Abstract
Mouse models with liver-specific expression of firefly luciferase were developed that enable a noninvasive and longitudinal assessment of small-interfering RNA (siRNA)–mediated gene silencing in hepatocytes of live animals via bioluminescence imaging. Using these models, a set of lipid nanoparticles (LNPs) with different compositions of cationic lipids, polyethylene glycol (PEG), and cholesterol, were tested for their abilities in delivering a luciferase siRNA to the liver via systemic administration. A dose-dependent luciferase knockdown by LNP/siRNA assemblies was measured by in vivo bioluminescence imaging, which correlated well with the results from parallel ex vivo analyses of luciferase mRNA and protein levels in the liver. RNA interference (RNAi)–mediated target silencing was further confirmed by the detection of RNAi-specific target mRNA cleavage. A single dose of LNP02L at 3 mg/kg (siRNA) caused 90% reduction of luciferase expression and the target repression lasted for at least 10 days. With identical components, LNPs containing 2% PEG are more potent than those with 5.4% PEG. Our results demonstrate that these liver-luciferase mouse models provide a powerful tool for a high-throughput evaluation of hepatic delivery platforms by noninvasive imaging and that the molar ratio of PEG lipid can affect the efficacy of LNPs in silencing liver targets via systemic administration.
Introduction
Small-interfering RNAs (siRNAs) can repress gene expression specifically and efficiently by triggering intracellular RNA interference (RNAi) and thus have a great potential to become a new modality of therapeutics.1 To harness siRNAs for broad therapeutic applications, an effective and safe delivery of siRNAs to target organs and cells via systemic administration is required, which represents a major challenge.2,3 Although systemic delivery of siRNA to a target organ involves multiple steps including siRNA biodistribution, intracellular uptake and endosomal release etc., target gene repression is the pharmacodynamic end point and can be used to evaluate the functionality of delivery platforms. Considering that systemic delivery of siRNA cannot be recapitulated in any in vitro system, the discovery and development of siRNA delivery platforms rely on an accurate assessment of siRNA-mediated gene knockdown in target organs or cells of an intact animal. Therefore, animal models that enable a quantitative and high-throughput evaluation of siRNA-induced gene silencing in a specific organ or tissue are highly desired.
The luciferase transgenic mouse models proved to be instrumental for monitoring certain biological processes including gene expression via bioluminescence imaging.4,5,6,7,8 A mouse model with ubiquitous expression of firefly luciferase was previously described for studying siRNA pharmacodynamics.9 Although such a model offers the opportunity to assess siRNA-induced luciferase silencing in multiple organs via ex vivo analyses, it cannot be used for noninvasive assessment of luciferase knockdown in any organ by in vivo bioluminescence imaging, owing to a high background resulting from the expression of luciferase in skin, muscle, and adjacent organs. On the contrary, mouse models with organ-specific expression of luciferase would not suffer from this drawback, in which siRNA-mediated knockdown of luciferase in the organ of interest can be readily measured by noninvasive bioluminescence imaging in living animals. The liver is a major organ targeted by siRNA-based therapeutics for the treatment of liver-associated diseases and delivery of siRNA to hepatocytes has been a focus of many studies.10,11,12,13,14 Mouse models with liver-specific expression of luciferase can greatly facilitate the discovery and preclinical development of hepatic delivery platforms by significantly enhancing screening throughput and enabling kinetic assessment of target silencing. In addition, these models offer a useful tool for studying siRNA activities in vivo.
Here, we report the generation of mouse models with liver specific expression of a firefly luciferase reporter from the Rosa26-LoxP-stop-LoxP-luciferase (Rosa26-LSL-Luc) strain15 using two different methods, namely by intravenous (IV) administration of Cre-expressing recombinant adenoviruses to Rosa26-LSL-Luc mice, or by crossing the Rosa26-LSL-Luc strain to an albumin-Cre (Alb-Cre) mouse line. We show that in these liver-luciferase mouse models, hepatic luciferase expression can be monitored by noninvasive bioluminescence imaging, which correlates closely with luciferase mRNA and protein measures, validating these models for evaluation of luciferase knockdown by noninvasive imaging. Using these mouse models, a group of lipid nanoparticles (LNPs), under different compositions of cationic lipid, polyethylene glycol (PEG) lipid, and cholesterol or at different molar ratios of identical components, were tested for systemically delivering a luciferase siRNA to the liver and their structure–activity relationships were explored.
Results
Generation of mouse models with liver-specific expression of luciferase
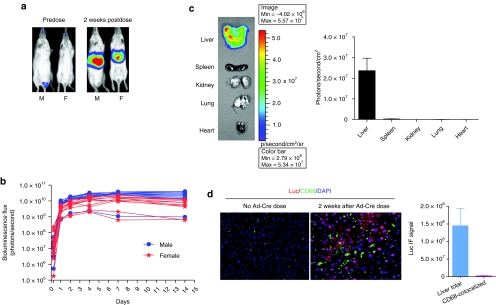
The Rosa26-LSL-Luc mouse line generated by Safran et al. is a conditional luciferase knock-in mouse strain in which the complementary DNA encoding the firefly luciferase protein, preceded by a LoxP-stop-LoxP cassette, was introduced into the Rosa26 locus, which drives transcription ubiquitously and constitutively.15 In this strain, the expression of luciferase transgene is dependent on Cre recombinase–mediated removal of the upstream floxed STOP sequence. Thus, mouse models with organ-specific expression of luciferase can be generated from the Rosa26-LSL-Luc strain either by administration of Cre-expressing recombinant adenoviruses (Ad-Cre) to the organ of interest or by crossing this strain with a tissue-specific Cre-expressing mouse line. We used both approaches to generate liver-specific luciferase mouse models. Due to a profound liver tropism, IV administered recombinant adenoviruses were shown to locate predominantly in the liver and transduce hepatocytes.16,17,18 As a quick way to produce liver-specific luciferase mice, we administered Ad-Cre to Rosa26-LSL-Luc mice via tail-vein injection and then monitored luciferase expression via noninvasive bioluminescence imaging at different times over 4 weeks. As shown in Figure 1, luciferase expression in the liver region was induced after IV administration of Ad-Cre with luciferase levels increased over time, peaked by day 7 and then persisted stably (Figure 1a,b). Follow-up biweekly imaging over 3 months showed a constant and stable expression of luciferase in the liver region (data not shown). Different levels of luciferase expression among mice receiving Ad-Cre were observed (Figure 1b). This might be due to the difference in liver size. To confirm that the luciferase expression induced by an IV dose of Ad-Cre is liver-specific, we examined luciferase expression in major organs by ex vivo imaging of individual organs 2 weeks after dosing of Ade-Cre and found that luciferase signal could be detected only in the liver, not in other organs (Figure 1c). Moreover, in situ immunofluorescent analysis of the luciferase protein in liver tissues showed that luciferase was predominantly expressed in liver parenchymal cells, whereas few Kupffer cells, stained by CD68 antibody, exhibited luciferase expression (Figure 1d). Because Kupffer cells reside in sinusoids and are movable, the lack of Ad-Cre-transduced Kupffer cells might be due to the clearance of transduced Kupffer cells from the liver. These data demonstrated that liver-specific luciferase mice can be generated from the Rosa26-LSL-Luc strain by a single IV dose of Ad-Cre. These Ad-Cre-treated mice are named Adeno-liver-Luc mice.
Figure 1.
Generation of hepatic luciferase-expressing mice by intravenous (IV) administration of Ad-Cre to Rosa26-LSL-Luc mice. (a) Mice were imaged before and 2 weeks after IV administration of Ad-Cre. The representative images (ventral view) from one male and one female are presented, showing the induction of luciferase signal in the liver region after a single IV dose of Ad-Cre. (b) Male and female mice (n = 8) were imaged at different times (0.25, 1, 2, 4, 7, and 14 days) after IV administration of Ad-Cre. Bioluminescence flux emanated from the liver region was counted and presented. (c) Two weeks after IV administration of Ad-Cre, four mice (female) were killed and ex vivo bioluminescence imaging of individual organs was conducted. A representative image of organs from one animal and the bioluminescence counts (per unit square) from different organs of four animals are presented. (d) Two weeks after IV administration of Ad-Cre or vehicle (phosphate-buffered saline), mice (female, n = 4) were killed and sections of liver lateral lobe were made and stained for the luciferase protein (red), CD68, a marker of Kupffer cells (green), and nuclei by 4′,6-diamidino-2-phenylindole (blue). Representative immunofluorescent (IF) images (×20) are shown. In addition, the total luciferase IF signal and the luciferase signal colocalized with CD68 staining from Ad-Cre-treated mice were quantified using the Slidebook software and nine randomly chosen fields from each sample were analyzed. Bars indicate SEM. F, female; M, male.
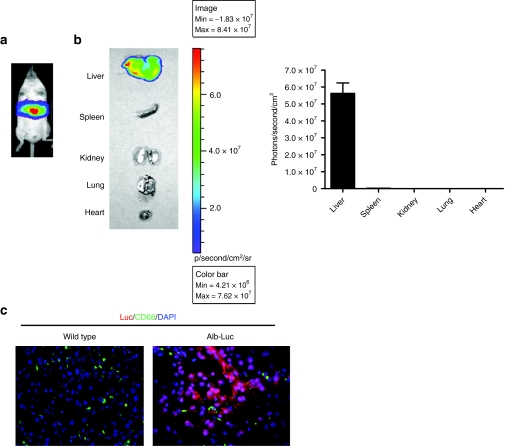
To circumvent the need of administering recombinant adenoviruses for producing liver-specific luciferase mice, we generated a mouse line with liver-specific expression of luciferase, termed Alb-Luc, by crossing the Rosa26-LSL-Luc strain with an Alb-Cre mouse line where Cre expression is controlled by the albumin promoter. Because the albumin promoter is active only in postnatal hepatocytes,19 Cre-dependent activation of luciferase expression occurs exclusively in hepatocytes in Alb-Luc mice. As shown in Figure 2, liver-specific expression of luciferase was detected by noninvasive imaging in Alb-Luc mice, which was further confirmed by ex vivo imaging of individual organs and in situ detection of the luciferase protein. The hepatic expression of luciferase is slightly higher in Alb-Luc mice than in Adeno-liver-Luc mice (Figures 1c and 2b, Supplementary Figure S1).
Figure 2.
Characterization of transgenic mice with hepatocyte-specific expression of luciferase (Alb-Luc), generated by crossing Alb-Cre males to Rosa26-LSL-Luc females. (a) In vivo bioluminescent imaging of an Alb-Luc mouse (ventral view), showing liver-restricted luciferase signal. (b) Ex vivo bioluminescent imaging of individual organs from an Alb-Luc mouse and the quantification of bioluminescence per unit square emanated from individual organs of four Alb-Luc mice. Bars indicate SEM. (c) Immunofluorescent analysis of luciferase expression in the liver of Alb-Luc and wild-type mice as described in Figure 1d. Representative images (×20) are shown. Red, green, and blue signals represent luciferase, CD68, and nucleus stains, respectively. DAPI, 4′,6-diamidino-2-phenylindole.
Validation of mouse models with liver-specific expression of luciferase for evaluating systemic delivery of siRNA to the liver
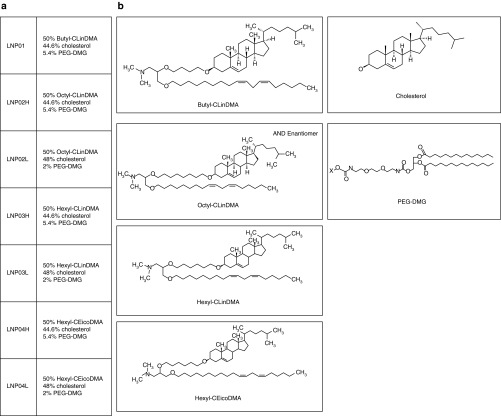
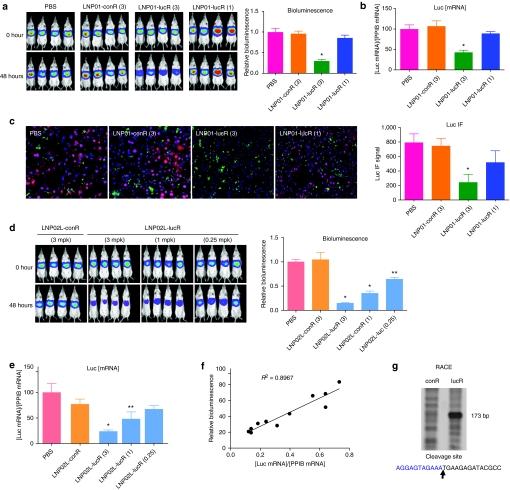
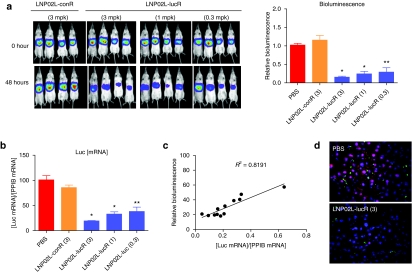
To determine whether Adeno-liver-Luc and Alb-Luc mouse models can be used for evaluating siRNA delivery to the liver via noninvasive bioluminescence imaging, we assessed LNP/siRNA assembly–mediated repression of luciferase by both noninvasive imaging and ex vivo analyses of luciferase mRNA and protein levels in the liver to examine the correlation between in vivo imaging results and the ex vivo assay data. A chemically modified luciferase siRNA (lucR) with an IC50 of 0.45 nmol/l in inhibiting luciferase expression when transfected into a HeLa-Luc cell line was selected as the payload for assembling with LNPs for systemic administration. The compositions of different LNPs and lipid structures are shown in Figure 3. LNP was assembled with lucR as described.20 The mean particle diameter is <150 nm and the siRNA encapsulation efficiency is 81–92% (Supplementary Table S1). LNP01- and LNP02L-formulated lucR (LNP01-lucR and LNP02L-lucR) were used in the validation experiments. Adeno-liver-Luc mice, in which hepatic expression of luciferase had become constant and stable for at least 2 weeks after receiving Ad-Cre, were IV administered with 3 or 1 mg/kg of LNP01-lucR (siRNA dose) or 3 mg/kg of LNP01-formulated control siRNA (LNP01-conR). Hepatic expression of luciferase before administration of LNP/siRNA assembly (basal levels), and at different times postadministration was measured by noninvasive bioluminescence imaging, and in parallel mice were killed at corresponding time points after dosing for determination of luciferase mRNA and protein levels in the liver by Taqman assay and in situ immunofluorescent analysis. Because Adeno-liver-Luc mice might display different basal levels of luciferase signal as described above (Figure 1b), bioluminescence readings from each animal at each time point after LNP treatment was normalized by its basal levels and presented as a ratio relative to the mean of vehicle (phosphate-buffered saline)-treated group at the corresponding time point (defined as relative bioluminescence). The results collected at 48 hours after LNP/siRNA treatment are presented in Figure 4a–c, showing that LNP01-lucR caused a dose-dependent repression of luciferase detected by both noninvasive imaging and ex vivo analyses with good correlations. Similarly, at 24, 72, and 96 hours after dosing, noninvasive imaging measures of luciferase knockdown well correlated with the data from parallel ex vivo analyses (data not shown). To further examine the correlation between noninvasive imaging measures and luciferase mRNA levels, Adeno-liver-Luc mice were IV dosed with LNP02L-lucR at 3, 1, and 0.25 mg/kg and luciferase expression was determined by both in vivo imaging and ex vivo Taqman assay at 48 hours. As shown in Figure 4d–f, a dose-dependent reduction of luciferase signal measured by noninvasive imaging was consistent with luciferase mRNA knockdown. In addition, LNP02L-lucR-mediated RNAi in the liver of Adeno-liver-Luc mice was confirmed by the detection of RNAi-specific luciferase mRNA cleavage by 5′-RACE (Figure 4g).
Figure 3.
LNPs: lipid compositions and structures. (a) Lipid compositions of LNPs. Molar ratios (%) are indicated. (b) Lipid structures. LNP, lipid nanoparticle.
Figure 4.
Evaluation of hepatic luciferase knockdown in Adeno-liver-Luc mice by in vivo bioluminescence imaging and ex vivo analyses. After full induction and stabilization of luciferase expression for at least 2 weeks following intravenous (IV) administration of Ad-Cre, Adeno-liver-Luc mice (n = 4) were imaged before LNP treatment (0 hour) and then dosed with LNP/small-interfering RNA (siRNA) as indicated. At 48 hours postadministration of LNP/siRNA assemblies, mice were imaged and then killed immediately to collect liver tissues for ex vivo analyses. The numbers in brackets indicate doses of siRNA (mg/kg). Bars indicate SEM. (a) In vivo imaging of Adeno-liver-Luc mice before (0 hour) and 48 hours after IV administration of LNP/siRNA assemblies or PBS (vehicle). Counts of bioluminescence measured at 48 hours were normalized by the 0 hour readings for each animal, and presented as ratios relative to the mean of PBS-treated mice (relative bioluminescence). *P < 0.005 versus PBS. (b) At 48 hours after LNP/siRNA treatment, luciferase and Ppib (a house keeping gene) mRNA levels in the liver were quantified by quantitative reverse transcription-PCR. The quantities of luciferase mRNA relative to Ppib mRNA levels are presented. *P < 0.005 versus PBS. (c) The luciferase protein levels in the liver was assessed by immunofluorescent 48 hours after LNP/siRNA treatment. Representative images (×20) are shown. Red, green, and blue signals represent luciferase, CD68, and nucleus stains, respectively. The luciferase signal was quantified using the Slidebook software and nine randomly chosen fields from each sample were analyzed. *P < 0.005 versus PBS. (d) Images and quantification of in vivo bioluminescence imaging as described in a. *P < 0.005, **P < 0.05 versus PBS. (e) Quantification of luciferase mRNA in the liver as described in b. *P < 0.005, **P < 0.05 versus PBS. (f) The correlation of luciferase bioluminescence reduction versus luciferase mRNA knockdown at 48 hours postadministration of LNP02L-lucR (n = 4). (g) Detection of RNA interference (RNAi)-specific cleavage product and site of luciferase mRNA by 5′-RACE. Liver samples collected from conR or lucR-treated mouse were subject to 5′-RACE analysis. RNAi-specific cleavage product was detected in lucR-treated samples and sequencing confirmed the cleavage site as predicted. LNP, lipid nanoparticle; PBS, phosphate-buffered saline.
Using a similar approach, we showed that in Alb-Luc mice LNP01-lucR and LNP02L-lucR caused dose-dependent repression of luciferase and that luciferase levels measured by noninvasive imaging and ex vivo assays were consistent (Figure 5a–c). In situ immunofluorescent analysis of luciferase protein in liver tissues revealed that luciferase was expressed specifically in hepatocytes as expected (Figure 5d). Taken together, both Adeno-liver-Luc and Alb-Luc mouse models can be used for evaluating siRNA delivery to the liver and studying siRNA activities in vivo by noninvasive bioluminescence imaging.
Figure 5.
Evaluation of hepatic luciferase knockdown in Alb-Luc mice by in vivo bioluminescence imaging and ex vivo analyses. Alb-Luc mice (n = 4) were imaged before LNP treatment (0 hour) and then dosed with LNP/small-interfering RNA (siRNA) as indicated. At 48 hours postadministration of LNP/siRNA assemblies, mice were imaged and then killed immediately to collect liver tissues for ex vivo analyses. The numbers in brackets indicate doses of siRNA (mg/kg). Bars indicate SEM. (a) Images and quantification of in vivo bioluminescence imaging as described in Figure 4a. *P < 0.001, **P < 0.01 versus PBS. (b) Quantification of luciferase mRNA in the liver as described in Figure 4b. *P < 0.001, **P < 0.01 versus PBS. (c) The correlation of luciferase bioluminescence reduction versus luciferase mRNA knockdown at 48 hours postadministration of LNP02L-lucR (n = 4). (d) Representative images from immunofluorescent of liver tissues. Treatment with LNP02L-lucR reduced the luciferase signal (red). LNP, lipid nanoparticle; PBS, phosphate-buffered saline.
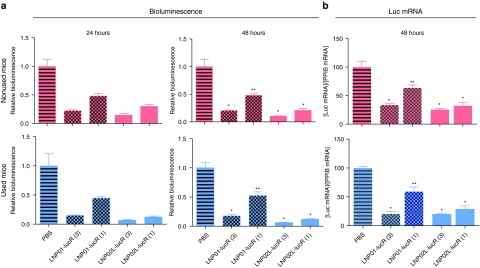
Because there is no need to kill animals for noninvasively evaluating hepatic luciferase levels by using these mouse models, we also determined whether used mice, namely the mice previously treated with LNP/siRNA assemblies, could be reused for evaluating additional LNPs after luciferase expression recovered and remained stable and constant. We compared LNP01- and LNP02L-lucR-mediated luciferase silencing between used and naive (nonused) cohorts of Adeno-liver-Luc mice. The used cohort had been dosed once with LNP01-lucR and luciferase expression in these mice had recovered and remained stable as assessed by in vivo imaging before being reused. After administration of LNP01- or LNP02-lucR, both used and naive cohorts were imaged at 24 and 48 hours and then killed at 48 hours for ex vivo assessment of luciferase mRNA levels in the liver. As shown in Figure 6, results collected from the used cohort match those generated from naive mice, indicating that these mice can be reused for LNP evaluation by noninvasive imaging.
Figure 6.
Comparison of LNP/small-interfering RNA (siRNA)-induced luciferase repression between naive (nonused) and used Adeno-liver-Luc mice. A cohort of used mice (n = 4), which had been treated once with LNP01-lucR and in which hepatic luciferase expression had recovered as assessed by in vivo imaging, and a cohort of naive mice (n = 4) were IV dosed with LNP/lucR assemblies as indicated. Hepatic luciferase expression at 24 and 48 hours after LNP/lucR treatment were assessed by in vivo imaging. Also, immediately after 48 hours imaging, liver samples were collected for ex vivo analysis of luciferase mRNA levels. (a) Qualification of bioluminescence imaging (relative bioluminescence) as described in Figure 4a. (b) Qualification of luciferase mRNA in the liver as described in Figure 4b. *P < 0.001, **P < 0.01 versus PBS. Bars indicate SEM. LNP, lipid nanoparticle; PBS, phosphate-buffered saline.
Evaluation of LNPs for delivering siRNA to hepatocytes in Adeno-liver-Luc and Alb-Luc mice by noninvasive imaging
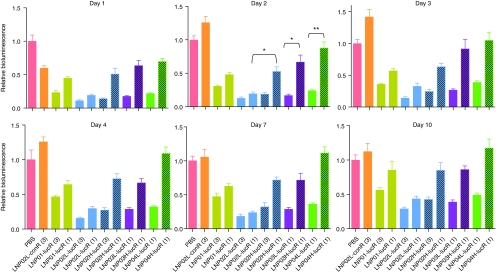
To explore the structure–activity relationships of LNPs, a set of LNPs under different compositions or at different molar ratios of identical components (Figure 3) were evaluated for their abilities in delivering lucR to hepatocytes in Adeno-liver-Luc mice. After IV administration of LNP/lucR assemblies at different doses, hepatic luciferase expression was monitored by noninvasive imaging at multiple times. A single dose of LNP02L-lucR at 3 mg/kg caused >90% target knockdown and a significant target suppression (>70%) lasted for at least 10 days (Figure 7). With identical components, LNPs containing 2% PEG (LNP02L, LNP03L, and LNP04L) were more efficacious than those containing 5.4% PEG (LNP02H, LNP03H, and LNP04H) (Figure 7). Additionally, based on the results from this study, it appears that under an identical molar ratio of CLinDMA:PEG:cholesterol (50%:5.4%:44.6%), the head linker length of CLinDMA (butyl-ClinDMA in LNP01, octyl-CLinDMA in LNP02H, and hexyl-CLinDMA in LNP03H) showed no significant effect on LNP efficacy (Figure 7). In contrast to the robust and persistent knockdown of luciferase induced by LNP/lucR assemblies, LNP02L-conR, the control assembly, at a high dose (3 mg/kg) caused a transient (<48 hours) and mild (<40%) reduction of luciferase signal (Figure 7). Similar results were obtained with another control siRNA sequence under the identical formulation (data not shown). This appears to be a nonspecific interference with luciferase expression, which might be caused by LNP-associated acute toxicity. Importantly, RNAi-mediated luciferase mRNA cleavage was only detected in LNP/lucR-treated mice, not in LNP/conR-treated mice (Figure 4g and data not shown).
Figure 7.
Evaluation of LNPs for delivering a luciferase small-interfering RNA (siRNA) (lucR) to the liver in Adeno-liver-Luc mice by noninvasive bioluminescence imaging. Adeno-liver-Luc mice (n = 4) were imaged before LNP/siRNA treatment and then intravenously dosed with PBS or one of LNP/siRNA assemblies as indicated. At different times after dosing, bioluminescence flux was measured by in vivo bioluminescent imaging and presented as relative bioluminescence as described in Figure 4a. The numbers in brackets indicate doses of siRNA (mg/kg). *P < 0.001, **P < 0.01. Bars indicate SEM. LNP, lipid nanoparticle; PBS, phosphate-buffered saline.
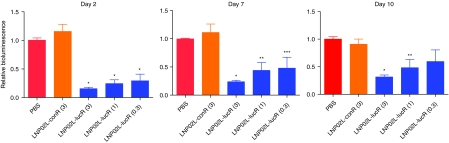
The kinetics of luciferase silencing in hepatocytes induced by LNP02L-lucR was also evaluated in Alb-Luc mice by noninvasive imaging (Figure 8). The results are consistent with those obtained from Adeno-liver-Luc mice shown in Figure 7.
Figure 8.
Evaluation of the kinetics of hepatic luciferase knockdown induced by LNP02L-lucR in Alb-Luc mice by noninvasive bioluminescence imaging. Alb-Luc mice (n = 4) were imaged before LNP/siRNA treatment and then intravenously dosed with PBS, LNP02L-conR, or LNP02L-lucR at different doses. At different times after dosing, bioluminescence flux was measured by in vivo bioluminescent imaging and presented as relative bioluminescence as described in Figure 4a. The numbers in brackets indicate doses of small-interfering RNA (mg/kg). *P < 0.001, **P < 0.01, ***P < 0.05 versus PBS. Bars indicate SEM. LNP, lipid nanoparticle; PBS, phosphate-buffered saline.
Discussion
Evaluation of systemic delivery of siRNA to a particular organ entails animal models in which a target gene expression in the organ of destination can be quantitatively measured in an efficient way. Bioluminescence imaging has proved to be a robust, sensitive, and cost-effective means to monitor luciferase expression in animals with no inherent background, superior to fluorescent imaging.21,22 Ubiquitous expression of a luciferase reporter in mice hinders noninvasive assessment of luciferase levels in a target organ by in vivo bioluminescence imaging, although it allows an ex vivo examination of luciferase expression in multiple organs.9 A prominent advantage of mouse models with organ-specific expression of a luciferase reporter is that these models enable noninvasive and longitudinal assessment of siRNA efficacy in the target organ, thereby offering a valuable tool for screening delivery vehicles and testing siRNA activities in vivo with a greatly increased throughput. A combination of the constitutively and ubiquitously active endogenous Rosa26 promoter and a very low, if any, leakage of the floxed STOP cassette located upstream of the firefly luciferase gene makes the Rosa26-LSL-Luc strain an ideal parental line from which organ/tissue-restricted luciferase-expressing mouse models can be generated.15 The ability of Ad-Cre to transduce both dividing and nondividing cells17,23,24,25,26,27 and to activate luciferase expression in transduced cells in the Rosa26-LSL-Luc strain offers a convenient way to generate organ-specific luciferase mice.15 Taking advantage of the finding that liver is the primary target after peripheral IV injection of recombinant adenoviruses,16,17,18 we generated liver-luciferase mice by tail-vein injection of Ad-Cre. Although Ad-Cre mediated Cre expression is episomal and transient, a transient expression of Cre following a single dose of Ad-Cre is able to cause a constant and permanent expression of luciferase (Figure 1) by removing the floxed STOP sequence through recombination. Both ex vivo imaging of individual organs and in situ detection of the luciferase protein confirmed that luciferase expression was induced predominantly in liver parenchymal cells and that the majority, if not all, hepatocytes expressed luciferase following a single IV dose of Ad-Cre (Figure 1). Using a similar approach, we generated lung-luciferase mice by inhalant administration of Ad-Cre (data not shown). A major drawback for generating organ-specific luciferase mice by administrating Ad-Cre is that luciferase expression induced by Ad-Cre in these mice is not inheritable and thus an administration of Ad-Cre is always required to produce luciferase-expressing mice. To circumvent this flaw, we generated a mouse line (Alb-Luc) with hepatocyte-specific expression of luciferase by crossing the Rosa26-LSL-Luc strain with an albumin promoter-Cre strain where Cre expression is hepatocyte specific. As expected, luciferase expression was detected only in hepatocytes in Alb-Luc mice (Figure 2).
There are two major concerns over the approach to use luciferase bioluminescence as a surrogate for measuring siRNA-induced luciferase mRNA knockdown: (i) How can we control for the different basal levels of luciferase among animals and the potential variations from different bioluminescence measurements, and (ii) does luciferase-dependent bioluminescence always correlate with luciferase mRNA levels? To address the first issue, we determined basal levels of luciferase in each animal 24 hours before LNP treatment and all later measures were normalized by its basal levels from the same animal. In addition, the bioluminescence measures at each time point were presented as a ratio relative to the mean of vehicle-treated animals at the corresponding time to control for potential variations between bioluminescence measurements. To examine the correlations between bioluminescence measures and mRNA levels, we determined the luciferase knockdown caused by two LNPs at different doses using different methods. Our results validated the correlation between noninvasive bioluminescence measures and luciferase mRNA levels in both Adeno-liver-Luc and Alb-Luc mice (Figures 4 and 5).
Using liver-luciferase mice, we evaluated a set of LNPs for their abilities to deliver siRNA to the liver by noninvasive bioluminescence imaging to explore the impact of molar ratio of PEG-DMG and the head linker length of CLinDMA on hepatic delivery activity (Figures 3 and 7). The activities of these LNPs were differentiated by the bioluminescence imaging results. We found that molar ratio of PEG lipid could affect LNP efficacy. This is not unanticipated. Coating with PEG lipid could provide “stealthness” to mitigate clearance by the reticular endothelial system and reduce particle size (Supplementary Table S1), whereas it might interfere with LNP-mediated intracellular delivery.28,29,30 Therefore, an optimal ratio of PEG lipid should be selected to reach a balance between the pros and cons conferred by PEGylation. Our observation that LNPs containing 2% PEG-DMG (LNP02L, LNP03L, and LNP04L) were more efficacious than the corresponding LNPs with identical components but containing 5.4% PEG-DMG (LNP02H, LNP03H, and LNP04H) indicated that this is the case. Also, our results from this model showed that the head linker length (butyl, hexyl, or octyl) of CLinDMA did not have significant effect on delivery activity under an identical molar ratio (50%:44.6%:5.4%) of CLinDMA:cholesterol:PEG-DMG. Taken together, this report shows that mouse models with liver-specific expression of luciferase can be used to screen liver delivery vehicles, study structure–activity relationships of delivery platforms and test siRNA structure–activity relationships by noninvasive imaging.
Materials and Methods
Mice. All mouse studies were conducted at AAALAC-accredited Merck Research Laboratories' animal facility located at West Point, PA, and all study protocols were approved by Merck West Point Institutional Animal Care and Use Committee. All mice were mated and bred at Taconic Farms (Hudson, NY). The Rosa26-LSL-Luc strain was licensed from Dana-Farber Cancer Research Institute. Alb-Cre mice (purchased from the Jackson Laboratory, Bar Harbor, ME) were mated with C57BL6TRYC2J mice (albino C57BL6; Jackson Laboratory) and then backcrossed onto the C57BL6TRYC2J background for 10 generations. N10 Tyrc, Alb-Cre mice were subsequently crossed with Rosa26-LSL-Luc mice and then intercrossed. Offsprings homozygous for Rosa26-LSL-Luc, Tyrc and Alb-cre (Alb-Luc mice) were identified by genotyping and the line was maintained on an FVB/C57BL6TRYC2J background. Adeno-liver-Luc mice were generated by tail-vein injection of Ad-Cre recombinant adenoviruses (Vector Biolabs, Eagleville, PA) at a dose of 5E10 viral particles and induction of luciferase expression was monitored by noninvasive imaging.
Bioluminescence imaging. Bioluminescence imaging was performed with an IVIS Spectrum imaging system (Caliper Life Sciences, Almeda, CA). For noninvasive imaging, mice were administered intraperitoneally with 𝒹-luciferin (Caliper Life Sciences) at a dose of 90 mg/kg. Five minutes after receiving 𝒹-luciferin, mice were anesthetized in a chamber with 3% isoflurane and placed on the imaging platform while being maintained on 3% isoflurane via a nose cone. Mice were imaged at 10–15 minutes postadministration of 𝒹-luciferin using an exposure time of 1 second or longer to make sure that the signal acquired is within effective detection range (above noise levels and below CCD saturation limit). Bioluminescence values were quantified by measuring photon flux (photons/second) in the region of interest where bioluminescence signal emanated using the Living IMAGE Software provided by Caliper (Hopkinton, MA). For ex vivo imaging of individual organs, mice were killed 5 minutes postadministration of 𝒹-luciferin, and organs were immediately collected and imaged. Bioluminescence values were calculated by measuring photons/second for each organ.
LNP/siRNA assemblies. A chemically modified siRNA including 2′-F pyrimidine, 2′-Ome, or deoxy purines at ribose and inverted abasic end caps at the passenger strand as described11,20 for inhibiting firefly luciferase and an irrelevant siRNA with similar chemical modifications20 for control were synthesized at Merck (San Francisco, CA). The guide (antisense) strand sequence of the luciferase siRNA (lucR) is as follows:
lucR: 5′-UAUCUCUUCAUAGCCUUAUUU-3′
siRNAs were encapsulated into LNPs to produce LNP–siRNA assemblies as described.20 Particle size was measured by dynamic light scattering using a Zetasizer (Malvern Instruments, Westborough, MA). siRNA encapsulation efficiency was determined by a RiboGreen assay (Invitrogen, Carlsbad, CA). The potential endotoxin contamination was examined using a chromogenic limulus amebocyte lysate assay (Lonza, Basel, Switzerland) and in all liposomal siRNA preparations used in our animal studies, the endotoxin levels at the highest dose of LF01-siRNA were below the endotoxin release limit for humans (5 EU/kg), defined by US Federal Food and Drug Administration and World Health Organization.
Quantification of mRNA. Quantitative reverse transcription-PCR assays were used to quantify luciferase mRNA levels relative to the housekeeping gene Ppib in lysates prepared from tissues using ABI7900 (Applied Biosystems, Foster City, CA). The primers and probe for measuring luciferase mRNA are as follows:
Forward primer: 5′-CCGGCGCCATTCTATCC-3′
Reverse primer: 5′-TCTTCATAGCCTTATGCAGTTGCT-3′
Probe: 5′-FAM-TGGAAGATGGAACCGC-3′
Immunofluorescent assay. Portions of mouse liver (left lateral lobe) were embedded in OCT and 5-µm cryosections were prepared on plus slides (Fisher Scientific, Waltham, MA). The sections were fixed in freshly prepared 4% paraformaldehyde and endogenous peroxides were blocked with 0.3% H2O2 in methanol. Sections were incubated with rabbit anti-luciferase antibody (L-0159; Sigma, St Louis, MO) diluted 1:300 in 10% normal goat serum for 1 hour at room temperature, followed by incubation with goat anti-rabbit antibody conjugated to Alexa 488 (Invitrogen) at 1:50 for 30 minutes at room temperature. Each of the above steps were followed with three washes in phosphate-buffered saline, 5 minutes each. Slides were cover-slipped with Vectashield containing 4′,6-diamidino-2-phenylindole (Vector Biolabs) and imaged on an Olympus BX51 Fluorescent microscope fitted with a prior motorized stage and a Hamamatsu Digital CCD camera (Center Valley, PA). Image analysis was completed using the Slidebook software suite (version 4.2; Intelligent Imaging Innovations, Denver, CO).
5′-RACE. The RNAi-specific mRNA cleavage product was determined using the GeneRacer Kit (Invitrogen). RACE analysis was carried out according to the supplied protocol, omitting the dephosphorylation step. The total liver RNA was ligated to the GeneRacer adaptor, reverse transcribed using a gene-specific primer (5′-GTAGGCTGCGAAATGCCCATACT-3′) and amplified by PCR using a primer complementary to the adaptor and a luciferase-specific primer (5′-CGCAACTGCAACTCCGATAAAT-3′). The PCR product was further amplified using the nested PCR primer for the adaptor, and a luciferase primer (5′-TTGTATTCAGCCCATATCGTTTCA-3′). This173-bp fragment was cloned into the pCR4-TOPO plasmid (Invitrogen) and sequenced.
Statistics. Statistic significance (P value) was determined by Student's t-test (unpaired, one-tailed). R2 was determined using linear regression.
SUPPLEMENTARY MATERIAL Figure S1. Bioluminescence flux emanated from the liver region of Alb-Luc mice and Adeno-liver-Luc mice. Alb-Luc and Adeno-liver-Luc (2 weeks post IV administration of Ad-Cre) mice (n=20) were imaged to evaluate the total photons emitted from the liver region. Mean ± SEM are presented. Bars indicate SEM. Table S1. The particle sizes of LNP/lucR assemblies and siRNA encapsulation efficiency.
Acknowledgments
We thank Myung Kyun Shin and Thomas Vogt for advice and assistance in generating Alb-Luc mice. We thank Weimin Wang and the departments of Medicinal Chemistry and Pharmaceutical Research and Development for analyzing and providing LNP/siRNA nanoparticles. We also thank Tsing-bau Chen, Xianzhi Mao, and the Laboratory of Animal Resources for technical support. We are very grateful to Alan Sachs, Paul Burke, Cyrille Sur, and David Williams for helpful discussions.
Supplementary Material
Bioluminescence flux emanated from the liver region of Alb-Luc mice and Adeno-liver-Luc mice. Alb-Luc and Adeno-liver-Luc (2 weeks post IV administration of Ad-Cre) mice (n=20) were imaged to evaluate the total photons emitted from the liver region. Mean ± SEM are presented. Bars indicate SEM.
The particle sizes of LNP/lucR assemblies and siRNA encapsulation efficiency.
REFERENCES
- Dorsett Y., and, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM., and, Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126:231–235. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp-Lorenzino L., and, Ruddy M. Challenges and opportunities for local and systemic delivery of siRNA and antisense oligonucleotides. Clin Pharmacol Ther. 2008;84:628–632. doi: 10.1038/clpt.2008.174. [DOI] [PubMed] [Google Scholar]
- Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, et al. Bioluminescence imaging of myeloperoxidase activity in vivo. Nat Med. 2009;15:455–461. doi: 10.1038/nm.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman B, Hall DE, Bhojani MS, Hamstra DA, Chenevert TL, Ross BD, et al. Noninvasive real-time imaging of apoptosis. Proc Natl Acad Sci USA. 2002;99:16551–16555. doi: 10.1073/pnas.252644499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Prior JL, Luker GD., and, Piwnica-Worms D. Generation of a highly inducible Gal4–>Fluc universal reporter mouse for in vivo bioluminescence imaging. Proc Natl Acad Sci USA. 2008;105:15932–15937. doi: 10.1073/pnas.0801075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Kim WY, O'Connell F, Flippin L, Günzler V, Horner JW, et al. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci USA. 2006;103:105–110. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S., and, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods. 2005;2:607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- Svensson RU, Shey MR, Ballas ZK, Dorkin JR, Goldberg M, Akinc A, et al. Assessing siRNA pharmacodynamics in a luciferase-expressing mouse. Mol Ther. 2008;16:1995–2001. doi: 10.1038/mt.2008.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Kim WY, Kung AL, Horner JW, DePinho RA., and, Kaelin WG., Jr Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- Herz J., and, Gerard RD. Adenovirus-mediated transfer of low density lipoprotein receptor gene acutely accelerates cholesterol clearance in normal mice. Proc Natl Acad Sci USA. 1993;90:2812–2816. doi: 10.1073/pnas.90.7.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford-Perricaudet LD, Makeh I, Perricaudet M., and, Briand P. Widespread long-term gene transfer to mouse skeletal muscles and heart. J Clin Invest. 1992;90:626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TA, Mehaffey MG, Kayda DB, Saunders JM, Yei S, Trapnell BC, et al. Adenovirus mediated expression of therapeutic plasma levels of human factor IX in mice. Nat Genet. 1993;5:397–402. doi: 10.1038/ng1293-397. [DOI] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- Abrams MT, Koser ML, Seitzer J, Williams SC, Dipietro MA, Wang W, et al. Mol Ther. epub ahead of print; 2009. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag PR, Olomu IN, Stevenson DK., and, Contag CH. Bioluminescent indicators in living mammals. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- Weissleder R., and, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- Van Doren K., and, Gluzman Y. Efficient transformation of human fibroblasts by adenovirus-simian virus 40 recombinants. Mol Cell Biol. 1984;4:1653–1656. doi: 10.1128/mcb.4.8.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akli S, Caillaud C, Vigne E, Stratford-Perricaudet LD, Poenaru L, Perricaudet M, et al. Transfer of a foreign gene into the brain using adenovirus vectors. Nat Genet. 1993;3:224–228. doi: 10.1038/ng0393-224. [DOI] [PubMed] [Google Scholar]
- Cristiano RJ, Smith LC, Kay MA, Brinkley BR., and, Woo SL. Hepatic gene therapy: efficient gene delivery and expression in primary hepatocytes utilizing a conjugated adenovirus-DNA complex. Proc Natl Acad Sci USA. 1993;90:11548–11552. doi: 10.1073/pnas.90.24.11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragot T, Vincent N, Chafey P, Vigne E, Gilgenkrantz H, Couton D, et al. Efficient adenovirus-mediated transfer of a human minidystrophin gene to skeletal muscle of mdx mice. Nature. 1993;361:647–650. doi: 10.1038/361647a0. [DOI] [PubMed] [Google Scholar]
- Li Q, Kay MA, Finegold M, Stratford-Perricaudet LD., and, Woo SL. Assessment of recombinant adenoviral vectors for hepatic gene therapy. Hum Gene Ther. 1993;4:403–409. doi: 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- Immordino ML, Dosio F., and, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- Jain A., and, Jain SK. PEGylation: an approach for drug delivery. A review. Crit Rev Ther Drug Carrier Syst. 2008;25:403–447. doi: 10.1615/critrevtherdrugcarriersyst.v25.i5.10. [DOI] [PubMed] [Google Scholar]
- Ogris M, Steinlein P, Carotta S, Brunner S., and, Wagner E. DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3:E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioluminescence flux emanated from the liver region of Alb-Luc mice and Adeno-liver-Luc mice. Alb-Luc and Adeno-liver-Luc (2 weeks post IV administration of Ad-Cre) mice (n=20) were imaged to evaluate the total photons emitted from the liver region. Mean ± SEM are presented. Bars indicate SEM.
The particle sizes of LNP/lucR assemblies and siRNA encapsulation efficiency.