and TATA box elements. This in vivo DNA methylation pattern suggests that the enhancer function of SFFV remains active (or is selected for in vivo), despite the fact that the majority of eGFP expression has become silenced due to extensive DNA methylation of the core promoter region. Indeed, the low ratio of eGFP-expressing cells to VCN (Figure 4a and Table 1) is a clear indication of eGFP transgene silencing. The high-level mean fluorescent intensity (range 595–769) of residual eGFP+ cells (Figure 4a) is also an indication of potent enhancer activity, which contrasts with the very low-level mean fluorescent intensity (range 18–40) in P19 cells where the Sp1/enhancer core region of this element is completely methylated (Figure 1b, underlined numbers). In addition, a comparison of the average ratio of methylated to nonmethylated CpG residues among all SFFV-eGFP vector copies analyzed at 3 (24 vector copies) and 6 months (17 vector copies) after transplantation revealed a significant increase in methylation over time (P value = 0.0039 determined using the Wilcoxon rank-sum test) (Figure 4b). This progressive DNA methylation in vivo correlates with the decreased ratio of eGFP+ cells to VCN (Table 1).
Figure 4.
DNA methylation analysis of the SFFV promoter in vivo following gene transfer to mouse hematopoietic stem cells. Mouse bone marrow lin-selected stem cells were transduced with SIN-LV SFFV-eGFP virus and engrafted into lethally irradiated animals. Bone marrow from transduced mice was isolated at 3 (n = 3) and 6 (n = 2) months after transduction and subjected to DNA methylation analysis. (a) DNA methylation analysis of SFFV proviral sequences from five transduced mice. Note: the SFFV promoter is densely methylated in all five mice. Methylated CpG sites were concentrated around the CAAT/TATA box core promoter elements in contrast to the Sp1/enhancer core region. MFI, mean fluorescence intensity; VCN, vector copies per cell. (b) Comparison of the average ratio of methylated: nonmethylated CpG dinucleotides among vector copies analyzed at 3 months and 6 months after transduction. Methylated CpG sites increased significantly over this time period (The P values were determined using the Wilcoxon rank-sum test).
Table 1. Summary of the ratio of transgene EGFP expression to vector copies in mice transduced with lentiviral vectors.
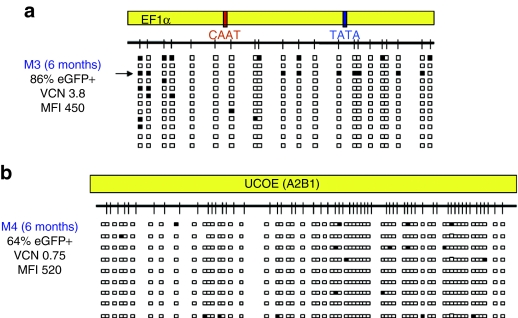
The EF1α promoter was subject to less CpG methylation overall than the SFFV LTR in engrafted hematopoietic cells, although isolated vector copies from each transplanted mouse did show significant (40–60%) methylation (Figure 5a, Supplementary Figure S7). However, these were relatively infrequent, and therefore unlikely to affect overall transgene expression levels. In contrast to the SFFV LTR and EF1α promoter, the A2UCOE displayed little or no methylated CpG residues in all vector copies, in all mice analyzed (Figure 5b; Supplementary Figures S4b and S8), with the ratio of eGFP-expressing cells to VCN essentially close to one in all cases (Table 1).
Figure 5.
DNA methylation analysis of the EF1α and the A2UCOE regulatory elements in vivo following gene transfer to mouse hematopoietic stem cells. Methylation analysis of DNA from total bone marrow cells isolated from mice transduced with the (a) EF1α-eGFP vector (N = 5) and (b) A2UCOE-eGFP vector (N = 5) vectors at 3 and 6 months described as in Figure 4. The result from a representative transduced mouse (M3 and M4) at 6 months from each vector group is shown (data from other transduced mice in the same cohort are shown in the Supplementary Figures S7 and S8). The arrows indicate the vector copies with 40–60% methylated CpGs. MFI, mean fluorescence intensity; VCN, vector copies per cell.
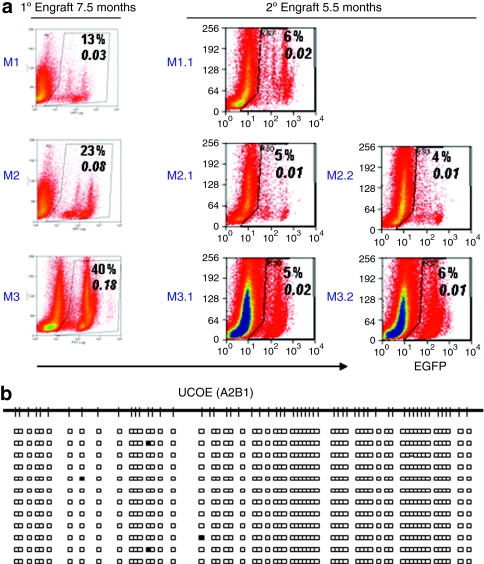
In order to assess whether the resistance to DNA methylation is preserved in long-term HSC, mouse bone marrow transduced with the A2UCOE vector was recovered from primary recipients at 7.5 months after transplantation and injected into a second cohort of lethally irradiated animals. Bone marrow from secondary recipients was harvested after a further 5.5 months for analysis of transgene expression and DNA methylation (Figure 6). The pattern of eGFP expression remained remarkably consistent between primary and secondary recipients (Figure 6a) indicating that the A2UCOE retained reliable expression in the absence of silencing. The decline in the proportion of eGFP+ cells observed in secondary recipients was accompanied by reduced overall VCN, and was therefore most likely due to dilution of transduced HSC. Furthermore, analysis confirmed lack of CpG methylation of the A2UCOE region from secondary recipients (Figure 6b) as observed in primary recipients, illustrating that this element displays a true resistance to this epigenetic modification in long-term HSC.
Figure 6.
eGFP expression and DNA methylation status of A2UCOE-eGFP transduced hematopoietic stem cell in secondary engrafted recipients. (a) Left panel: flow cytometry analysis of eGFP expression from primary (1°) recipients at 7.5 months after transplantation. Middle and right panels: analysis of eGFP expression in secondary (2°) recipients at 5.5 months after transplantation. The percentage of eGFP+ cells is shown in each plot. Italicized numbers represent vector copy number per cell determined by Q-PCR. Note: eGFP expression profiles remained remarkably similar between 1° and 2° recipients as identified by discrete populations of cells expressing different levels of eGFP. (b) DNA methylation analysis on A2B1 region of A2UCOE from a representative (M1.1) secondary recipient. Note: there are only scattered methylated CpG sites that are found on A2UCOE proviral vector copies.
Discussion
The efficiency and therefore efficacy of gene transfer in numerous previous studies has been compromised by instability (high variability and silencing) of transgene expression, mostly from viral promoters.13,14,15,16,17,18,19,20,21,28 A possible solution to problems of transgene silencing is to employ a UCOE to drive or regulate expression. UCOEs are structurally defined as consisting of closely spaced divergently transcribed promoters of housekeeping genes encompassed by a methylation-free CpG island and functionally characterized by a dominant chromatin opening capability, being able to confer reproducible and stable transgene expression, even from within centromeric heterochromatin integration sites.26,27 In addition, we have previously demonstrated that transgenes within a SIN-LV under the control of the A2UCOE derived from the HNRPA2B1-CBX3 locus are expressed at highly stable, reproducible, and homogeneous levels both in vitro and in vivo.28 Here, we have investigated whether the stability and consistency of transgene expression from the A2UCOE is by virtue of its resistance to DNA methylation–mediated silencing and that the loss of expression from other promoters is a direct result of DNA methylation.
Our data show for the first time that the molecular basis for the stability of A2UCOE-driven transgene expression from within a SIN-LV is due, at least in part, to its resistance to DNA methylation (Figures 2d, 5b, and 6b; Supplementary Figures S2c, S4, and S8). The resistance to DNA methylation of the A2UCOE was demonstrated in vitro using a hypermethylation system based on P19 cells and in vivo following ex vivo transplantation of transduced murine bone marrow HSC. In contrast, the EF1α promoter (at least in the absence of positive selection) and more prominently, the SFFV LTR may be prone in certain contexts to DNA methylation–mediated silencing. Importantly, we show that resistance to DNA methylation and stable A2UCOE-eGFP vector expression was achieved with an average VCN per cell of one compared to the high VCNs observed in cells transduced with the EF1α-eGFP, in particular with the SFFV-eGFP constructs (Figure 1b; Table 1).
The human housekeeping elongation factor 1-α gene (EF1α) promoter has become a popular choice to regulate retroviral transgene expression.34,35,36 However, there is evidence that this promoter is prone to silencing.32,37 A 1.5 kb EF1α promoter fragment extending into the first intron used to express a nuclear localized β-galactosidase reporter gene within a SIN-LV construct showed a decline in transgene expression associated with DNA methylation and histone deacetylation in P19 cells.37 Using the same cell line, we now show that the substantial loss of EF1α-eGFP transgene expression directly correlates with levels of DNA methylation of this promoter (Figures 1b and 2c and Supplementary Figures S2b and S3c) regardless of the level of VCN in transduced cells. However, we found relatively few instances of EF1α promoter methylation when the same vector was introduced into mouse HSCs in vivo and that this did not appear to have an overall negative impact on numbers of eGFP-expressing cells (Figure 5a and Supplementary Figure S7). We also note that a relatively high VCN is seen in cells transduced with the EF1α-eGFP vector both in vitro and in vivo (Figure 1b and Table 1) compared to the A2UCOE construct (Figure 1b and Table 1). As the ratio of numbers of eGFP+ cells to VCN is greater than one in all mice analyzed, it appears that at least a proportion of transgenes driven by the EF1α promoter are not transcribed in vivo, especially where a high VCN (2.4–3.8 copies per cell) is seen (Table 1). As this is taking place on the basis of low levels of DNA methylation (Figure 5a and Supplementary Figure S7), this might suggest that another silencing mechanism in addition to DNA methylation can play a role in the regulation of expression. This could include repeat-induced gene silencing triggered by the presence of multiple transgene copies,38 and establishment of repressive histone modification marks.
The SFFV LTR has also been widely used to regulate transgene expression within retroviral vectors, especially due to its high activity in hematopoietic cells.39,40 However, to date, the stability of expression from the SFFV promoter within SIN-LV especially in vivo has not been thoroughly assessed. In a previous investigation, we found a highly significant reduced ratio of transgene-expressing cells to VCN at 3 months after transplantation of HSC in vivo, giving a clear indication of silencing of this element.28 By directly assessing DNA methylation, in this study, we show that this element becomes extensively methylated both in vitro and in vivo (Figures 2b and 4, Supplementary Figures S2a and S3a,b). In P19 cells, the SFFV promoter is subject to a high density of DNA methylation, which is associated with a 95% reduction in eGFP transgene–expressing cells and a severe attenuation of mean fluorescent intensity (Figure 1b). In particular, we observed intense methylation of CpG sites around the Sp1/enhancer core region, which are crucial components responsible for maintaining its high transcriptional activation capability.38 However, unlike the DNA methylation pattern seen in P19 cells, extensive methylation of CpG residues in vivo occurred mostly around the CAAT and TATA box elements of the SFFV core promoter, with little or no methylation at the Sp1/enhancer region (Figure 4a). This suggests that in vivo SFFV enhancer function remains largely intact (or is selected for through mutagenesis), although in the vast majority of cases, eGFP transgene expression is silenced due to proximal promoter methylation (Figure 4a; Table 1). The DNA methylation pattern of the SFFV promoter/enhancer region that we observe in vivo is very similar to what has been described in a gene therapy clinical trial for chronic granulomatous disease employing a non-SIN gammaretroviral vector incorporating the SFFV LTR to regulate expression of a gp91phox transgene.11 In this trial, vector integration within the growth promoting MDS1-EVI1, PRDM16, and SETBP1 gene cluster led to their upregulation via the unmethylated and hence active SFFV enhancer. This in return resulted in the clonal expansion of cells harboring these integration events and hence initial efficacy. However, progressive methylation of the SFFV core promoter region led to transgene silencing and treatment failure.11 Our findings are clearly consistent, and provide additional molecular evidence that the enhancer region of the SFFV LTR even in the context of a SIN-LV is not prone to DNA methylation in vivo and therefore may induce enhancer-mediated insertional mutagenesis even when promoter activity and hence transgene expression are silenced.
However, linking in cis of a core A2UCOE fragment as little as 1.2 kb in length upstream of the SFFV LTR successfully negates silencing through almost complete prevention of DNA methylation (Figure 3b,c; Supplementary Figure S5). This is consistent with previous findings, which showed that the A2UCOE enhances and stabilizes expression from the human cytomegalovirus promoter/enhancer in stably transfected tissue culture cells.27 In addition, combining the A2UCOE with tissue-specific regulatory elements such as the human desmin promoter/enhancer41 or the β-globin locus control region and promoter (G.E. Talbot and M.N. Antoniou, unpublished results) can augment their function but without perturbing their specificity. Nevertheless, these data provide strong evidence that the A2UCOE can generate an extended transcriptionally permissive chromatin environment allowing efficient expression from not only the inherent UCOE promoters but also adjacent regulatory elements.
The molecular mechanisms by which the A2UCOE (and other similar elements) can establish and maintain an open and active chromatin structure are not, as yet, well understood. However, a two component model of UCOE function has been proposed. This consists of (i) a large region of methylation-free DNA that extends well beyond the boundaries of the CpG island in association with active histone modification marks, and (ii) bidirectional transcription with an inherent chromatin opening function from the UCOE closely spaced dual divergent promoters.26,27 Recently, a comprehensive epigenetic analysis of the native genomic A2UCOE found that this region is associated with an extended (~5 kb) region of methylation-free DNA, in combination with a distinct pattern of active histone modification markers.42 The reproduction of this epigenetic signature at transgene integration sites may be a significant contributory factor in the A2UCOE's ability to negate DNA methylation, heterochromatin formation and prevent transcriptional silencing.
The two component model of UCOE function may provide an explanation for the observed orientation dependent ability of the A2UCOE to prevent silencing of linked SFFV (Figure 3 and Supplementary Figure S5) or human cytomegalovirus27 promoter and enhancer regions. Efficient stabilizing function was evident only with the CBX3 end of the A2UCOE placed directly adjacent to the SFFV LTR (Figure 3 and Supplementary Figure S5). The opposite orientation was previously found to be more effective in negating silencing of the human cytomegalovirus promoter in stably transfected Chinese hamster ovary tissue culture cells.27 A recent study, which cataloged all human housekeeping genes by the most stringent criteria (principally level and degree of uniformity of expression between 42 different tissue types) has shown transcription from the HNRPA2B1 promoter to be not only well above average but to possess the lowest coefficient of variation of any of the 1,522 genes listed.43 In marked contrast, transcription from the CBX3 promoter is highly variable between different cell types. Therefore, depending on the cellular background, the divergent transcription from the CBX3 and HNRPA2B1 promoters of the A2UCOE can be balanced (for example, within hematopoietic cells; see ref. 41) or disproportionally high or low in either HNRPA2B1or CBX3 directions. According to our current model of UCOE function, poor divergent transcription away from the expression cassette would be expected to compromise its dominant chromatin remodeling capability. Thus, depending on the expression profile of the A2UCOE in a given cell type, the orientation with the strongest divergent transcription may be more efficacious in stabilizing heterologous promoter function. The stability of expression from the 2.2 kb A2UCOE in P19 cells, albeit with divergent CBX3 transcription, is in all likelihood due to the fact that this element extends to the end of the methylation-free CpG island on the side of CBX3.
We have previously shown that the A2UCOE lacks classical enhancer function28 and thus should not possess the ability to mediate long-range transcriptional activation from heterologous promoters at LV integration sites via now established chromatin looping mechanisms.44,45 However, the A2UCOE does have the potential to disturb host gene function and cause insertional mutagenesis resulting from divergent transcription from within the LV and into the surrounding gene-containing genomic region. This read-through transcription can in principle lead to either an activating or negative interference effect on endogenous gene function or may lead to the production of aberrant transcripts and protein products, a feature common to all LVs. Experiments are currently underway to directly assess the insertional mutagenesis potential of A2UCOE LVs via these mechanisms with additional refinements in vector design to minimize their likelihood.
In summary, this study investigated the DNA methylation status of three different internal promoters (SFFV, EF1α, and A2UCOE) within SIN-LVs in vitro, using P19 cells, and in vivo, following ex vivo gene transfer into mouse HSCs. Our study provides convincing evidence that DNA methylation indeed plays a direct role in regulating transgene expression in a SIN-LV system. In comparison with the SFFV and EF1α promoters, we show that stable transgene expression from the A2UCOE within an LV context in vitro and in vivo is due, at least in part, to its resistance to DNA methylation. Furthermore, for application to hematopoietic disorders, the A2UCOE resists methylation in long-term HSC. This reinforces its potential use as an excellent regulatory element for gene transfer and therapy applications.
Materials and Methods
Plasmid and LV construction. Lentiviral A2UCOE-eGFP and SFFV-eGFP used in this study are as previously described.33 The elongation factor 1-α (EF1α) LV EF1α-eGFP-WPRE was constructed as follows. The short (~250 bp) EF1α promoter was released from SRS11 EFS-γC vector36 and inserted in place of the SFFV promoter LTR within p′HR-cppt-SEW.46 The A2UCOE-SFFV-eGFP LVs were constructed by linking in both orientations either a core 1.5 kb BsmBI fragment extending from within the first intron of CBX3 and HNRPA2B1 (ref. 26) or a 1.2 kb subfragment of the 1.5 kb element extending from the BsmBI site of CBX3 to a TthIII I site within exon I of HNRPA2B1 immediately upstream of the SFFV promoter in the SFFV-eGFP LV.
Maintenance of tissue culture cell lines. Mouse embryonic teratocarcinoma stem P19 cells were obtained from the European Collection of Cell Cultures and maintained in alpha MEM medium (Sigma-Aldrich, Poole, UK) supplemented with 2 mmol/l glutamine, 1% nonessential amino acids, 2.5% fetal bovine serum, 7.5% calf serum, and 10 µg/ml each of penicillin and streptomycin. Human HeLa and HEK293T cells were maintained in DMEM medium (Invitrogen, Paisley, UK) containing 10% fetal bovine serum (Sigma-Aldrich) plus 2 mmol/l glutamine and 10 µg/ml each of penicillin and streptomycin.
LV preparation and transduction of cell lines. LVs were produced by standard transient co-transfection of HEK293T cells with three plasmids: the LV, pMD.G2 (envelop plasmid) and pCMVΔ8.91 (packaging plasmid), with polyethylenimine (Sigma-Aldrich) as previously described.33 The viral vector titer of eGFP-containing preparations was determined by transducing HeLa cells with serial dilutions of virus and monitoring expression after 3 days by flow cytometry. Lentiviruses containing eGFP under control of the A2UCOE, EFα1, and SFFV promoters were used to transduce P19 cells at different multiplicities of infection and grown under the conditions described above. Transduced cells were collected every 3–5 days for a total period of 42 days of continuous culture, and eGFP reporter gene expression was analyzed by flow cytometry.
Ex vivo analysis of transduced murine HSCs. Bone marrow lineage negative (lin−) HSCs were isolated from the tibia and femur of C57BL/6J mice at ~10 weeks of age, as previously described.33 HSCs were seeded at 1 × 106/ml in StemSpan medium (StemCell Technologies, London, UK) and transduced with viral vector added at a multiplicity of infection of 20–25 in the presence of cytokines. Transduced cells were cultured overnight and then injected into lethally irradiated recipient mice via the tail vein (2.5–5 × 105 cells/mouse). Peripheral blood and bone marrow cells were analyzed 3 and 6 months after transplantation by flow cytometry for eGFP transgene expression and Q-PCR for VCN. For secondary transplantation, total bone marrow cells from primary transplanted mice were isolated 7.5 months after transplantation and injected into a second cohort of lethally irradiated C57BL/6J mice (at least 1 × 107 cells per mouse).
DNA methylation by bisulfite conversion and sequencing
Sodium bisulfite treatment of genomic DNA: Genomic DNA was isolated from cells using the DNeasy kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. Sodium bisulfite treatment of genomic DNA was performed to convert unmethylated cytosine to thymine residues using the EpiTect bisulfite kit (Qiagen) according to the manufacturer's instructions. In brief, 500 ng of genomic DNA was mixed with bisulfite reagent and incubated at different temperatures. Genomic DNA was then purified by passing through the column supplied in the kit after completion of the conversion reaction.
Nested PCR amplification of bisulfite-treated genomic DNA: PCR primers were designed based on converted sequences. Reverse primers were chosen from the eGFP sequence to avoid the amplification of endogenous copies of EF1α and A2UCOE. The forward primers were chosen to cover the full length of the SFFV (containing 23 CpG sites) and EF1α (containing 21 CpG residues) promoters. The forward primers for the A2UCOE were chosen to amplify 650 bp of the HNRPA2B1 region including the promoter, which contains 60 CpG dinucleotides. Primers were also designed to amply 827 bp of the divergent CBX3 promoter region of the A2UCOE, which included a total of 86 CpG sites. PCR conditions were as follows: first round, annealing temperature 58 °C, 33 cycles; second round, annealing temperature 56 °C, 32 cycles.
The PCR primer sequences used were as follows: (i) SFFV promoter: Forward-1, 5′TAG AAA AAG GGG GGA ATG AAA; Reverse-1, 5′AAA CAA CTC CTC ACC CTT ACT CAC; Forward-2, 5′GGG GGA ATG AAA GAT TTT ATT TG; Reverse-2, 5′ACC CTT ACT CAC CAT AAT TTC AAC C. (ii) EF1α promoter: Forward-1, 5′TAA AAG AAA AGG GGG GAT TGG; Reverse-2, 5′AAA CAA CTC CTC ACC CTT ACT CAC; Forward-2, 5′GGG GTA TAG TGT AGG GGA AAG AAT AG; Reverse-2, 5′ACC CTT ACT CAC CAT AAT TTC AAC C. (iii) HNRPA2B1: Forward-1, 5′ACA CCA CCC CAA TAA ACA ACT C; Reverse-1, 5′GGG TTA GGG GTG GGT TAT TG; Forward-2, 5′GAT TTG GGT GTT TTT GGA AGG; Reverse-2, 5′ACC CTT ACT CAC CAT AAT TTC AAC C. (iv) CBX3: Forward-1, 5′GTT GTT TTT TGG GAG GTT TAA GTT; Reverse-1, 5′TAA TTT TTA AAC TCC CAA CTC CC; Forward-2, AGG GGG AAA TGT GTT TTG TTT; Reverse-2, CTC CCC CAA CAT CTA AAA AAT TC.
Sequence analysis of PCR products: The single PCR product band was purified after resolution by agarose gel electrophoresis using the Qiagen gel extraction kit, and ligated into the PCR 2.1-TOP vector (Invitrogen) according to the manufacturer's instructions. The plasmid DNAs from randomly selected colonies were sequenced using M13 primers compatible with the plasmid vector backbone. Sequence comparison was performed using Vector NTI software (Invitrogen).
Determination of LV copy number. Genomic DNA was isolated from cells using the DNeasy kit (Qiagen). Real-time quantitative PCR was used to determine LV copy number employing the ABI 7000 Sequence Detection System (ABI, Applied Biosystems, Cheshire, UK). PCR primers targeting eGFP transgene sequences common to all vectors were as follows: Forward-5′GCTACCCCGACCACATGAAG3′ Reverse-5′CGGGCATGGCGGACTT3′. eGFP probe sequence: 5′-FAM-CAGCACGACTTCTTC-NFQ. Primer for the endogenous reference Ttn sequences were as described above. The Ttn probe sequence was as follows: 5′-FAM–TGCACGGAATCTCGTCTCAGTC—TAMRA-3′.
SUPPLEMENTARY MATERIAL Figure S1. Flow cytometry histograms of eGFP expression in P19 cells transduced with lentiviruses shown in Figure 1a, at different time points shown on Figure 1b. Figure S2. A2UCOE gives rise to stable eGFP expression in P19 cells compared to SFFV and EF1α. Figure S3. DNA methylation analysis on SFFV and EF1α promoters in P19 cells following the lentivirus transduction. Figure S4. DNA methylation analysis of the CBX3 region of the A2UCOE in P19 cells and in vivo following lentiviral transfer to mouse HSCs. Figure S5. Negation of silencing of SFFV by incorporating a 1.5 kb A2UCOE upstream of SFFV in P19 cells. Figure S6. DNA methylation analysis on A2B1 region of A2UCOE linked upstream of the SFFV LTR. Figure S7. DNA methylation analysis of the EF1α promoter in vivo following gene transfer to mouse HSCs. Figure S8. DNA methylation analysis of the A2UCOE regulatory element in vivo following gene transfer to mouse HSCs.
Acknowledgments
This work was supported by the Wellcome Trust (A.J.T., M.P.B.) and the UK Biotechnological and Biological Sciences Research Council (F.Z.).
Supplementary Material
Flow cytometry histograms of eGFP expression in P19 cells transduced with lentiviruses shown in Figure 1a, at different time points shown on Figure 1b.
A2UCOE gives rise to stable eGFP expression in P19 cells compared to SFFV and EF1α.
DNA methylation analysis on SFFV and EF1α promoters in P19 cells following the lentivirus transduction.
DNA methylation analysis of the CBX3 region of the A2UCOE in P19 cells and in vivo following lentiviral transfer to mouse HSCs.
Negation of silencing of SFFV by incorporating a 1.5 kb A2UCOE upstream of SFFV in P19 cells.
DNA methylation analysis on A2B1 region of A2UCOE linked upstream of the SFFV LTR.
DNA methylation analysis of the EF1α promoter in vivo following gene transfer to mouse HSCs.
DNA methylation analysis of the A2UCOE regulatory element in vivo following gene transfer to mouse HSCs.
REFERENCES
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- Gaspar HB, Bjorkegren E, Parsley K, Gilmour KC, King D, Sinclair J, et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol Ther. 2006;14:505–513. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Schmidt M, et al. Gene therapy for chronic granulomatous disease: current status of the German clinical study. Hum Gene Ther. 2008;19:1097. [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraunus J, Schaumann DH, Meyer J, Modlich U, Fehse B, Brandenburg G, et al. Self-inactivating retroviral vectors with improved RNA processing. Gene Ther. 2004;11:1568–1578. doi: 10.1038/sj.gt.3302309. [DOI] [PubMed] [Google Scholar]
- Klug CA, Cheshier S., and, Weissman IL. Inactivation of a GFP retrovirus occurs at multiple levels in long-term repopulating stem cells and their differentiated progeny. Blood. 2000;96:894–901. [PubMed] [Google Scholar]
- Bestor TH. Gene silencing as a threat to the success of gene therapy. J Clin Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, et al. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 2000;19:5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenqvist N, Hård Af Segerstad C, Samuelsson C, Johansen J., and, Lundberg C. Activation of silenced transgene expression in neural precursor cell lines by inhibitors of histone deacetylation. J Gene Med. 2002;4:248–257. doi: 10.1002/jgm.268. [DOI] [PubMed] [Google Scholar]
- Parera M, Ibañez A, Clotet B., and, Martinez MA. Lack of evidence for protease evolution in HIV-1-infected patients after 2 years of successful highly active antiretroviral therapy. J Infect Dis. 2004;189:1444–1451. doi: 10.1086/382485. [DOI] [PubMed] [Google Scholar]
- Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- Chang AH, Stephan MT., and, Sadelain M. Stem cell-derived erythroid cells mediate long-term systemic protein delivery. Nat Biotechnol. 2006;24:1017–1021. doi: 10.1038/nbt1227. [DOI] [PubMed] [Google Scholar]
- Mok HP, Javed S., and, Lever A. Stable gene expression occurs from a minority of integrated HIV-1-based vectors: transcriptional silencing is present in the majority. Gene Ther. 2007;14:741–751. doi: 10.1038/sj.gt.3302923. [DOI] [PubMed] [Google Scholar]
- Minoguchi S., and, Iba H. Instability of retroviral DNA methylation in embryonic stem cells. Stem Cells. 2008;26:1166–1173. doi: 10.1634/stemcells.2007-1106. [DOI] [PubMed] [Google Scholar]
- Jakobsson J, Rosenqvist N, Thompson L, Barraud P., and, Lundberg C. Dynamics of transgene expression in a neural stem cell line transduced with lentiviral vectors incorporating the cHS4 insulator. Exp Cell Res. 2004;298:611–623. doi: 10.1016/j.yexcr.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Gaszner M., and, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Nielsen TT, Jakobsson J, Rosenqvist N., and, Lundberg C. Incorporating double copies of a chromatin insulator into lentiviral vectors results in less viral integrants. BMC Biotechnol. 2009;9:13. doi: 10.1186/1472-6750-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbinati F, Arumugam P, Higashimoto T, Perumbeti A, Mitts K, Xia P, et al. Mechanism of reduction in titers from lentivirus vectors carrying large inserts in the 3′LTR. Mol Ther. 2009;17:1527–1536. doi: 10.1038/mt.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou M, Harland L, Mustoe T, Williams S, Holdstock J, Yague E, et al. Transgenes encompassing dual-promoter CpG islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics. 2003;82:269–279. doi: 10.1016/s0888-7543(03)00107-1. [DOI] [PubMed] [Google Scholar]
- Williams S, Mustoe T, Mulcahy T, Griffiths M, Simpson D, Antoniou M, et al. CpG-island fragments from the HNRPA2B1/CBX3 genomic locus reduce silencing and enhance transgene expression from the hCMV promoter/enhancer in mammalian cells. BMC Biotechnol. 2005;5:17. doi: 10.1186/1472-6750-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J, et al. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Jones-Villeneuve EM, Edwards MK., and, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Jones-Villeneuve EM, McBurney MW, Rogers KA., and, Kalnins VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D., and, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Zhang Y, Zieth CR., and, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cihák A. Biological effects of 5-azacytidine in eukaryotes. Oncology. 1974;30:405–422. doi: 10.1159/000224981. [DOI] [PubMed] [Google Scholar]
- Baum C, Hegewisch-Becker S, Eckert HG, Stocking C., and, Ostertag W. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumas DB, Spangrude GJ, Brooks DM, Williams CD., and, Chesebro B. High-frequency cell surface expression of a foreign protein in murine hematopoietic stem cells using a new retroviral vector. Blood. 1996;87:509–517. [PubMed] [Google Scholar]
- Thornhill SI, Schambach A, Howe SJ, Ulaganathan M, Grassman E, Williams D, et al. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yang Q., and, Chang LJ. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J Virol. 2005;79:13497–13508. doi: 10.1128/JVI.79.21.13497-13508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Mai T, Yang X., and, Jardine K. Evidence for repeat-induced gene silencing in cultured mammalian cells: inactivation of tandem repeats of transfected genes. Exp Cell Res. 2002;274:1–8. doi: 10.1006/excr.2001.5443. [DOI] [PubMed] [Google Scholar]
- Baum C, Itoh K, Meyer J, Laker C, Ito Y., and, Ostertag W. The potent enhancer activity of the polycythemic strain of spleen focus-forming virus in hematopoietic cells is governed by a binding site for Sp1 in the upstream control region and by a unique enhancer core motif, creating an exclusive target for PEBP/CBF. J Virol. 1997;71:6323–6331. doi: 10.1128/jvi.71.9.6323-6331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert HG, Stockschläder M, Just U, Hegewisch-Becker S, Grez M, Uhde A, et al. High-dose multidrug resistance in primary human hematopoietic progenitor cells transduced with optimized retroviral vectors. Blood. 1996;88:3407–3415. [PubMed] [Google Scholar]
- Talbot GE, Waddington SN, Bales O, Tchen RC., and, Antoniou MN. Desmin-regulated lentiviral vectors for skeletal muscle gene transfer. Mol Ther. 2010;18:601–608. doi: 10.1038/mt.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl Allen M., and, Antoniou M. Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE) Epigenetics. 2007;2:227–236. doi: 10.4161/epi.2.4.5231. [DOI] [PubMed] [Google Scholar]
- She X, Rohl CA, Castle JC, Kulkarni AV, Johnson JM., and, Chen R. Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics. 2009;10:269. doi: 10.1186/1471-2164-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W., and, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Miele A., and, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry histograms of eGFP expression in P19 cells transduced with lentiviruses shown in Figure 1a, at different time points shown on Figure 1b.
A2UCOE gives rise to stable eGFP expression in P19 cells compared to SFFV and EF1α.
DNA methylation analysis on SFFV and EF1α promoters in P19 cells following the lentivirus transduction.
DNA methylation analysis of the CBX3 region of the A2UCOE in P19 cells and in vivo following lentiviral transfer to mouse HSCs.
Negation of silencing of SFFV by incorporating a 1.5 kb A2UCOE upstream of SFFV in P19 cells.
DNA methylation analysis on A2B1 region of A2UCOE linked upstream of the SFFV LTR.
DNA methylation analysis of the EF1α promoter in vivo following gene transfer to mouse HSCs.
DNA methylation analysis of the A2UCOE regulatory element in vivo following gene transfer to mouse HSCs.