Abstract
Dystrophin, the cytoskeletal protein whose defect is responsible for Duchenne muscular dystrophy (DMD), is normally expressed in both muscles and brain. Genetic loss of brain dystrophin in the mdx mouse model of DMD reduces the capacity for type A γ-aminobutyric acid (GABAA)-receptor clustering in central inhibitory synapses, which is thought to be a main molecular defect leading to brain and cognitive alterations in this syndrome. U7 small nuclear RNAs modified to encode antisense sequences and expressed from recombinant adeno-associated viral (rAAV) vectors have proven efficient after intramuscular injection to induce skipping of the mutated exon 23 and rescue expression of a functional dystrophin-like product in muscle tissues of mdx mice in vivo. Here, we report that intrahippocampal injection of a single dose of rAAV2/1-U7 can rescue substantial levels of brain dystrophin expression (15–25%) in mdx mice for months. This is sufficient to completely restore GABAA-receptor clustering in pyramidal and dendritic layers of CA1 hippocampus, suggesting exon-skipping strategies offer the prospect to investigate and correct both brain and muscle alterations in DMD. This provides new evidence that in the adult brain dystrophin is critical for the control of GABAA-receptor clustering, which may have an important role in activity-dependent synaptic plasticity in hippocampal circuits.
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder caused by mutations in the dystrophin gene that prevent expression of this membrane-bound cytoskeleton-associated protein normally expressed in both muscle and brain. Brain dystrophin is detected in the postsynaptic densities of pyramidal neurons at inhibitory synapses in brain structures involved in cognitive functions, such as hippocampus, neocortex, and cerebellum. Specific cognitive deficits have been characterized in all DMD patients, suggesting an important contribution of full-length dystrophin loss,1 whereas the severe mental retardation described in a sub-population of patients likely implies additional inactivation of other DMD-gene products derived from distinct internal promoters, such as Dp71 and Dp140.2 Dystrophin role in critical brain functions is supported by studies in the dystrophin-deficient mdx mouse reporting a range of brain alterations,3,4 including changes in hippocampal synaptic plasticity,5 altered density and morphology of hippocampal synapses,6 associated with behavioral and cognitive deficits.5,7 Several reports suggest that a 40–70% reduction in the number of type A γ-aminobutyric acid (GABAA) receptor clusters is a primary brain defect associated with genetic loss of dystrophin in hippocampus,8 cerebellum,9 and amygdala,10 suggesting dystrophin may stabilize these receptors at inhibitory synapses, perhaps by limiting their lateral diffusion outside the synapse.11,12 Yet, compelling evidence for direct or indirect interaction between dystrophin and GABAA receptors is still missing.
Dystrophin is a large (427 kd) protein expressed from a 2.5-Mb gene with 79 exons encoding a 14-kb mRNA transcript. The protein contains an N-terminal domain with an actin-binding site, a central rod domain with 24 triple-helical spectrin-like repeats, and cystein-rich and C-terminal domains interacting with several membrane-bound and cytosolic proteins. Whereas out-of-frame mutations in the DMD gene result in the absence of dystrophin expression, mutations that maintain the reading frame are associated with the milder Becker muscular dystrophy and produce dystrophin proteins that only miss some of the repeats of the central rod domain and retain some functional activity.13 Cognitive deficits are less frequent in Becker muscular dystrophy and likely result from mutations specifically affecting the C-terminal domain of dystrophin.14 Antisense oligonucleotides have been recently used to redirect splicing of the dystrophin pre-mRNA and induce skipping of the mutated exon 23 that contains a stop codon in mdx mice, thus generating a transcript of smaller size with restored open reading frame.4,15 We have shown in mdx mice that intramuscular injections of a single dose of recombinant adenovirus-associated vector (rAAV2/1 serotype) expressing antisense sequences linked to a modified U7 small nuclear RNA can skip exon 23, drive sustained therapeutic levels of rescued dystrophin-like protein and correct muscular dystrophy.16
The rAAV2/1 serotype that drives long-term dystrophin expression with low immunogenicity in muscle tissues also displays good neuronal tropism and transduction efficiency in rodent brain tissue in vivo, suggesting it may also have therapeutic efficacy in the dystrophin-deficient brain.17,18 Our goal here was to examine whether stable and widespread protein expression can be obtained in the mature brain of mdx mice after a single intrahippocampal injection of the rAAV2/1-U7 system, and to determine whether this may restore the clustering of GABAA receptors in hippocampus.
Results
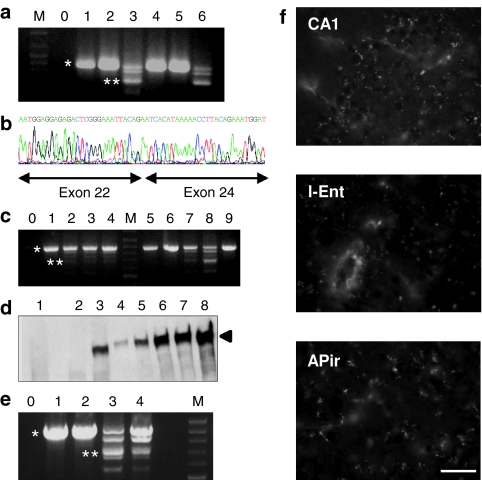
A single dose of the U7-SD23/BP22 rAAV2/1 vector16 was administered to mdx mice by bilateral intrahippocampal microinjections, and brains were analyzed after 4 months to examine rescue of the skipped dystrophin mRNA and protein. Reverse transcription-PCR using primers in exons 20 and 26 of the dystrophin mRNA yielded a 901-bp product corresponding to the native mRNA species and a 688-bp product corresponding to exon 23 skipping (Figure 1a,b). The profile of the PCR products in mdx mice injected with AAV vectors (mdx-AAV) was similar to that found in injected skeletal muscle,16 with intermediate-sized bands reflecting the formation of heteroduplex between the native and exon 23-skipped products (Figure 1a,c,e), as demonstrated by DNA sequencing showing a mixture of sequences with and without the skipped exon.19 The amount of the 688-bp product and its distribution away from the injection site was variable among mdx-AAV mice. Yet, skipping events could be detected bidirectionally (rostro-caudally) >1 mm away from the injection site (Figure 1c). Therefore, a single AAV2/1 injection resulted in exon 23 skipping 2–3 mm alongside the dorsal hippocampus for at least 4 months. A dystrophin-like protein was also consistently detected in hippocampus of the treated mice by the DYS1 antibody both by western blotting (Figure 1d) and immunofluorescence (Figures 1f and 2a). The rescued protein was detected ~1 mm away from the injection site, thus confirming vector diffusion estimated from RNA analysis. In addition, both the skipped mRNA (Figure 1e) and the dystrophin-like protein (Figure 1f) could be detected in the entorhinal cortex, consistent with retrograde axonal transport of rAAV2/1 particles to this hippocampal afferent structure.18 Dystrophin immunoreactivity (IR) was also present in the amygdalopiriform transition area, an extensive region of the temporal lobe that lies at the junction of piriform and entorhinal cortices. Although amygdalopiriform transition area and entorhinal cortex are interconnected, amygdalopiriform transition area also provides inputs to the ventral subiculum and hippocampal CA1,20 suggesting that dystrophin expression in this area also results from retrograde transport from hippocampus.
Figure 1.
Characterization of the exon 23-skipped brain dystrophin. (a) Detection of exon 23-skipped dystrophin mRNA. RNA samples were taken from WT hippocampus, cortex, and whole brain (lanes 1, 2, 4), from mdx whole brain (lane 5), from AAV-treated mdx hippocampus (lane 3) and from skeletal muscle of another mdx mouse injected intramuscularly with the same AAV vector (lane 6). The 901-bp fragment amplified from the native dystrophin mRNA (*) detected in WT and mdx mice was largely replaced by the 688-bp fragment from the dystrophin transcript lacking exon 23 (**) in AAV-treated mdx mice. (b) DNA sequence of the 688-bp band demonstrating loss of exon 23. (c) Expression of dystrophin transcripts analyzed by nested RT-PCR alongside the dorsal hippocampus in two AAV-treated mdx mice (lanes 1–4; 5–8) and in the whole hippocampus of one saline mdx mouse (lane 9). Sections in AAV-treated mice are from various distances from injection site: −1 to −1.8 mm rostral (lanes 1, 5), −0.5 to −1 mm rostral (lanes 2, 6), −0.5 rostral to +0.5 mm caudal (lanes 3, 7), 0.5–0.85 mm caudal (lanes 4, 8). The 901-bp fragment (*) was only detected in the saline mdx mouse (lane 9) and in distal tissue samples in AAV-treated mdx mice (lanes 1, 5, and 6). The 688-bp band corresponding to exon 23-skipped mRNA (**) was found at various distances from the injection site in AAV-treated mutants (lanes 2, 3, 4, 7, and 8). (d) Western blot of total protein extracted from hippocampus of mdx (lane 1, protein load 300 µg), mdx-AAV (lanes 2: −0.5 to −1 mm rostral; lane 3: +0.5 to 0.85 mm caudal from injection site; protein load 300 µg) and WT mice (lanes 4–8; incremental protein loads: 10, 25, 50, 75, and 100 µg) stained with DYS1 antibody. The arrow indicates full-length 427-kd dystrophin. In this mdx-AAV mouse, hippocampal expression of rescued dystrophin was estimated 10% of WT. (e) Expression of the 688-bp fragment (**) in mdx-AAV mouse hippocampus (lane 3) and entorhinal cortex (lane 4). Only the full-length 901-bp fragment (*) was found in entorhinal cortex of WT (lane 1) and saline mdx mice (lane 2). M: marker, 0: H2O control. (f) Dystrophin immunoreactivity (IR) in CA1 hippocampus (CA1), lateral entorhinal cortex (l-Ent), and amygdalopiriform transition area (APir), as detected 4 months after single intrahippocampal injection of rAAV2/1 vector in the mdx mouse. Dystrophin-IR, typically characterized by intensely stained fluorescent puncta in WT mice, were absent in sections from saline-treated mdx mice. Each panel represents the projection of stacks of five images. Bar = 40 µm. rAAV, recombinant adeno-associated virus; WT, wild type.
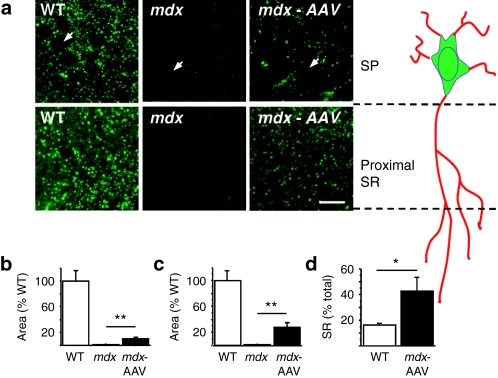
Figure 2.
Immunoreactivity of the rescued dystrophin in CA1 hippocampus. (a) Dystrophin-IR in stratum pyramidale (SP) and proximal stratum radiatum (SR) of CA1 hippocampus in WT and mdx mice injected with PBS and in AAV-treated mdx mice. Each panel represents projection of five-image stacks. Arrows: pyramidal-cell bodies in SP. Bar = 20 µm. (b) Area covered by dystrophin-IR in SP as % of WT. (c) Area covered by dystrophin-IR in SR as % of WT. (d) Fraction area covered by dystrophin-IR in the SR (% of total SP + SR). *P < 0.05; **P < 0.001. AAV, adeno-associated virus; IR, immunoreactivity; PBS, phosphate-buffered saline; WT, wild type.
In brain sections, immunohistochemical staining with the DYS1 antibody revealed a typical pattern of dystrophin-IR in the hippocampus, characterized in wild-type (WT) mice by intensely stained puncta along the somatic membrane and in the proximal dendritic layer of the CA1/CA3 pyramidal-cell layers, but not in dentate gyrus, whereas as expected there was no staining in CA1 from mdx mice8,21 (Figures 1f and 2a). Sections taken <1 mm away from the injection site in mdx-AAV mice were clearly positive for dystrophin-IR in CA1–CA3 hippocampal subfields, but not in the dentate gyrus, indicating that the skipping process induced dystrophin expression in the same hippocampal layers as in WT mice. However, the number of immunopositive puncta was lower in mdx-AAV mice. To estimate the expression level of dystrophin in mdx-AAV mice, we quantified dystrophin-IR in CA1 (WT, n = 6; mdx, n = 5; mdx-AAV, n = 5) using semiquantitative imaging procedures.22 Whereas mdx mice showed complete absence of dystrophin-IR except for minimal background staining (<2% of WT levels), mdx-AAV mice displayed significant levels of dystrophin-IR in both stratum pyramidale (SP) (10.23 ± 2.6% of WT levels, range 3–20%, P < 0.001, Figure 2b) and stratum radiatum (SR) (27.74 ± 7.09%, range 5–60%, P < 0.01, Figure 2c) of the dorsal hippocampus, as assessed by quantification of the total area covered by dystrophin-IR within a total tissue surface of 10,000 µm2 (Figure 2b,c). Hence, the number of dystrophin-positive puncta was substantial in both layers in mdx-AAV mice (SP: 14.15 ± 2.6% of WT, P < 0.0001, range 3–27%; SR: 25.71 ± 4.8%, range 13–41%, P < 0.001). The relative area covered by dystrophin-IR in the SR (% of total SP + SR, Figure 2d) was 42.44 ± 10.97% in mdx-AAV mice against 16.24 ± 1.21% in WT mice (P < 0.03), suggesting that a larger fraction of dystrophin proteins was targeted to dendritic processes in AAV-treated mdx mice compared to WT mice.
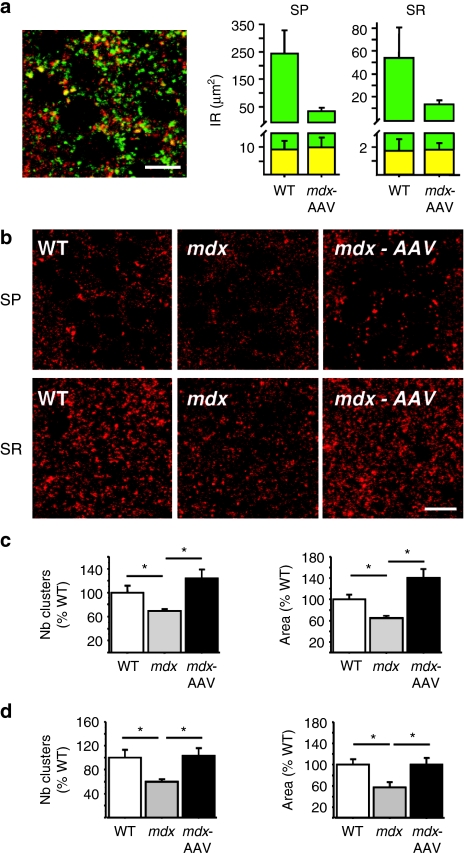
Partial colocalization of dystrophin clusters with a subset of α2 subunit-containing GABAA-receptor clusters has been demonstrated in hippocampal area CA1.8 In our own study, α2 subunit-IR typically appeared as discrete and intensely labeled clusters around somata and proximal dendritic layer in CA1–CA3, which paralleled the pattern of expression of dystrophin (Figure 3a,b). As expected, double-labeling experiments revealed partial juxtaposition and sometimes colocalization of dystrophin clusters with the α2 subunit-containing GABAA-receptor clusters (Figure 3a). Perfect overlap between dystrophin-IR and α2 subunit-IR in SP was estimated only 3.73 ± 1.36% in WT mice, whereas it raised to 28.36 ± 3.91% of total dystrophin expression in mdx-AAV mice (P < 0.001). A larger ratio in mdx-AAV than in WT mice was also observed in the SR (13.36 versus 3.18%). As shown in Figure 3a, this in fact reflects comparable amounts of dystrophin proteins colocalized with α2 subunit-IR in the two groups of mice, despite the lower level of dystrophin in the AAV-treated mdx mice. Indeed, a comparable fraction of GABAA-receptor clusters colocalized with dystrophin-IR in the two groups in both hippocampal layers (SP: WT, 10.27 ± 3.1%; mdx-AAV, 15.96 ± 4.59%, NS; SR: WT, 1.43 ±.62%; mdx-AAV, 2.6 ± 2.26%, NS). As a functional readout, we therefore examined whether moderate levels of dystrophin-like protein expression in mdx-AAV mice were sufficient to impinge on the expression of GABAA-receptor clusters. A reduction in GABAA-receptor clustering, characterized by a 40% decrease in the number of α2 subunit-containing clusters and total area covered by α2 subunit-IR, has been initially described in area CA1 of mdx mice.8 We confirmed this in phosphate-buffered saline-treated mdx mice, showing a significant reduction in the number of clusters and area covered by α2 subunit-IR in both CA1 SP and SR layers (P < 0.05, compared to WT mice; Figure 3b–d). In striking contrast, the number of clusters and area covered by α2 subunit-IR were significantly larger in AAV-treated mdx mice compared to mdx mice (P < 0.05), reaching a level that was no longer different from that of WT mice (Figure 3b–d). As previously reported,8 the size of α2 subunit clusters was slightly reduced in area CA1 of mdx mice (86.4% of WT). However, the size of clusters in mdx-AAV mice was strictly comparable to that of WT mice (100.99% of WT, P = 0.9) and significantly larger than in control mdx mice (P < 0.05). In all, our quantitative analysis of α2 subunit-IR therefore revealed comparable expression levels of α2 subunit-containing GABAA-receptor clusters in WT and AAV-treated mdx mice, suggesting complete recovery of GABAA-receptor clustering after exon skipping-mediated partial restoration of dystrophin expression.
Figure 3.
Immunoreactivity of GABAA-receptor α2-subunit in CA1 hippocampus. (a) Partial colocalization of dystrophin-IR (green) with that of GABAA-receptor α2 subunits (red) in CA1 hippocampus. Left, representative sample confocal laser scanning microscope image showing superimposition of two stack projections (five images for each fluorochrome taken simultaneously); yellow dots are overlaps between dystrophin and α2-subunit IR. Right, histograms showing quantification of total dystrophin IR (green) and fraction colocalized with α2-subunit IR (yellow), expressed as area covered by IR (µm2) in SP and SR. Note that despite lower dystrophin expression levels in mdx-AAV mice, comparable amounts of proteins colocalized with α2 subunits were found in the two groups of mice. (b) Specific GABAA-receptor α2-subunit-IR in stratum pyramidale (SP) and stratum radiatum (SR) of CA1 hippocampus. (c) Number of clusters and area covered by α2 subunit-IR in SP as % of WT. (d) Number and area covered by α2 subunit-IR in SR as % of WT. Bars = 10 µm in a, 20 µm in b. *P < 0.05. AAV, adeno-associated virus; GABAA, type A γ-aminobutyric acid; IR, immunoreactivity.
Discussion
AAV vectors have recently emerged as powerful tools for gene transfer into the brain for both functional explorations and therapeutic purposes.23 In this study, we have used an AAV vector expressing antisense sequences to modulate splicing of the dystrophin pre-mRNA in the brain of mdx mice. A major advantage of this approach over the ectopic expression of a dystrophin complementary DNA is that physiological levels of functional mRNA can be restored, allowing a more relevant phenotypic analysis. However, exon skipping may only restore semifunctional dystrophin expression when mutations affect critical functional domains of the protein and it may not be applicable in cases of large genomic deletions.15 We show here that exon-skipping rescues stable expression (4 months postinjections) of a dystrophin-like protein to 15–25% of normal dystrophin levels in area CA1 of the hippocampus of mdx mice, and enables a complete recovery of α2 subunit-containing GABAA-receptor clustering in this brain region. This indicates that the synaptic molecular alteration associated with dystrophin loss is reversible.
Several studies have shown that rAAV2/1 vectors target primarily neurons in the central nervous system and can support long-term and widespread distribution of transduction with minimal or no toxicity.18 Our data confirm that rAAV2/1 can transduce CA1–CA3 hippocampal cells effectively and stably. The recovered expression of dystrophin in adjacent, afferent, cortical areas such as the entorhinal cortex likely reflects efficient retrograde transport of rAAV2/1 after a single intrahippocampal injection. This indicates that rAAV2/1 has the potential to provide therapeutic effects to various brain regions involved in cortico-hippocampal circuits, including remote brain structures difficult to access from a single intracerebral injection site. The injection of a 3-µl volume of rAAV2/1 (corresponding to 6.3 × 109 vector genome) did not result in observable tissue lesions, but was able to rescue detectable levels of dystrophin-like protein in part of the hippocampus (~2–3 mm of dorsal hippocampus). Such a dose of AAV vector has been shown to diffuse efficiently in the mouse brain (e.g., see ref. 24), and the partial correction observed here is likely due to a minimum threshold of modified U7 small nuclear RNAs (U7-SmOPT) expression needed for efficient exon skipping. This could be improved by exploring other serotypes or improving U7 cassettes for neuronal expression. Although we have no indication about a possible indirect interaction of the viral vector itself with GABAA receptors, it seems unlikely that this could explain the convergent changes of several specific markers commonly associated with dystrophin loss. Rather, our data suggest that the rescued expression of dystrophin-like proteins was preferentially targeted to inhibitory synapses and GABAA receptors, leading to comparable colocalization levels as in WT mice and to complete normalization of both the number and size of GABAA-receptor clusters. Functional studies are needed to determine whether the restored capacity for GABAA-receptor clustering in a limited portion of hippocampus is sufficient to correct synaptic and cognitive dysfunctions in mdx mice. Interestingly, it has been shown that modest (~27%) but potentially widespread expression of such a dystrophin-like protein through chronic brain administration of morpholino antisense oligonucleotides can compensate for a noncognitive amygdala-dependent behavioral disturbance in mdx mice, enhanced defensive freezing responses after restraint. Because this behavioral deficit may rely on GABAergic alteration in the amygdala,10 to the extent that the two structures may be comparable it is possible that this result was also due to a mechanism of GABAA-receptor clustering rescue, as demonstrated here in the case of the hippocampus. Taken together, the current data suggest that improvement of both molecular and behavioral alterations associated with dystrophin loss can be achieved in mature tissues by vectorized antisense-mediated exon skipping. The etiology of the cognitive impairment in DMD is however complex and far from being fully elucidated, as it likely relies on cumulative effects of inactivation of several DMD-gene products involved in distinct cellular brain functions. Understanding the specific mechanisms affected by loss or dysfunction of these distinct proteins is a prerequisite to direct appropriate therapeutic interventions and correct the brain alterations associated with DMD.4
Beyond the putative therapeutic outcomes, exon skipping-mediated dystrophin recovery is a promising and powerful tool to locally manipulate protein expression and examine its physiological role in synaptic function. The few studies addressing this question revealed that the initial clustering of GABAA receptors and gephyrin at central inhibitory synapses is independent from that of dystrophin and associated protein complex.25,26 It is suggested that dystrophin is not involved in synaptogenesis but may have an important role in modulating the subsequent maintenance and stabilization of postsynaptic GABAA-receptor clusters.11 Our present results provide new arguments to support this hypothesis. We report a relatively low level of colocalization between dystrophin and α2 subunit-containing GABAA receptors in the CA1 hippocampal region (<5% in WT mice), in agreement with previous studies suggesting that dystrophin is only expressed in a subset of inhibitory synapses, juxtaposed rather than colocalized with GABAA-receptor subunits. A remaining dystrophin pool could be putatively involved in trafficking/targeting processes, expressed in “empty” synapses transiently devoid of GABAA receptors, or associated with other undefined partners in specific synapse subtypes.11,25 In AAV-treated mdx mice, however, a larger fraction of the dystrophin-like protein colocalized with α2 subunit-IR (~28%). This in fact resulted in comparable net amounts of dystrophin/GABAA-receptor clusters colocalization in WT and AAV-treated mdx mice, which may explain why small amounts of re-expressed dystrophin-like protein were sufficient to completely restore GABAA-receptor clustering. Although dystrophin loss may affect different subunits of the GABAA receptor in distinct brain structures, e.g., α1 in cerebellum and α2 in hippocampus, the present data place α2 subunit-containing GABAA-receptor clusters as a primary functional target when small amounts of dystrophin are re-expressed in the hippocampus. Moreover, a larger fraction of dystrophin-like protein was targeted to dendritic processes in AAV-treated mdx mice compared to WT mice. This supports ultrastructural and electrophysiological data suggesting that dystrophin and α2 subunit-containing GABAA-receptor clusters play a major role in dendritic signal integration and inhibitory drive rather than in somatic GABAergic inhibition.6
In line with the idea that dystrophin is not essential during early GABAergic synaptogenesis but rather modulates or confers specialized properties to a subset of inhibitory synapses at a later stage, we show that the mechanisms required for GABAA-receptor clustering can be modulated, perhaps promoted, by induction of dystrophin-like protein expression in the brain of mature mdx mice originally devoid of this protein since ontogeny. Whereas kainic acid injections in the adult hippocampus can modulate in parallel the number of GABAA-receptor and dystrophin clusters,22 no clear direct or indirect interactions between dystrophin and GABAA-receptors have been demonstrated. Here, we provide new evidence to consider GABAA-receptor clustering as a dynamic process controlled by dystrophin expression in the adult brain, which may have an important, but yet undefined, role in activity-dependent synaptic plasticity in hippocampal circuits.
Materials and Methods
Animals and microinjections. C57BL/10ScSn-Dmdmdx/J (mdx) dystrophin-deficient and C57BL/10J littermate (WT) male mice were obtained by mating heterozygous mdx females with C57BL/10J males. Siblings were kept in the same cage (two to five per cage) under a 12-h light-dark cycle (light on: 7.00 ) with food and water ad libitum. Intrahippocampal injections of the U7-SD23/BP22 AAV2 vector packaged into an AAV1 capsid (AAV2/1) were made in 8-week-old anesthetized mdx and WT littermate mice (ketamin, 95 mg/kg; xylazin, 24 mg/kg; intraperitoneal). Recombinant AAV2/1 viral vector (rAAV2/1) or saline (phosphate-buffered saline, 0.1 mol/l) solutions were injected bilaterally into SR of dorsal hippocampus (−2 mm from bregma; 1.5 mm lateral; −1.5 mm from dura). A total volume of 3 µl was infused in each site at a rate of 0.2 µl/minute, which corresponded to 6.3 × 109 vector genome for AAV (vector titration: 2.1 × 1012 vector genome/ml). Mice were given a 4-month recovery period before being sacrificed for mRNA and protein analyses. Mouse phenotype was verified by postmortem histological analysis of quadriceps muscle.5
All experiments were conducted under appropriate biological containment in accordance with European Communities Council Directive (CEE 86/609) for animal care and experimentation, and were conducted following the guidelines of the animal facility in Orsay (France) approved by the national direction of veterinary services (Direction des Services Vétérinaires, France, agreement no. B91-471-104).
RNA and protein analyses. Brains were quickly excised after sacrifice by cervical dislocation and were frozen in powdered dry ice. Coronal 20-µm thick cryostat sections were collected from fresh-frozen brains and mounted onto superfrost slides. Hippocampal and cortical tissue samples were excised from intermediate cryosections using a scalpel and then pooled for RNA and protein analyses.
RNA analyses. Total RNA extracts were isolated using TRIzol-reagent (Life Technologies, Carlsbad, CA) and 200 ng was used for nested reverse transcription-PCR using the Access Reverse Transcription-PCR system (Promega, Charbonnieres-Les-Bains, France) as previously described.16 The first reaction was performed with one pair of primers in exons 20 (Ex20ext) and 26 (Ex26ext) for 30 cycles (94 °C/30 seconds; 55 °C/1 minute; 72 °C/2 minutes). Then 2 µl of the first reaction were amplified for 23 cycles with a second pair of primers (Ex20int and Ex26int). PCR products were analyzed using 1%-agarose gel electrophoresis in Tris-acetate-EDTA; specific bands were purified using the QIAquick gel extraction kit protocol (Qiagen, Courtaboeuf, France) for sequence analysis.
Western blotting. Proteins were first extracted by treating tissue samples with specific buffer (1% sodium dodecyl sulfate, 125 mmol/l Tris-HCl pH 6.8, 4 mol/l urea, 1% β-mercaptoethanol, 1% glycerol, 0.001% bromophenol blue) and then an aliquot was precipitated using the Compat-Able Protein Assay Preparation Reagent Set and proteins quantified with the BCA Protein Assay kit (Pierce). Proteins were resolved on 4.4% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrotransferred to polyvinylidene fluoride membranes and probed with 1:50 dilution of NCL-DYS1 (monoclonal antibody to dystrophin R8 repeat; NovoCastra, Newcastle upon Tyne, UK) followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:3,000) and ECL Analysis System (Amersham, Saclay, France).
Semiquantitative immunohistochemistry. Cryosections were processed and semiquantitative immunohistochemical analyses of GABAA-receptor clusters were performed as described elsewhere22,27,28 with some modifications. Slides were thawed for 5 minutes at room temperature, immersed in an acetone/methanol fixative solution (1:1) for 2 minutes at −20 °C, washed in three baths of phosphate-buffered saline, incubated first in a blocking solution for 40 minutes (2% normal goat serum, 0.3% Triton X-100, 1% bovine serum albumin), then overnight at 4 °C in a mixture of primary antibodies diluted in blocking solution (monoclonal antidystrophin NCL-DYS1, diluted 1:5, NovoCastra; polyclonal anti-GABAA-receptor α2 subunit, diluted 1:250; Sigma-Aldrich, St Louis, MO), washed and incubated with secondary antibody conjugated to Cy3 or Alexa 488 (Jackson Immunoresearch, Suffolk, UK) diluted 1:500 for 1 hour at room temperature. Controls were prepared by omitting primary antibody; in these controls, no specific staining could be detected.
Images were collected using a laser scanning confocal microscope with simultaneous dual-channel recording of double-labeled sections. They were taken at equivalent locations and exposure times and the intensity of the excitation lines (488 and 568 nm) was adjusted to eliminate possible cross-excitation of the fluorochromes. Typically, stacks of 7–8 images (512 × 512 pixels) spaced by 1 µm were recorded at a magnification of 225 nm/pixel. For quantification of GABAA-receptor and dystrophin clusters, digital images were processed with the WCIF ImageJ imaging system (http://www.uhnresearch.ca/facilities/wcif/imagej/) as follows: The punctate IR for GABAA receptors and dystrophin representing presumptive postsynaptic clusters was analyzed semiquantitatively in SP and SR of the CA1 subfield of dorsal hippocampus. Quantification of GABAA-receptor and dystrophin clusters was performed on single confocal images taken randomly along the CA1 layer (230 × 230 µm, resolution: 225 nm/pixel), with a minimal size of clusters arbitrarily set to 0.1 µm2, corresponding to two adjacent pixels at the magnification used. A threshold segmentation algorithm was used for automatic detection of clusters. The relative area covered by clusters, the number and size of the clusters were analyzed within a total tissue surface of 10,000 µm2 derived from 5–6 brain sections per animal, for both sampled hippocampal subregions. Clusters double-labeled for dystrophin and the α2 GABAA-receptor subunit were identified using a colocalization algorithm based on Mander's coefficient in the JACoP plug-in of ImageJ.28 Data are presented as means ± SEM. One-way analysis of variances were used for group comparisons. P values <0.05 were considered statistically significant.
Acknowledgments
This work was supported by grants from AFM (Association Française contre les Myopathies, France) to Cyrille Vaillend (grant no. DdT1 2006 & MED2 2008). The authors are grateful to Nathalie Samson and Pascale Veyrac (CNRS, France) and to Sandra Vandergeenst (Université Paris Sud, France) for animal care. We thank Régine Robichon (CNRS, France) and Serge Marty (CNRS, France) for advice on immunohistochemistry, and Ruben Miranda (University of Balearic Islands, Spain) and Fabrice P. Cordelières (Institut Curie, France) for advice on immunofluorescence quantification.
REFERENCES
- Hinton VJ, De Vivo DC, Nereo NE, Goldstein E., and, Stern Y. Poor verbal working memory across intellectual level in boys with Duchenne dystrophy. Neurology. 2000;54:2127–2132. doi: 10.1212/wnl.54.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desguerre I, Christov C, Mayer M, Zeller R, Becane HM, Bastuji-Garin S, et al. Clinical heterogeneity of duchenne muscular dystrophy (DMD): definition of sub-phenotypes and predictive criteria by long-term follow-up. PLoS ONE. 2009;4:e4347. doi: 10.1371/journal.pone.0004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Head SI, Rae C., and, Morley JW. Brain function in Duchenne muscular dystrophy. Brain. 2002;125 Pt 1:4–13. doi: 10.1093/brain/awf012. [DOI] [PubMed] [Google Scholar]
- Perronnet C., and, Vaillend C.2010Dystrophins, utrophins and associated scaffolding complexes: Role in mammalian brain and implications for therapeutic strategies J Biomed Biotechin press. [DOI] [PMC free article] [PubMed]
- Vaillend C, Billard JM., and, Laroche S. Impaired long-term spatial and recognition memory and enhanced CA1 hippocampal LTP in the dystrophin-deficient Dmd(mdx) mouse. Neurobiol Dis. 2004;17:10–20. doi: 10.1016/j.nbd.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Miranda R, Sébrié C, Degrouard J, Gillet B, Jaillard D, Laroche S, et al. Reorganization of inhibitory synapses and increased PSD length of perforated excitatory synapses in hippocampal area CA1 of dystrophin-deficient mdx mice. Cereb Cortex. 2009;19:876–888. doi: 10.1093/cercor/bhn135. [DOI] [PubMed] [Google Scholar]
- Vaillend C, Rendon A, Misslin R., and, Ungerer A. Influence of dystrophin-gene mutation on mdx mouse behavior. I. Retention deficits at long delays in spontaneous alternation and bar-pressing tasks. Behav Genet. 1995;25:569–579. doi: 10.1007/BF02327580. [DOI] [PubMed] [Google Scholar]
- Knuesel I, Mastrocola M, Zuellig RA, Bornhauser B, Schaub MC., and, Fritschy JM. Short communication: altered synaptic clustering of GABAA receptors in mice lacking dystrophin (mdx mice) Eur J Neurosci. 1999;11:4457–4462. doi: 10.1046/j.1460-9568.1999.00887.x. [DOI] [PubMed] [Google Scholar]
- Grady RM, Wozniak DF, Ohlemiller KK., and, Sanes JR. Cerebellar synaptic defects and abnormal motor behavior in mice lacking α- and β-dystrobrevin. J Neurosci. 2006;26:2841–2851. doi: 10.1523/JNEUROSCI.4823-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi M, Zushida K, Yoshida M, Maekawa M, Kamichi S, Yoshida M, et al. 2009A deficit of brain dystrophin impairs specific amygdala GABAergic transmission and enhances defensive behaviour in mice Brain 132Pt 1): 124–135. [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Schweizer C, Brünig I., and, Lüscher B. Pre- and post-synaptic mechanisms regulating the clustering of type A γ-aminobutyric acid receptors (GABAA receptors) Biochem Soc Trans. 2003;31 Pt 4:889–892. doi: 10.1042/bst0310889. [DOI] [PubMed] [Google Scholar]
- Craig AM., and, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Torelli S., and, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- Daoud F, Angeard N, Demerre B, Martie I, Benyaou R, Leturcq F, et al. Analysis of Dp71 contribution in the severity of mental retardation through comparison of Duchenne and Becker patients differing by mutation consequences on Dp71 expression. Hum Mol Genet. 2009;18:3779–3794. doi: 10.1093/hmg/ddp320. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang CM, Clark KR., and, Sferra TJ. Recombinant AAV serotype 1 transduction efficiency and tropism in the murine brain. Gene Ther. 2003;10:1528–1534. doi: 10.1038/sj.gt.3302011. [DOI] [PubMed] [Google Scholar]
- Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Santiago AC., and, Shammah-Lagnado SJ. Afferent connections of the amygdalopiriform transition area in the rat. J Comp Neurol. 2005;489:349–371. doi: 10.1002/cne.20637. [DOI] [PubMed] [Google Scholar]
- Lidov HG, Byers TJ., and, Kunkel LM. The distribution of dystrophin in the murine central nervous system: an immunocytochemical study. Neuroscience. 1993;54:167–187. doi: 10.1016/0306-4522(93)90392-s. [DOI] [PubMed] [Google Scholar]
- Knuesel I, Zuellig RA, Schaub MC., and, Fritschy JM. Alterations in dystrophin and utrophin expression parallel the reorganization of GABAergic synapses in a mouse model of temporal lobe epilepsy. Eur J Neurosci. 2001;13:1113–1124. doi: 10.1046/j.0953-816x.2001.01476.x. [DOI] [PubMed] [Google Scholar]
- Danos O. AAV vectors for RNA-based modulation of gene expression. Gene Ther. 2008;15:864–869. doi: 10.1038/gt.2008.69. [DOI] [PubMed] [Google Scholar]
- Desmaris N, Verot L, Puech JP, Caillaud C, Vanier MT., and, Heard JM. Prevention of neuropathology in the mouse model of Hurler syndrome. Ann Neurol. 2004;56:68–76. doi: 10.1002/ana.20150. [DOI] [PubMed] [Google Scholar]
- Brünig I, Suter A, Knuesel I, Lüscher B., and, Fritschy JM. GABAergic terminals are required for postsynaptic clustering of dystrophin but not of GABA(A) receptors and gephyrin. J Neurosci. 2002;22:4805–4813. doi: 10.1523/JNEUROSCI.22-12-04805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévi S, Grady RM, Henry MD, Campbell KP, Sanes JR., and, Craig AM. Dystroglycan is selectively associated with inhibitory GABAergic synapses but is dispensable for their differentiation. J Neurosci. 2002;22:4274–4285. doi: 10.1523/JNEUROSCI.22-11-04274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoè-Pognetto M, Panzanelli P, Sieghart W., and, Fritschy JM. Colocalization of multiple GABA(A) receptor subtypes with gephyrin at postsynaptic sites. J Comp Neurol. 2000;420:481–498. doi: 10.1002/(sici)1096-9861(20000515)420:4<481::aid-cne6>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Bolte S., and, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224 Pt 3:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]