Abstract
Despite rapid progress in the development of potent and selective small interfering RNA (siRNA) agents for skin disorders, translation to the clinic has been hampered by the lack of effective, patient-friendly delivery technologies. The stratum corneum poses a formidable barrier to efficient delivery of large and/or charged macromolecules including siRNAs. Intradermal siRNA injection results in effective knockdown of targeted gene expression but is painful and the effects are localized to the injection site. The use of microneedle arrays represents a less painful delivery method and may have utility for the delivery of nucleic acids, including siRNAs. For this purpose, we developed a loadable, dissolvable protrusion array device (PAD) that allows skin barrier penetration. The PAD tips dissolve upon insertion, forming a gel-like plug that releases functional cargo. PAD-mediated delivery of siRNA (modified for enhanced stability and cellular uptake) resulted in effective silencing of reporter gene expression in a transgenic reporter mouse model. PAD delivery of luciferase reporter plasmids resulted in expression in cells of the ear, back, and footpad skin as assayed by intravital bioluminescence imaging. These results support the use of PADs for delivery of functional nucleic acids to cells in the skin with an efficiency that may support clinical translation.
Introduction
A number of skin disorders involve inappropriate or mutant gene expression1 and would benefit from the development of effective small interfering RNA (siRNA) therapeutics. Delivery of siRNA to keratinocytes in vitro and in vivo can efficiently silence pathological expression of mutant genes involved in monogenic skin diseases; for example, mutations in the keratin 6, 16, and 17 genes lead to the rare skin disorder pachyonychia congenita.2,3 An siRNA targeting the N171K mutation in keratin 6a has been identified that is highly specific and does not affect expression of the wild-type gene, which differs by only a single nucleotide.4,5 It is this exquisite specificity, as well as potency, that make siRNA therapeutics attractive, but development of therapeutic siRNA for diseases of the skin, and other disorders, has been limited by the lack of efficient “patient-friendly” delivery methodologies.4,6 In the treatment of skin diseases, the stratum corneum comprises a significant barrier to therapeutic molecules and is particularly impermeable to compounds above ~500 Da with high charge densities, such as nucleic acids.7,8 A variety of transdermal delivery methods have been explored, but to date intradermal injections continue to be the most effective.6,9
Arrays of tiny, needle-like structures, generally called microneedles, have emerged as a potentially viable method for nucleic acid delivery to skin.10 Microneedles of appropriate length theoretically allow similar access to skin layers as with hypodermic intradermal injections, and although smaller volumes of material would typically be delivered (due to the smaller size and loading capacity of the microneedles), the large number of simultaneous microinjections delivered by a microneedle array lead to a more uniform application density. Because the length of the microneedles can be controlled to limit penetration and delivery to the epidermis, and not the innervated dermis, these delivery methods tend not to be painful.11 These features make microneedle delivery very attractive for treating dermatological diseases.
In this study, we fabricated a protrusion array device (PAD), made of polyvinyl alcohol (PVA) polymer, which has the delivery features of microneedles and the sustained release characteristics of biocompatible polymers. To evaluate the performance of PAD prototypes for the delivery of nucleic acids to the skin, three lines of experimentation were utilized. First, fluorescently labeled siRNAs, including Accell siRNAs that are modified to facilitate cellular uptake, were loaded into microneedles and applied to the skin on mouse paws and siRNA distribution assessed using fluorescence microscopy of skin sections. Second, footpad skin of transgenic mice (Tg CBL/hMGFP), expressing luciferase (red click beetle luciferase [CBL] and humanized Montastrea green fluorescent protein [hMGFP]) in the epidermis,9 was treated with PADs loaded with siRNAs targeting the reporter; gene silencing was analyzed by reverse transcription quantitative PCR, fluorescence microscopy of skin sections and in vivo fluorescence imaging. Finally, to further study the ability of PADs to deliver larger nucleic acid cargos, firefly luciferase expression was analyzed by intravital bioluminescence imaging in skin following application of PADs loaded with reporter plasmid DNA.
Results
A major obstacle to developing siRNA-based therapeutics for skin disorders is overcoming the stratum corneum barrier. The natural function of the stratum corneum is to prevent water loss and exclude external agents from entering the body with a molecular cutoff of ~500 Da. Microneedles can effectively circumvent this barrier by direct skin penetration, and microneedles, comprised of biocompatible polymers as in PADs, have the added benefit of the tips remaining in the epidermis and/or dermis (depending on design and length) and can act as reservoirs, releasing the cargo as they dissolve. The PAD design tested in this study targeted the epidermis for delivery of functional nucleic acids.
PAD fabrication and loading
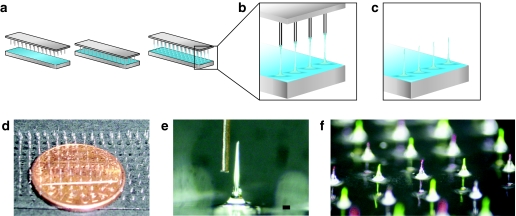
PADs were produced by bringing a pin template into contact with a thin film of PVA solution (Figure 1a, left and middle panels), and withdrawing the template under a controlled air flow to produce fiber-like structures with loadable channels (Figure 1a, right panel and Figure 1b). The dried needle structures are separated from the template, and mechanically trimmed to a uniform height with sharp beveled tips (Figure 1c,e,f). Typical fabrication and drying of PAD structures occurs at or below 50 °C (as low as 30 °C, data not shown), below established siRNA degradation temperatures,12 such that the nucleic acids remain functional. The device is removed from the substrate as a regular array of dissolvable microneedles with an integral polymer backing (Figure 1d). Typical PAD microneedles are <100-µm thick, with a sharp tip only microns across (Figure 1e). PADs can be loaded by immersing the needles in aqueous solutions or suspensions of payload materials, such as nucleic acids, drugs, vaccines, or other therapeutic agents. Furthermore, multiple payloads can be delivered from separate needle populations on the same array (Figure 1f). To show this flexibility of loading, phycobiliprotein R-phycoerythrin (RPE) and fluorescein were alternately loaded on array microneedles, demonstrating that multiple cargos can be delivered with a single device (Figure 1f).
Figure 1.
Schematic of fabrication of protrusion array device (PAD) and example images. (a) Pin template and glass slide covered with a thin film of 20% polyvinyl alcohol (PVA) solution (left panel). The pin template is placed in contact with the PVA solution (middle panel). Microneedles are produced by withdrawing the pins as the film is drying, forming fiber-like structures (right panel). (b) Enlarged view of fibers. (c) Protrusions are subsequently trimmed to the desired length and tip shape. (d) PAD supported by a glass substrate, with a penny to show scale. (e) Micrograph of one microneedle after trimming to 1 mm length, showing beveled structure that facilitates skin penetration, with a human hair to show scale (bar = 80 µm). (f) PAD needles loaded alternately with fluorescein (green) and R-phycoerythrin (red).
PADs deliver siRNA cargo across the stratum corneum
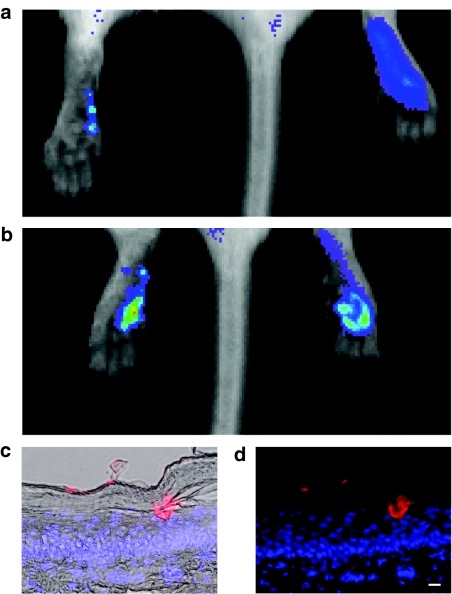
PADs were loaded with a fluorescently tagged siRNA mimic (siGLO Red) and applied to the left paw footpad. As a control, siGLO Red in phosphate-buffered saline solution was injected intradermally into the right paw. Localized fluorescence corresponding to individual microneedle penetration sites could be visualized in vivo (Figure 2a). After four applications of arrays (each containing four microneedles corresponding to a total deliverable dose of ~160–320 ng), the signal detected in the left paw (5.89 × 1010 photons/second/cm2/steradian) was similar to the signal detected following intradermal injection of 500 ng into the right paw (5.55 × 1010 photons/second/cm2/steradian) (Figure 2b). Individual microneedles loaded with siGLO Red could be observed penetrating the stratum corneum barrier to form localized depots in both the epidermis (Figure 2c,d, and Supplementary Figure S1) and dermis (Supplementary Figure S1).
Figure 2.
Imaging of individual microneedle penetration sites in vivo and microneedle plug visualization in skin sections. (a) PADs were loaded with siGLO Red (a fluorescently tagged siRNA mimic, ~20 ng/microneedle) and applied to the left footpad. As a control, 0.5 µg of siGLO Red (in 50 µl PBS) was injected intradermally into the right footpad. Mice were immediately intravitally imaged for fluorescence using the Xenogen IVIS 200 system. Localized fluorescence corresponding to individual microneedle penetration sites was observed following PAD application. (b) The application of four (4 × 1) arrays to the left foot resulted in detection of red fluorescent signal of similar magnitude to the right foot. (c,d) Fluorescence microscopy of 10 µm frozen skin sections showing a microneedle depot loaded with siGLO Red [longer exposure time (data not shown) shows initial release of siGLO Red] demonstrating drug release to the epidermis. Sections were stained with DAPI (blue) to visualize nuclei (bar = 10 µm). (c) Overlay with brightfield image. (d) No brightfield overlay. DAPI, 4,6-diamidino-2-phenylindole; PAD, protrusion array device; PBS, phosphate-buffered saline; siRNA, small interfering RNA.
Distribution of Accell Red siRNA following PAD delivery
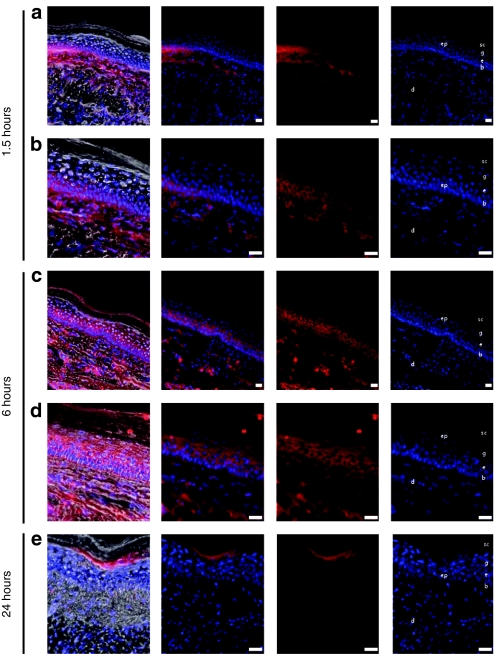
Mouse footpads were treated with PADs loaded with fluorescently tagged nontargeting Accell siRNA that was modified for greater stability and uptake. Following a 1.5- or 6-hour incubation, mice were sacrificed and frozen footpad skin sections analyzed by fluorescence microscopy. Following PAD application, fluorescently tagged siRNA distributed throughout the epidermis and dermis near the treatment site (Figure 3). The siRNA delivered in this manner exhibited cytoplasmic perinuclear localization in epidermal keratinocytes (Figure 3), suggesting cellular uptake. The siRNA was initially accumulated in the lower layers of the epidermis within 1.5 hours following PAD application (Figure 3a,b), and was widely dispersed throughout the epidermis and dermis within 6 hours (Figure 3c,d). At 24 hours post-application, the signal was detected mainly in the granulosum/corneum layers of the epidermis (Figure 3e). siGLO Red distribution followed a similar pattern, but unlike the modified siRNA, siGLO siRNA did not appear to readily bind to and/or be taken up by the keratinocytes (i.e., no perinuclear distribution, see Supplementary Figure S1) and its distribution was generally restricted to defined regions of the epidermis at or near the site of injection (data not shown).
Figure 3.
Fluorescence microscopic analysis of Accell Red siRNA distribution in mouse footpad skin. Transgenic CBL/hMGFP mouse footpads were treated with two (3 × 5) PADs loaded with Accell Red (DY-547-labeled) nontargeting siRNA. Mice were sacrificed at (a,b) 1.5, (c,d) 6, and (e) 24 hours after PAD application and footpad skin sections removed for analysis. Accell Red siRNA (red fluorescence, middle panels) distributes through dermis (d) and epidermis (ep). With this needle design and length, the red fluorescence signal is detected in the basal (b) and spinosum (s) layers at (a,b) 1.5 hours and reaches the granular layer (g) and stratum corneum (sc) at (c,d) 6 hours. Sections were stained with DAPI to visualize nuclei (right panel). Brightfield fluorescence overlay (left panel). Bar = 20 µm. CBL, click beetle luciferase; DAPI, 4,6-diamidino-2-phenylindole; hMGFP, humanized Montastrea green fluorescent protein; PAD, protrusion array device; siRNA, small interfering RNA.
Silencing of CBL/hMGFP reporter gene in transgenic mouse epidermis
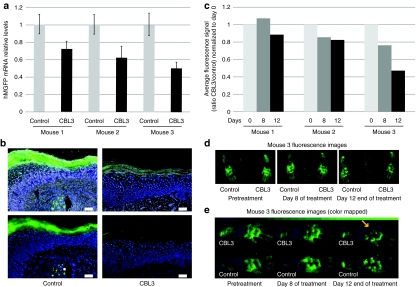
We previously demonstrated the ability of unmodified CBL3 siRNA, delivered by intradermal injection, to silence reporter gene (hMGFP) expression in the epidermis of a transgenic mouse (Tg CBL/hMGFP) skin model.9 In order to test the ability of PAD to deliver functional Accell CBL3 siRNA, mouse footpads were treated and the effect on reporter gene expression was analyzed. Three PADs (3 × 5 microneedle arrays) were applied to paw skin every other day for 12 days. On day 13, the mice were sacrificed, palm skin excised, and CBL/hMGFP reporter mRNA levels were measured by quantitative PCR (Figure 4a). A noticeable reduction (25–50%) in expression was found in all three CBL3 siRNA-treated palms compared to the counterpart palms treated with nonspecific control siRNA. Little or no reduction was found in footpads treated with nonspecific siRNAs and footpads treated with empty PADs (data not shown), although variability was observed between individuals. In addition to mRNA reduction, decreased signal from reporter protein was also observed by fluorescence microscopy of footpad sections (Figure 4b) of skin treated with CBL3 Accell siRNA (right panel) but not nonspecific control siRNA (left panel). The reduced reporter mRNA levels on each mouse analyzed correlated well with the amount of protein reduction measured by in vivo fluorescence imaging (Figure 4c). Representative images (mouse 3) demonstrating inhibition are shown in Figure 4d. Because visual discrimination of low-intensity treated areas is difficult in the default linear black-to-green false color representation in Figure 4d, Figure 4e shows the same data using a binned black–blue–green false color to better resolve these regions.
Figure 4.
CBL3 Accell siRNA-loaded PADs inhibit hMGFP expression in mouse footpad skin. Transgenic CBL/hMGFP mouse footpads were treated every 2 days with three (3 × 5) PADs loaded with either CBL3 or nonspecific control Accell siRNA for 12 days. (a) RT-qPCR analysis of Tg CBL/hMGFP mice treated with CBL3 siRNA. Total RNA, isolated from paw palm skin of three mice treated with CBL3 siRNA (right footpad) or a nonspecific control siRNA (left footpad), was reverse transcribed and hMGFP mRNA levels quantified by qPCR. The hMGFP levels were normalized to K14 levels (endogenous control). Each bar corresponds to the mean of three replicates. Bars indicate SE. (b) Fluorescence microscopy of frozen skin sections prepared from treated mice. Mice were sacrificed and frozen skin sections (10 µm) prepared. hMGFP expression (or lack thereof) was visualized by fluorescence microscopy of samples from mouse footpads treated with nonspecific (left panel) or CBL3 (right panel) Accell siRNA. Upper panel shows brightfield overlay and bottom panel shows fluorescence only. Bar = 50 µm. Nuclei are visualized by DAPI stain (blue). (c) In vivo quantification of hMGFP fluorescence. Three mice were treated with CBL3 (right paw) or nonspecific (left paw) Accell siRNA and imaged with the CRi Maestro imaging system during treatment. Quantification of the region of interest adjusted to the palm of each mouse was performed using Maestro quantification software after background subtraction. The ratio in the average signal (counts/second/mm2) of CBL3-treated palm and nonspecific-treated palm normalized to day 0 is reported in the graph. (d) hMGFP expression in mouse three imaged during treatment with the CRi Maestro imaging system. The right paw was treated with CBL3 Accell siRNA whereas the left paw was treated with nonspecific control. Images were taken using autoexpose settings and unmixed using previously defined spectra and autofluorescence. The images are pseudocolored green. (e) Enlarged view of mouse three treated paws, color-mapped to a binned black–blue–green color scheme to facilitate comparison of low-intensity regions (see Materials and Methods section). Arrow shows area of signal reduction. Color bar at top shows lookup table assignments corresponding to pixel bit values in increments of 10. CBL, click beetle luciferase; DAPI, 4,6-diamidino-2-phenylindole; hMGFP, humanized Montastrea green fluorescent protein; PAD, protrusion array device; RT-qPCR, reverse transcription quantitative PCR; SE, standard error; siRNA, small interfering RNA; Tg, transgenic.
Delivery of luciferase expression plasmid to mouse skin and visualization of expressed reporter protein by intravital imaging
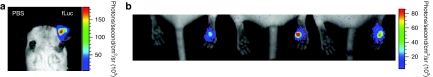
PADs containing firefly luciferase expression plasmid were applied to mouse ear, paw, and flank skin. Following luciferin administration 24 hours later, luciferase expression was assayed by in vivo bioluminescence imaging. Luminescence was detected in all the treated skin types (Figure 5a,b and Supplementary Figure S2), whereas no expression was found in skin treated with PADs loaded with phosphate-buffered saline vehicle alone. To test the consistency of plasmid delivery and expression in mouse paw skin, 12 microneedles (each containing a deliverable payload of ~12 ng pGL3-CMV-Luc/microneedle) were applied per mouse paw on day 1 and again on day 2, and imaged on day 3 by in vivo bioluminescent imaging (Figure 5b). We observed consistent luciferase expression using this method (Figure 5b and data not shown; i.e., experiment was repeated five times with similar results), demonstrating that PADs are able to reliably deliver functionally active plasmid DNA to skin cells.
Figure 5.
Analysis of fLuc reporter gene expression following PAD-mediated delivery of expression plasmids to mouse skin. (a) Ear delivery. The ear on the right was treated with a PAD loaded with ~12 ng/microneedle of pGL3-CMV-Luc plasmid (12 microneedles). The ear on the left was treated with the delivery device loaded with PBS vehicle alone. PADs were inserted into the ear for 20 minutes. After 24 hours, luciferase expression was determined following IP luciferin injection by whole animal imaging using the Xenogen IVIS 200 in vivo system (red is the highest expression level, blue lowest). (b) Footpad delivery. Right footpads were treated with PADs (12 microneedles) loaded with luciferase expression plasmid for two consecutive days and analyzed as described above. Left footpads were treated with PADs loaded with PBS vehicle alone (control). fLuc, firefly luciferase; IP, intraperitoneal; PAD, protrusion array device; PBS, phosphate-buffered saline.
Discussion
The lack of effective delivery tools for small inhibitory RNA has been a significant impediment for their use as therapeutics and development of such tools is the focus of a tremendous research effort. Each tissue target will have specific delivery requirements. The skin offers unique opportunities for delivery given its accessibility; however, its natural role as a barrier to xenobiotics presents particular challenges for nucleic acid-based therapies. The primary barrier in the skin is the stratum corneum and this barrier is readily breached with hypodermic needles, but the large surface area of the skin would require multiple injections. The use of microneedle arrays, designed specifically for appropriate depth of penetration and distribution, offer opportunities for more uniform and less painful delivery of therapeutic nucleic acids to larger areas of skin.
Since siRNAs have the potential to target any mRNA, including mutant mRNAs that differ from wild type by as little as a single nucleotide,5,13,14,15,16 these molecules are ideally suited for treating monogenic skin disorders. Recently, TD101, an siRNA targeting a mutant version of keratin 6a responsible for the dominant-negative monogenic skin disorder pachyonychia congenita,3,17 was administered by intradermal injection in a phase 1b clinical trial, with encouraging results.6 The outcome of this first siRNA clinical trial in skin included improvement in symptoms at the site of siRNA treatment, but not in the paired control injection site on the opposite foot that received vehicle alone. Importantly, no adverse events were observed. Although direct injection of “naked” nucleic acids into the skin has been suggested as the most simple, safe, and efficient delivery method,18 its efficiency is limited to a highly localized region of the epidermis coincident to the injection site, and a large number of injections may be needed to achieve a therapeutic outcome.4,6,9 Moreover, in the clinical trial, intradermal injections of either siRNA or vehicle alone were accompanied by severe pain,6 necessitating nerve blocks as well as oral pain medication at the time of injection, underscoring the need for development of “patient-friendly” delivery technologies (i.e., effective delivery of functional siRNA with little or no pain).
In an attempt to decrease the pain associated with hypodermic needle administration, while improving transdermal delivery and minimizing undesirable reactions, a variety of micron-scale devices have been developed.10 A number of research groups have tested prototype rigid needles either as pretreatments to create channels for transdermal delivery or as delivery systems themselves.10 However, typical silicon or steel microneedles used in early studies create inherent risk due to needle breakage during skin insertion, and potential complications due to the deposited foreign bodies. A significant innovation that addresses these problems and that adds additional functionality, is the use of dissolvable microneedles, in which needles hydrate after penetrating skin to form a degradable temporary drug depot.19 However, the temperatures required for current melt-cast methods (typically above 100 °C) used to produce soluble microneedles would likely cause degradation of nucleic acid payloads.12 In this study, we evaluated our recently developed PAD technology, in which dissolvable microneedles are directly formed from a film of PVA, a biocompatible polymer with a history of safe injection in humans, and an approved ingredient in the US Food and Drug Administration inactive ingredient database (http://www.accessdata.fda.gov/scripts/cder/iig/index.cfm). Importantly, the fabrication of these microneedles can be done at relatively low temperatures (≤50 °C) at which many biomolecules, including siRNAs, are stable.12
In the recent symposium “Achieving Successful Delivery of Nucleic Acids to Skin,”20 many promising technologies aiming to deliver nucleic acids to skin were presented and discussed, and the general conclusion was that the ideal delivery technique likely will be composed of multiple technologies: one to help nucleic acids traverse the stratum corneum, a second to facilitate nucleic acid uptake and efficient utilization by keratinocytes. The PAD technology presented here fulfills the first objective, i.e., delivery across the stratum corneum barrier, with deposition of siRNA (or plasmid) in the epidermis. In order to facilitate siRNA diffusion and uptake, we used modified siRNA (Accell21), that was developed to enable siRNA-specific gene silencing in the absence of carriers such as liposomes. This technology has been successfully utilized in vivo to reduce the levels of KLF6-SV1 protein, which has been associated with ovarian cancer progression and chemoresistance.22 Systemically administered siRNAs targeting KLF6-SV1 blocked tumor progression and resulted in improvement in median and overall survival in an ovarian cancer model.22 Our work in keratinocyte cell lines and human skin organotipic models (R.P. Hickerson, H. Ilves, M. Flores, D. Leake, S.A. Leachman and R.L. Kaspar, manuscript in preparation) demonstrated that these modified siRNAs were taken up by keratinocytes in the absence of other carriers and were functionally delivered to the cells. Therefore, we evaluated whether combining these two technologies (PAD to traverse the skin barrier and Accell to facilitate keratinocyte siRNA uptake) would result in functional knockdown of target. Indeed, we demonstrated reduced reporter gene expression in the Tg CBL/hMGFP mouse model via a number of complementary assays to evaluate knockdown (see Figure 4). Thus, the simultaneous use of both technologies may have the capability to functional deliver siRNA in a “patient-friendly” manner.
The ability of PADs to successfully deliver plasmid DNA payloads to mouse as well as human explant (data not shown) skin is demonstrated by detection of reporter activity, which can only occur following successful plasmid uptake and transcription and translation to luciferase protein, which then acts on its substrate luciferin to produce light that is visualized by bioluminescence imaging. It should be noted, however, that this process appears to be inefficient as only a small number of epidermal cells (probably keratinocytes due to their location, abundance, and morphology) were found to express the reporter in mouse skin as well as in ex vivo human skin using green fluorescent protein expressing plasmids (data not shown). Similar results have been previously reported by others using β-galactosidase-expressing plasmids delivered through microchannels created to increase skin permeability using silicon microneedles alone23,24 or in combination with hydrogel formulations to increase sustained release.25 Detection of β-galactosidase-expressing keratinocytes has been also used as proof of cellular viability of skin organ cultures over time.26 The limitation of gene expression to localized regions surrounding the microneedle insertion site23,24,25,26,27 suggests that a physical interaction between the microneedle and the cell may influence uptake. Other studies suggest that nucleic acids may require an additional force (e.g., pressure) to be taken up by keratinocytes.28 Therefore, in order to improve nucleic acid dispersion and uptake over wider regions of the skin, a combinatorial approach with other techniques may be beneficial.
siRNA may have advantages over plasmid DNA with respect to delivery through the stratum corneum barrier, dispersion through the epidermis, as well as internalization by cells due to its reduced size,29 which typically is over 100 times smaller than plasmid DNA. However, siRNA molecules (at ~13,000 Da) still exceed the size limit of diffusion through the stratum corneum and the negative charges from the phosphodiester backbone are an additional challenge. We have directly compared siGLO Red (an unmodified siRNA mimic) with Accell Red (an siRNA containing modifications designed to facilitate cellular uptake) following PAD delivery (see Figure 3 and Supplementary Figure S1 and data not shown). SiGLO Red was observed moderately distributed in the epidermis near the deposited “plug” following PAD administration but cellular uptake was not readily observed. In contrast, Accell Red siRNAs were widely distributed throughout the mouse paw epidermis, demonstrating improved distribution through the tissue compared to siGLO Red. Moreover, following Accell Red delivery, red fluorescence signal appeared localized mostly in the perinuclear region, suggesting uptake and cytoplasmic localization. Although further studies are needed to demonstrate the mechanism of uptake and incorporation into the RNA-induced silencing complex, the pattern of signal detected, together with the functional activity of Accell CBL3 siRNAs, is consistent with delivery of sufficient siRNA to RNA-induced silencing complex to result in the observed silencing.
The demonstration that endogenously expressing genes can be reduced by Accell siRNAs delivered by PADs opens new opportunities in the treatment of skin diseases such as pachyonychia congenita. The increased cellular uptake apparent with modified siRNA in combination with the administration and tissue delivery advantages offered by PADs address two primary obstacles in developing siRNA therapeutics (i.e., siRNA uptake by keratinocytes and overcoming the stratum corneum barrier, respectively) for skin disorders. Other physical techniques may be combined with these advances to further increase delivery at the cellular level including electroporation, iontophoresis, and sonoporation; experiments are underway to test the potential of these combinations.
Materials and Methods
Animals. Mice (FVB females, 6–8 weeks old or Crl:SKH1-h hairless) were purchased from Charles River Laboratories (Wilmington, MA). Tg CBL/hMGFP mice9 were obtained from our breeding colonies at Stanford University (Stanford, CA). Animals were treated according to the guidelines of both the National Institutes of Health and Stanford University.
PAD preparation for delivery and application
PAD production. Protrusion arrays were produced using an assembly of commercially available “pin headers” (header strip 80 pin dual row 1 mm spacing, PED-80S-P2; Pan Pacific, Santa Cruz, CA) as templates. The fabrication of the microneedles was accomplished by bringing the template pattern of projections into contact with a 1-mm thick film of 20% PVA (Spectrum, Gardena, CA) polymer solution on a glass substrate (microscope slides #1324L; Globe Scientific, Paramus, NJ). Following contact with the polymer film, the projections were withdrawn a distance of 1 cm over 13 seconds under a uniform airflow of 3.0 m/s at 45 °C. Under these conditions, needle structures can be formed in the film with a hollow interior or exterior grooves, which can be subsequently loaded through “capillary action.”
PAD loading. The hollow channels in the microneedles form capillary-like structures that self-load when brought into contact with a liquid that efficiently wets the surface. The needles were loaded manually from a micropipette tip charged with nucleic acid solution. As a microneedle enters the meniscus of an aqueous solution, the needle surface rapidly wets with solution, and solution is drawn into the loadable channel. Simultaneously, the polymer hydrates and swells, imbibing the solution. If contact with the solution is short (e.g., <30 seconds), the needle softens but does not deform noticeably, and readily redries to incorporate the payload within and external to the stiff polymer matrix.
Microneedles were produced and mechanically trimmed to a nominal 1.0-mm length and 45° bevel, and loaded with “cargos” including nucleic acids (siRNAs and plasmids, see below), proteins (2 ng/microneedle of phycobiliprotein RPE [generously provided by Quantaphy, Santa Cruz, CA]) or dyes (2 nl/needle of a 1% fluorescein solution [Sigma, St Louis, MO]). Subsequent to loading, PADs were incubated in a 50 °C vacuum oven (Isotemp Vacuum Oven Model 280A; Fisher Scientific, Waltham MA) evacuated to −18 inches mercury pressure for 4 hours to “harden” microneedles to facilitate skin penetration. (The thin backing layer remains flexible, allowing conformation to skin contours during applications.)
PAD application. PADs were applied manually to subjects to allow the needle tips to pierce the outermost skin surface (by giving to the back of the array a single flick with the finger) and were left in place for 20 minutes. During this period, the needle tips hydrated below the skin surface and softened to form a viscous gel plug. After this hydration period, the dry outer portion was removed, leaving the hydrated portions, with their cargo, embedded in the application site.
Although PVA films are flexible, even when dry, the morphology of the needle structure provides sufficient rigidity to allow effective skin penetration. The tips are cut to a bevel point that allows efficient piercing of the stratum corneum, and in practice a variety of application methods have been successful. Because arrays support simultaneous application of multiple needles, the device is self-orienting, allowing pressure to be uniformly applied normal to the skin surface, which minimizes off-axis bending. The needle tips, softened in hydrated tissues, form a viscous gel plug that remains in the tissue after removal of the arrays. The cargo is released to the hydrated compartment as the needle dissolves.
Cargo release. The PAD devices deliver ~10% of the loaded dose to subject skin, or about 2 ng siRNA/needle. This estimate is based on a small study of 100 similar needles loaded with RPE, a fluorescent protein. In the RPE study (data not shown), needles were loaded by momentary contact with an RPE solution in a manner similar to that used in the present work. Dried needles were individually photographed and assayed for RPE content en-masse by fluorescence imaging, and then applied to human skin explant. The needles were then vacuum dried and individually reimaged and collective fluorescence analysis was repeated. Fluorescence images provided needle-specific quantitative data by region-of-interest analysis using Living Image software (Caliper Life Sciences, Almeda, CA), written as an overlay on Igor image analysis software (WaveMetrics, Lake Oswego, OR) showing pre- and postdelivery RPE content used to calculate delivery efficiency.
Analysis of siRNA distribution in mouse footpad skin. PAD devices were loaded with ~20 ng/microneedle of the DY-547 (a Cy3 analog; 557 nm excitation, 570 nm emission) fluorescently tagged siRNA mimic siGLO Red or Accell Red Nontargeting siRNA (Dharmacon Products, Thermo Fisher Scientific, Lafayette, CO). PADs were applied to the left paw of an FVB mouse using four consecutive applications (to maximize dose and coverage of skin) of arrays (1 × 4) containing four microneedles each. Each PAD was applied for 20 minutes to maximize hydration of the penetrating microneedles. As a positive control, the right paw received 0.5 µg of siGLO Red in 50-µl phosphate-buffered saline solution via intradermal injection. Mouse paws were imaged in an IVIS 200 Imaging System (Xenogen product from Caliper Life Sciences) using the DsRed filter set (excitation at 460–490 nm and 500–550 nm; emissions at 575–650 nm) during 1 second acquisition time. The resulting emitted light was quantified using LivingImage software as was described above. Background (DsRed setting) was subtracted and raw values were reported as photons/second/cm2/steradian. PADs in 3 × 5 (15 needles contained in a rectangular array of needles spaced at a 2 mm pitch) array format were also applied to FVB or Tg CBL/hMGFP mice. Following CO2 asphyxiation at the indicated times, skin tissue was dissected, embedded in O.C.T., and frozen sections analyzed using a red filter set (546 nm excitation; 580 nm emission) in an Axio Observer Inverted Fluorescence Microscope (Zeiss, Thornwood, NY) to visualize release from microneedle and biodistribution of siRNAs in skin. The images were taken with an AxioCam MRm (Zeiss) camera using AxioVs40 V4.6.3.0 software (Zeiss).
In vivo delivery of siRNAs using PADs and analysis of gene silencing. PADs containing ~20 ng/microneedle (estimated 10% to be released following array application) of Accell CBL3 and nonspecific control Accell siRNAs (Dharmacon) were applied for 20 minutes to right and left footpads, respectively, of Tg CBL/hMGFP mice anesthetized with 2% isofluorane. Mice were treated every 48 hours (to allow for recovery of treated paws) with three arrays (4 × 5 needle array 2 mm spacing between needles) per treatment for 12 days (treatment duration was determined based on previously published results,28 optimized to facilitate detection of green fluorescence reduction in the Tg hMGFP/CBL mouse model). The mice were sacrificed on day 13 of the experiment and treated footpad tissues removed. Footpad skin from one mouse per cohort was embedded in O.C.T. compound (Tissue-Tek, Torrance, CA) and frozen in dry ice. Vertical cross-sections (10 µm) were prepared and mounted with Hydromount (National Diagnostic, Highland Park, NJ) containing 1 µg/ml 4,6-diamidino-2-phenylindole (Sigma) for nuclear staining. Tissue sections were imaged with a green fluorescent protein filter set (470 nm excitation, 525 nm emission) in an Axio Observer Inverted Fluorescence Microscope equipped with an AxioCam MRm camera to visualize transgene fluorescence as described above. Prior to the initial treatment, isoflurane-anesthetized (2%) mice were intravitally imaged using the Maestro Optical imaging system (CRi, Woburn, MA) as previously described12 and again at days 8 and 12 of the treatment regimen. Images were taken with an excitation filter of 445–490 nm and a long-pass emission filter (515 nm). Images were automatically captured at 10 nm windows from 500 to 700 nm using the Maestro software (exposure times were automatically calculated). Spectral unmixing of the resulting cube image was performed using a user-defined hMGFP protocol. Each spectrum was set manually by unmixing autofluorescence from a negative non-hMGFP expressing mouse analyzed in parallel with a Tg CBL/hMGFP positive mouse. Standardized conditions and subject positioning for image acquisition facilitated meaningful comparison of data collected on different days. Regions of interest were drawn in the interior palm (avoiding the thicker and brighter PADs) of each paw and the average signal (counts/second/mm2) was calculated. The ratio of average signal in right (CBL3) versus left (nonspecific control) paws was calculated for each mouse and normalized with respect to the pretreatment analysis data. The unmixed signal was pseudocolored green. Maestro fluorescence images were saved in TIFF format, and further processed using ImageJ software. ImageJ was used to crop images to create a single master comparison image containing all test image data so that mapping would be applied equally to all paw images simultaneously. A binned black–blue–green lookup table was assigned to improve the visualization of low-intensity pixels, although no bit-values were changed for any pixels.
For quantitative PCR analysis, three mice were sacrificed and skin tissues removed, PAD and interior palm regions separated from the footpad, and frozen directly in dry ice. Tissue was homogenized in a “bead beater” instrument (FastPrep-24, FP24, from MP Biomedicals, Solon, OH) and RNA isolated as previously described.9 RNA was reverse transcribed using the Superscript III First Strand Synthesis system (Invitrogen, Carlsbad, CA) and quantitative PCR was run in the ABI 7500 Fast Sequence Detection system (Applied Biosystems, Foster City, CA) using standard procedures. hMGFP and mouse keratin 14 (Catalog #Mm00516876) Taqman Gene Expression Assays were used as previously described.9 All data points reported are the mean of three replicate assays and error is reported as the SE.
In vivo delivery of plasmid DNA using PADs. PADs (3 × 4) were loaded with ~12 ng/microneedle of pGL3-CMV-Luc plasmid and applied to ear and footpad skin of anesthetized (2% isofluorane) mice for 20 minutes. Twenty four and forty eight hours following the array application, the mice were intraperitoneally injected with luciferin (100 µl of 30 mg/ml luciferin; 150 mg/kg body weight) and the live anesthetized mice were imaged 10 minutes later in the IVIS 200 Imaging System (Caliper Life Sciences) as previously described.30 The resulting light emission was quantified using LivingImage software as described above.
SUPPLEMENTARY MATERIAL Figure S1. Fluorescence microscopy of mouse footpad skin sections demonstrates PAD-mediated siGLO Red (fluorescently labeled siRNA mimic) delivery to the epidermis (or dermis). Figure S2. Analysis of luciferase 2 (Luc2) reporter gene expression following PAD-mediated delivery of expression plasmids to mouse back skin.
Acknowledgments
This study was funded in part by grants from the National Institutes of Health [R44AR055881 and RC2AR058955 (R.L.K.)]. We thank Angela Detweiler and Heini Ilves for their technical support. E.G. is the recipient of a PC Project fellowship.
Supplementary Material
Fluorescence microscopy of mouse footpad skin sections demonstrates PAD-mediated siGLO Red (fluorescently labeled siRNA mimic) delivery to the epidermis (or dermis).
Analysis of luciferase 2 (Luc2) reporter gene expression following PAD-mediated delivery of expression plasmids to mouse back skin.
REFERENCES
- Uitto J., and, Pulkkinen L. The genodermatoses: candidate diseases for gene therapy. Hum Gene Ther. 2000;11:2267–2275. doi: 10.1089/104303400750035807. [DOI] [PubMed] [Google Scholar]
- Smith, FJD, Kaspar, RL, Schwartz, ME, McLean, WHI., and, Leachman, SA.2006. Pachyonychia congenita. GeneReviews < www.genetests.org/profiles/pc >.
- Leachman SA, Kaspar RL, Fleckman P, Florell SR, Smith FJ, McLean WH, et al. Clinical and pathological features of pachyonychia congenita. J Investig Dermatol Symp Proc. 2005;10:3–17. doi: 10.1111/j.1087-0024.2005.10202.x. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Hickerson RP, Hull PR, Smith FJ, Milstone LM, Lane EB, et al. Therapeutic siRNAs for dominant genetic skin disorders including pachyonychia congenita. J Dermatol Sci. 2008;51:151–157. doi: 10.1016/j.jdermsci.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson RP, Smith FJ, Reeves RE, Contag CH, Leake D, Leachman SA, et al. Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J Invest Dermatol. 2008;128:594–605. doi: 10.1038/sj.jid.5701060. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Hickerson RP, Schwartz ME, Bullough EE, Hutcherson SL, Boucher KM, et al. First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry BW. Novel mechanisms and devices to enable successful transdermal drug delivery. Eur J Pharm Sci. 2001;14:101–114. doi: 10.1016/s0928-0987(01)00167-1. [DOI] [PubMed] [Google Scholar]
- Brown MB, Martin GP, Jones SA., and, Akomeah FK. Dermal and transdermal drug delivery systems: current and future prospects. Drug Deliv. 2006;13:175–187. doi: 10.1080/10717540500455975. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gonzalez E, Ra H, Hickerson RP, Wang Q, Piyawattanametha W, Mandella MJ, et al. siRNA silencing of keratinocyte-specific GFP expression in a transgenic mouse skin model. Gene Ther. 2009;16:963–972. doi: 10.1038/gt.2009.62. [DOI] [PubMed] [Google Scholar]
- Arora A, Prausnitz MR., and, Mitragotri S. Micro-scale devices for transdermal drug delivery. Int J Pharm. 2008;364:227–236. doi: 10.1016/j.ijpharm.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill HS, Denson DD, Burris BA., and, Prausnitz MR. Effect of microneedle design on pain in human volunteers. Clin J Pain. 2008;24:585–594. doi: 10.1097/AJP.0b013e31816778f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson RP, Vlassov AV, Wang Q, Leake D, Ilves H, Gonzalez-Gonzalez E, et al. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides. 2008;18:345–354. doi: 10.1089/oli.2008.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R., and, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Ding H, Schwarz DS, Keene A, Affar el B, Fenton L, Xia X, et al. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Schlehuber LD, London IM., and, Lieberman J. Determinants of specific RNA interference-mediated silencing of human β-globin alleles differing by a single nucleotide polymorphism. Proc Natl Acad Sci USA. 2006;103:5953–5958. doi: 10.1073/pnas.0601309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FJ, Hickerson RP, Sayers JM, Reeves RE, Contag CH, Leake D, et al. Development of therapeutic siRNAs for pachyonychia congenita. J Invest Dermatol. 2008;128:50–58. doi: 10.1038/sj.jid.5701040. [DOI] [PubMed] [Google Scholar]
- Hengge UR, Walker PS., and, Vogel JC. Expression of naked DNA in human, pig, and mouse skin. J Clin Invest. 1996;97:2911–2916. doi: 10.1172/JCI118750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Park JH., and, Prausnitz MR. Dissolving microneedles for transdermal drug delivery. Biomaterials. 2008;29:2113–2124. doi: 10.1016/j.biomaterials.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar RL, McLean WH., and, Schwartz ME. Achieving successful delivery of nucleic acids to skin: 6th Annual Meeting of the International Pachyonychia Congenita Consortium. J Invest Dermatol. 2009;129:2085–2087. doi: 10.1038/jid.2009.220. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Eshraghi Y, Gliniak C., and, Gosain AK. Nonviral transfection of mouse calvarial organ in vitro using Accell-modified siRNA. Plast Reconstr Surg. 2010;125:494–501. doi: 10.1097/PRS.0b013e3181c82df1. [DOI] [PubMed] [Google Scholar]
- Difeo A, Huang F, Sangodkar J, Terzo EA, Leake D, Narla G, et al. KLF6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer Res. 2009;69:4733–4741. doi: 10.1158/0008-5472.CAN-08-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulman SA, Barrow D, Anstey A, Gateley C, Morrissey A, Wilke N, et al. Minimally invasive cutaneous delivery of macromolecules and plasmid DNA via microneedles. Curr Drug Deliv. 2006;3:65–75. doi: 10.2174/156720106775197510. [DOI] [PubMed] [Google Scholar]
- Birchall J, Coulman S, Pearton M, Allender C, Brain K, Anstey A, et al. Cutaneous DNA delivery and gene expression in ex vivo human skin explants via wet-etch micro-fabricated micro-needles. J Drug Target. 2005;13:415–421. doi: 10.1080/10611860500383705. [DOI] [PubMed] [Google Scholar]
- Pearton M, Allender C, Brain K, Anstey A, Gateley C, Wilke N, et al. Gene delivery to the epidermal cells of human skin explants using microfabricated microneedles and hydrogel formulations. Pharm Res. 2008;25:407–416. doi: 10.1007/s11095-007-9360-y. [DOI] [PubMed] [Google Scholar]
- Ng KW, Pearton M, Coulman S, Anstey A, Gateley C, Morrissey A, et al. Development of an ex vivo human skin model for intradermal vaccination: tissue viability and Langerhans cell behaviour. Vaccine. 2009;27:5948–5955. doi: 10.1016/j.vaccine.2009.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchall J, Coulman S, Anstey A, Gateley C, Sweetland H, Gershonowitz A, et al. Cutaneous gene expression of plasmid DNA in excised human skin following delivery via microchannels created by radio frequency ablation. Int J Pharm. 2006;312:15–23. doi: 10.1016/j.ijpharm.2005.12.036. [DOI] [PubMed] [Google Scholar]
- González-González E, Ra H, Spitler R, Hickerson RP, Contag CH., and, Kaspar RL.2010Increased interstitial pressure improves nucleic acid delivery to skin enabling a comparative analysis of constitutive promoters Gene Therepub ahead of print). [DOI] [PubMed]
- Geusens B, Sanders N, Prow T, Van Gele M., and, Lambert J. Cutaneous short-interfering RNA therapy. Expert Opin Drug Deliv. 2009;6:1333–1349. doi: 10.1517/17425240903304032. [DOI] [PubMed] [Google Scholar]
- Contag CH., and, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescence microscopy of mouse footpad skin sections demonstrates PAD-mediated siGLO Red (fluorescently labeled siRNA mimic) delivery to the epidermis (or dermis).
Analysis of luciferase 2 (Luc2) reporter gene expression following PAD-mediated delivery of expression plasmids to mouse back skin.