Abstract
Activation of proto-oncogenes by retroviral insertion is an important issue delaying clinical development of gene therapy. We have reported the nonrandom persistence of hematopoietic clones with vector insertions within the MDS1/EVI1 locus following transplantation of rhesus macaques. We now ask whether prolonged culture of transduced CD34+ cells before transplantation selects for clones with insertions in the MDS1/EVI11 or other proto-oncogene loci. CD34+ cells were transduced with standard retroviral vectors for 4 days and then continued in culture for an additional 6 days before transplantation. A 15% of insertions identified in granulocytes 6 months post-transplant were in MDS1/EVI11, significantly increased compared to the frequency in animals transplanted with cells immediately following transduction. MDS1/EVI1 clones became more dominant over time post-transplantation in one animal that was followed long term, accompanied by an increased overall copy number of vector-containing granulocytes, with one MDS1/EVI1 clone eventually accounting for 100% of transduced granulocytes and marrow colony-forming unit (CFU). This vector insertion increased the expression of Evi1 mRNA. There was no overrepresentation of MDS1/EVI1 insertions contributing to lymphoid lineages. Strategies involving prolonged ex vivo expansion of transduced cells may increase the risk of genotoxicity.
Introduction
Over the past decade gene therapy utilizing replication-incompetent retroviral vectors to target hematopoietic stem and progenitor cells (HSPCs) has proven to be an effective therapeutic modality in several clinical disorders.1,2 Although the risk of activating cellular proto-oncogenes by adjacent insertion of retroviral vector has always been a concern, until recently it was assumed to be primarily theoretical and very unlikely. However, a total of five patients enrolled in two gene therapy trials for X-linked severe combined immunodeficiency have now developed acute T-cell leukemia, linked to activation of LMO2 or other proto-oncogenes by the integrated vector enhancer.3 Investigators hypothesized that the X-linked severe combined immunodeficiency transgene, the γc signaling receptor, interacted with LMO2 specifically to induce leukemia.4 More recently, two patients receiving HSPC gene therapy for chronic granulomatous disease developed first oligoclonal and then monoclonal dominance and in vivo expansion of clones containing vector-activated MDS1/EVI1 or homolog PRDM16 genes in the myeloid lineage.5 Because the vector itself contained a transgene with no known growth-altering activities, insertional mutagenesis alone was implicated.
In our large-scale survey of standard Moloney murine leukemia vector (MLV) insertion patterns in the granulocyte progeny of transduced rhesus macaque HSPCs, we previously reported a total of 14 out of 702 total unique independent insertions in the first two introns of the MDS1/EVI1 gene locus, all identified in myeloid progeny cells.6,7 An EVI1 insertion was also reported in the first mouse developing acute myeloid leukemia following transplantation of bone marrow cells transduced with a replication-defective MLV vector, and MDS1/EVI1 was frequently activated by vector insertions in murine bone marrow cells that became immortalized in vitro following transduction with retroviral vectors containing only marker genes.8,9
It has been shown in primate transplantation and in other models that MLV vectors have a propensity to integrate near transcription start sites, CpG islands, and within or near coding regions of highly expressed genes.7,10,11 However, these factors alone cannot account for the remarkable overrepresentation of hematopoietic output from clones with insertions in or near MDS1/EVI1. Immediately following a brief in vitro transduction of CD34+ cells, MDS1/EVI1 was not identified as an overrepresented insertional “hot spot”.12 Therefore, we hypothesized that insertional activation of this locus resulted in immortalization of committed progenitors, specifically those with myeloid lineage potential, allowing them to engraft in vivo and contribute to hematopoiesis long term. To test this hypothesis, we investigated whether extending the ex vivo culture period following transduction would favor survival and eventual engraftment of clones with insertions in or near MDS1/EVI1, as compared to non-MDS/EVI1 clones.
Results
In order to investigate possible factors that favor increased representation of MDS1-EVI1 clones following transplantation with retrovirally transduced CD34+ cells, we sought to address whether more prolonged ex vivo culture of transduced CD34+ cells before transplantation would favor clones with insertions in or near MDS1/EVI1 or that of other proto-oncogenes. We cultured and transduced autologous rhesus CD34+ cells in the presence of stem cell factor, thrombopoietin, and flt3 ligand on retronectin-coated flasks for 4 days, identical to the standard retroviral transduction conditions we have utilized in previous studies. In this study, rather than immediately infusing the cells following transduction, they were expanded in vitro for an additional 6 days under the same culture conditions, as shown in Figure 1. The G1Na MLV vector containing a neomycin-resistance transgene was used for transductions, allowing comparison to our prior published data utilizing the same vector, target cells, and transduction conditions for 4 days without the subsequent expansion period.6,7 Two animals were transplanted with these expanded cells, and information on cell numbers collected, transduced, and transplanted are summarized in Table 1. Linear amplification–mediated PCR (LAM-PCR) was performed on small aliquots of cells removed before infusion and the retroviral insertion site identified (36 and 48 independent insertions in the two animals, respectively) are listed in Supplementary Table S1. None were shared between the two animals, and no common integration sites were identified in this small sample. Both animals recovered hematopoiesis rapidly and durably. Marking levels as assessed by quantitative real-time PCR for vector sequences in animal RQ4739 were 2–3% at 6 months following transplantation, and <1% in animal RQ6596, levels lower than generally achieved in rhesus macaques transplanted with cells immediately following the end of transduction with the same vector.
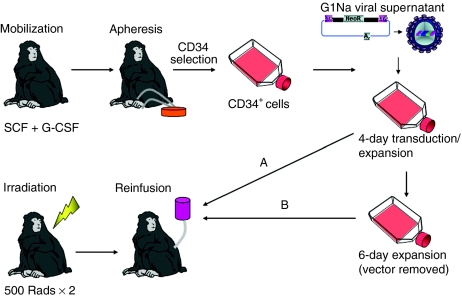
Figure 1.
Experimental design. Rhesus macaques were mobilized with G-CSF and SCF and cells were collected by apheresis and enriched for CD34+ cells via immunoselection. CD34+ cells were cultures in the presence of G1Na vector supernatants, SCF, FLT3 ligand, and TPO, in retronectin-coated flasks, with daily changes of vector and media for a total of 4 days. Cells were then either reinfused immediately (pathway A), or cultured without vector supernatant for and additional 6 days (pathway B) and reinfused. G-CSF, granulocyte colony–stimulating factor; SCF, stem cell factor; TPO, thrombopoietin.
Table 1. Autologous CD34+ cell collection, transduction, expansion, and transplantation.
At 6 months following transplantation, when stable long-term hematopoiesis had been established, integration sites in circulating granulocytes, T cells, and B cells were identified. LAM-PCR and shotgun cloning of integration sites was performed repetitively on DNA samples from granulocytes to retrieve insertions from all contributing clones (Supplementary Table S1). At 6 months, only 29 insertions from granulocytes could be retrieved from animal RQ4739 and 5 from animal RQ6596. In contrast, from 5 animals transplanted with 4-day cultured and transduced cells, a mean of 72 ± 31 granulocyte insertions were retrieved per animal following repetitive analyses of DNA samples at time points >6 months following transplantation. Animal RQ4739 was also studied in detail at 23, 43, and 48 months, at which time the clonal diversity in granulocytes decreased further, with only 10 unique insertions retrieved from granulocytes despite multiple replicates of LAM-PCR and shotgun cloning at 23 months, and only 2 at 43 months. No insertions identified in vivo from either animal at 6 months or later matched insertions identified in CD34+ cells at the time of infusion.
At 6 months post-transplant, there was a significant increase in the frequency of unique insertions in the MDS1/EVI1 locus on chromosome 2q in the rhesus (homologous to 3q26.2 in the human genome) in the two macaques receiving cells transduced for 4 days and then cultured for a total of 10 days before infusion (5 of 34 insertions), as compared to macaques transplanted with CD34+ cells infused following the 4-day transduction period (14 of 702 insertions) (P < 0.001). These insertions were in both forward and reverse orientations relative to the MDS1 and EVI1 genes, and were located within the first and second introns of the gene complex, similar to the general location of insertions in these loci MDS1 and EVI1 identified in our previous report regarding rhesus transplanted with cells immediately following a 4-day transduction6 (Figure 2). There were no other common integration sites identified.
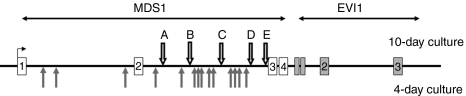
Figure 2.
Insertions within the MDS1/EVI1 locus: The rhesus MDS1/EVI1 locus is shown. Arrows A–E on the top show the location of MDS1/EVI1 insertions in the current study, in animals receiving cells cultured for 10 days. Grey arrows below show the location of insertions previously reported, in animals receiving cells cultured for 4 days.4
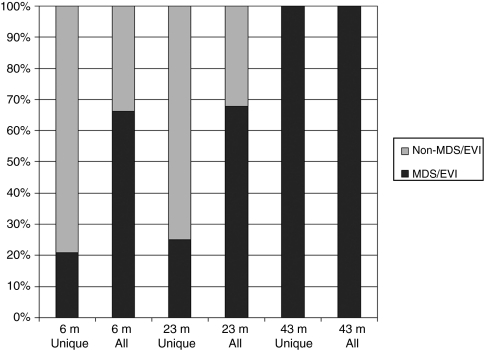
Besides the increased frequency of independent insertions in this locus, the dominance of clones with MDS1/EVI1 insertions was also more dramatic, as estimated based on the frequency of all MDS1/EVI1 insertions retrieved following shotgun cloning of LAM-PCR amplification products.5 Figure 3 shows the frequency of unique MDS1/EVI1 insertions in the granulocytes of 10-day animals as well as the overall frequency of MDS/EVI1 insertions compared to total valid insertions retrieved following shotgun cloning 6 months following transplantation, with MDS1/EVI1 accounting for 15% of unique insertions but 32% of all cloned insertions. One animal continued to be followed, and by 23 months following transplantation this dominance became more pronounced, with 20% of unique insertions being within MDS1/EVI1 and 68% of the total of all cloned insertions. By 43 months following transplantation, 100% of retrieved granulocyte RIS were in MDS/EVI1, with one of the MDS1/EVI1 insertions (clone D) first identified at 6 months and already dominant at 23 months accounting for 22/23 retrieved RIS, and a second MDS/EVI1 clone accounting for 1/23 insertions. Bone marrow was obtained at 48 months, and plated for colony-forming unit (CFU) assays in the presence of G418, with only those CFU containing vector able to survive neomycin selection. These colonies were analyzed by LAM-PCR, and 17/17 had the MDS1/EVI1 clone D insertion.
Figure 3.
Frequency of insertions into the MDS1/EVI1 locus in granulocytes. The relative frequency of insertions into the MDS1/EVI1 locus (shown in black) versus non-MDS/EVI1 insertions (shown in grey), as fractions of the total valid insertion sites indentified in granulocytes sampled at different time points following transplantation (m, months post-transplantation). “Unique” enumerates the number of individual unique clones detected contributing to a sample, no matter how many times the insertion is isolated via shotgun cloning of LAM-PCR product. “All” counts an insertion isolated via shotgun cloning every time it is isolated, thus allowing a rough measure of clonal dominance.
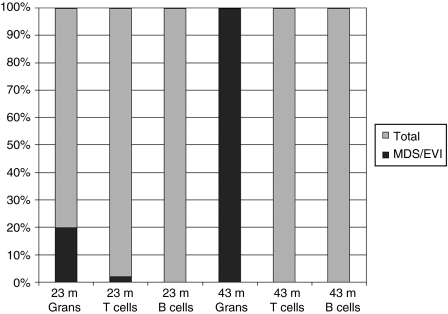
The increased frequency of cells with insertions in MDS1/EVI1 as compared to other genomic loci was detected in granulocytes, but not in T cells or B cells. At 23 and 43 months following transplantation of RQ4739, T and B cells from the peripheral blood were sorted to a purity of >5%. As shown in Figure 4, the frequency of MDS/EVI1 insertions in B and T cells was markedly lower than in granulocytes (P = 0.02, Fischer's exact test), similar to the very low frequency of MDS1/EVI1 insertions in T and B cells of the 4-day animals at approximately the same time point post-transplantation. The single MDS/EVI1 insertion found in T cells at 23 months was identical to one of the two dominant insertions identified in the granulocytes at the same time point, and may have resulted from granulocyte contamination of T cells following flow cytometric sorting, or alternatively from transduction of a multipotent progenitor or stem cell, but as previously reported by our own group and others, MDS/EVI1 insertions are almost completely restricted to myeloid progeny cells in all data analyzed to date.5,6,13,14 T cells were purified from peripheral blood at 48 months and expanded in vitro for 2 weeks. LAM-PCR on this population did not detect any MDS1/EVI1 clones, and clone-specific quantitative PCR for the dominant clone D insertion was completely negative.
Figure 4.
Frequency of vector insertions into the MDS1/EVI1 locus in myeloid and lymphoid lineages. The percentage of independent insertions into the MDS1/EVI1 locus as a fraction of the total number of independent insertions identified via linear amplification–mediated PCR and shotgun cloning of DNA extracted from purified granulocytes, T and B cells from RQ4739 at the time points indicated post-transplantation (m, months post-transplantation).
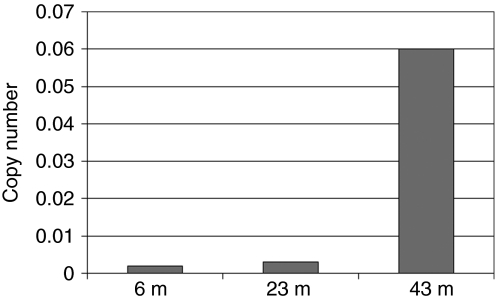
We next asked whether the progressive clonal dominance was accompanied by overall expansion of the clone's contribution to hematopoiesis. Allele-specific clone D primers were used for quantitative DNA PCR, and as shown in Figure 5, the copy number increased markedly in granulocytes, coincident with progression toward monoclonality by LAM-PCR, suggesting that progenitors containing the MDS/EVI1 insertion D were outcompeting untransduced hematopoietic progenitors.
Figure 5.
Real-time DNA PCR for MDS1/EVI1 insertion D in RQ4739 granulocytes. Absolute copy number of vector insertion MDS1/EVI1 D on a per cell basis measured via real-time PCR on DNA samples obtained from granulocytes of animal RQ4739 at the time points indicated (m, months post-transplantation).
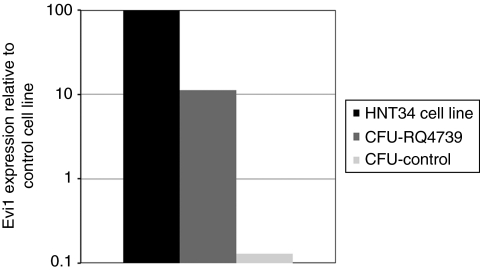
We used reverse transcriptase PCR to investigate whether Evi1 mRNA was overexpressed in myeloid cells, due to proviral activation of the gene from the strong enhancer located in the MLV proviral long terminal repeat. We could not detect increased expression in the granulocytes of these animals compared to control rhesus granulocytes at months 23 or 48 (data not shown). However, the overall level of marking in circulating granulocytes was <10%, potentially impairing detection of activated Evi1 expression in the background of nontransduced cells. Marrow obtained 52 months following transplantation was plated in neomycin, and vector-containing neomycin-resistant CFU-GM were harvested and pooled for RNA analysis. Evi1 expression was not detectable in control marrow CFU-GM, but the neo-resistant colonies from RQ4739 expressed Evi1 at an easily detectable level, significantly higher than nontransduced marrow CFU-GM, and in a range similar to expression in the human leukemic cell line HNT34, containing a translocation activating Evi1 expression (Figure 6).
Figure 6.
Expression of Evi1 mRNA. Real-time reverse transcriptase PCR was performed on RNA from the HNT34 leukemic cell line containing an EVI1 translocation, neomycin-resistant CFU-GM from RQ4739, and control nontransduced rhesus CFU-GM. Expression normalized to the cell line as 100% is shown.
At now almost 5 years following transplantation, RQ4739 remains healthy with normal blood counts and apparently normal maturation of all hematopoietic lineages in the bone marrow, with no evidence to date for progression to myelodysplasia or acute leukemia.
Discussion
Interest in the role of MDS1/EVI1 gene products in normal and abnormal hematopoiesis has been piqued by a number of recent complementary discoveries. This locus or its homolog PRDM16 have been found to be profoundly overrepresented as sites of MLV genomic insertions in vivo following transplantation of mice, monkeys, or humans with transduced HSPCs, and associated with generation of immortalized myeloid cell lines from bone marrow in vitro.5,6,8,15 Clones harboring these insertions initially appear to differentiate normally into daughter myeloid cells, however, activation of this locus as a single predisposing event appears to be sufficient to eventually progress to clonal dominance, myelodysplasia, and acute leukemia, including in two patients in a clinical trial for chronic granulomatous disease.6,9,13,14,16,17,18 No other loci have been found to be equivalently overrepresented, nor linked as single events to MLV-induced genotoxicity following transduction of hematopoietic cells with vectors expressing marker genes or nongrowth stimulating genes.
The overrepresentation of the MDS1/EVI1 locus as a target is unexplained. Either the locus is an extraordinary “hot spot” for MLV integration in HSPCs, or HPSCs that happen to have an MLV vector insert in this locus have a uniquely increased likelihood of engraftment, persistence, and myeloid differentiation in vivo. These explanations are not mutually exclusive, but we designed the experiments in this study to further investigate this issue. Surveys of MLV insertion site preferences in cell lines and primary cells indicate a moderate preference for integration within gene sequences and near transcription start sites.10 Genes that are highly expressed are also more common targets; however, the magnitude of this effect is only about twofold overall, and although MDS1/EVI1 genes are expressed in primitive CD34+ cells,19 insertion target preference characteristics seem unlikely to account for the many log greater overrepresentation of MDS1/EVI1 and PRDM16 insertions in vivo. Analysis of insertion sites in human CD34+ cells immediately following the end of transduction did not identify MDS1/EVI1 as a particular “hot spot” although a number of other sites in or near proto-oncogenes were overrepresented.12
The results of our study support the hypothesis that insertional activation of the MDS1/EVI1 locus results in selective survival or proliferation, as continued ex vivo culture before transplantation resulted in a significant enhancement of the relative frequency of clones with insertions in this locus compared to clones with insertions in other loci able to reconstitute in vivo. Because there was no further addition of vector following the initial 4-day transduction period, this finding could not be due to additional vector insertion events during more prolonged culture. We interpret our findings to indicate that many repopulating cells lost engraftment ability during the additional 6 days of culture. This is reflected in the significantly lower number of contributing individual transduced clones per animal, as defined by unique independent vector insertion sites, compared to the set of animals transplanted with cells immediately following the 4-day transduction period. CD34+ cells with insertions in the MDS/EVI1 locus were able to survive more prolonged culture, and retained engrafting ability, accounting for 15% of in vivo vector-containing clones at 6 months in the 10-day animals as compared to only 2% in the 4-day animals. These clones appeared to be more dominant, as assessed by the relative frequency of retrieval from shotgun cloning of amplified insertion fragments, and this dominance increased with time following transplantation, correlating with the findings from the chronic granulomatous disease clinical trial.5,18
The second major question raised by the results of long-term follow-up studies involving MLV vectors is why the contributions of MDS1/EVI1 clones, normal or malignant, appear to be limited to the myeloid lineage in murine, monkey and human models. The dogma, strongly supported by experimental data in murine models, is that only true multipotent long-term repopulating stem cells are able to self-renew and contribute to hematopoiesis long term. Why do MDS1/EVI1 clones persist in producing normal granulocytes for 2 years or more following transplantation without evidence for T- or B-cell production? Does activation and overexpression of MDS1/EVI1 gene products in a true HSC result in inhibition of lymphoid differentiation and production of only myeloid progeny cells? Or does an integration event activating the MDS1/EVI1 locus in a committed myeloid progenitor result in reacquisition of self-renewal capabilities, including the ability to self-renew and survive in vitro before transplantation? We believe our data support the second possibility. The frequency of long-term repopulating HSCs in large animals has been estimated using a number of different methods as ~5/10 million nucleated cells or 5/100,000 CD34+ cells.20,21 The number of CD34+ cells transduced therefore included ~5,000 HSCs, and with a transduction frequency of 1–10% using MLV vectors, most likely only 50–500 possible HSC targets. Based on random integration, each locus has a one in 100,000 likelihood of integration, and even with increased integration frequency due to the high expression level of MDS1/EVI1, it is difficult to understand how the degree of overrepresentation observed could be attributed to transduction of HSCs. However, if non-HSC committed progenitors could be targets, even random integration would be predicted to result in hundreds of clones with MDS1/EVI1 insertions. If these clones can then selectively survive prolonged culture and then engraft successfully, the observed overrepresentation of myeloid progeny cells with MDS1/EVI1 insertions could then be explained. In a murine model, Kustikova et al. only detected dominant Evi1 clones when cells with an HSC phenotype were utilized as targets, and were unable to show persistence of dominant clones when purified myeloid progenitors were transduced, suggesting that cell-intrinsic target cell characteristics are also important.22
How does overexpression or dysregulated expression of MDS1/EVI1 gene products in primitive HSPCs result in increased hematopoietic contributions and possible immortalization of committed myeloid progenitors? The Evi1 and Mds1-Evi1 proteins are zinc finger transcription factors. There is conflicting data regarding their impact on cell cycle progression and apoptosis, but the majority of evidence supports the acceleration of cycling when Evi1 is overexpressed. 23 Evi1 may protect primitive cells from transforming growth factor-β mediated apoptosis or growth arrest.24 Evi1−/− embryos die by day 10.5, and have greatly decreased numbers of cells with an HSC phenotype in the paraaortic splanchopleural region. These cells also proliferate poorly in vitro and have impaired long-term repopulating ability, suggesting that Evi1 is critical for self-renewal of hematopoietic cells.25 Dysregulated reactivation of Evi1 or MDS1/EVI1 via vector insertion could thus promote self-renewal, proliferation, and prevent apoptosis, both in vitro and in vivo. Further loss of normal regulatory controls via secondary genetic or epigenetic events could result in progression to leukemia. Expression of Evi1 has been linked to a specific subtype of acute myeloid leukemia, and a particularly poor prognosis, with or without actual chromosomal translocations involving the genomic MDS1/EVI1 locus.26 There is recent evidence that MDS1/EVI1 or EVI1 overexpression results in chromosomal instability, perhaps explaining the progression to monosomy 7 and clinical myelodysplasia/acute myeloid leukemia following prolonged expansion of clones with insertions activating this locus in two patients enrolled in a chronic granulomatous disease clinical gene therapy trial.18
Our studies have a number of implications for the design of clinical gene therapy protocols, laboratory experiments utilizing integrating retroviral vectors, and even ex vivo expansion of hematopoietic progenitors without transduction. It is clear that variables beyond the design of the vectors can impact on the potential for genotoxicity. We have shown that the length of time cells are maintained in culture following transduction can have very significant effects on the pattern of clonal contributions to hematopoiesis. Previously, we have also reported that ex vivo culture can impact on the quality of engraftment and immune reconstitution.27,28 We and others have found that maintenance or expansion of true long-term repopulating HSCs for >3–4 days ex vivo is very difficult, with most studies reporting a very significant decrease in competitive repopulating ability compared to fresh cells or cells cultured for briefer time periods.29,30 Besides loss of HSC numbers and function during prolonged culture, selection for cells with characteristics that permit survival and eventual engraftment can occur, including transduced cells with potentially genotoxic insertions, or even nontransduced cells that have acquired mutations. Shorter time periods in culture should minimize these selective pressures, and further optimization of an artificial microenvironment could potentially better mimic in vivo marrow niches better able to support normal hematopoiesis.
Materials and Methods
Collection, transduction, and transplantation of rhesus macaque CD34+ cells. Rhesus macaques were handled in accordance with protocols approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute. Mobilization with stem cell factor and granulocyte colony–stimulating factor, apheresis, and CD34-enrichment of rhesus macaque HSPCs were performed as previously described.31 CD34+ cells were placed in culture on RetroNectin (Takara Bio, Otsu, Japan)-coated flasks at a concentration of 2 × 105/ml in the presence of 100 ng/ml each of human Flt3 ligand, stem cell factor, thrombopoietin (R&D Systems, Minneapolis, MN), and 4 µg/ml protamine sulfate (Sigma, St Louis, MO).28,32 Cells were transduced every 24 hours with fresh supernatant collected from the MLV vector producer cell line G1Na.40 grown in Dulbecco's modified Eagle's medium (Gibco/Invitrogen, Carlsbad, CA) and 10% fetal calf serum (Thermo, Waltham, MA), for a total of 4 days exposure to vector. Transduced cells were then cultured for an additional 6 days under the same culture conditions and cytokines without addition of vector, and split into additional flasks and culture media as needed to keep the cell concentrations below 5 × 105 cells/ml. The cells were reinfused into autologous recipients following 500 rads times two total body irradiation and the animals were supported through engraftment and recovery as described.33
Colony-forming unit assays. To determine efficiency of transduction a portion of the transduced cells (3 × 104 cells/ml) were plated in methylcellulose medium (StemCell Technologies, Vancouver, British Columbia, Canada) supplemented with human erythropoietin (5 U/ml; Amgen, Thousand Oaks, CA), human granulocyte–macrophage colony–stimulating factor (10 ng/ml; R&D Systems), human interleukin-3 (10 ng/ml; R&D Systems), and human stem cell factor (100 ng/ml; Amgen) at 37 °C in 5% CO2. Between days 10 and 14, colonies of >50 cells were counted. Post-transplantation CD34+ marrow cells were plated under the same conditions, with or without addition of G418 (Mediatech, Herdon, VA) at a concentration of 1.5 mg/ml active.
Post-transplantation follow-up and sampling. Blood samples were collected monthly for the first 3 months, and then every 3 months or as required for supplemental analyses. Peripheral blood granulocytes, T and B cells were purified via Ficoll-hypaque-gradient separation and sorted by flow cytometry following staining with anti-CD20-PE (clone 2H7) and anti-CD3-FITC (clone SP34) (Becton-Dickinson, Franklin Lakes, NJ). DNA extraction was carried out on purified populations using the Qiagen DNeasy kit (Valencia, CA).
Measurement of vector and clone-specific copy number. Real-time PCR was performed as described on DNA samples extracted from peripheral blood mononuclear cells and granulocytes to determine vector copy number.34 Allele-specific real-time PCR for MDS1/EVI1 insertion site “D” (see Supplementary Table S1) was also performed using the following primers and probe: Forward 5′-GCTGTGGATTTCAAGTTAGAT-3′, Reverse 5′-CTAGCTTGCCAAACCTACA-3′, and probe 5′-FAM-ATTATTTTTTAGTTTGAAAGACCCCA-TAMRA-3′. PCR was performed with an ABI PRISM 7700 sequence detector (Applied Biosystems, Foster City, CA) and data were analyzed with Sequence Detection System software (Applied Biosystems, Foster City, CA).
Evi1 expression analysis. RNA was extracted from the human HNT34 leukemia cell line (ATCC, Manassas, VA), containing the t(3;3)(q21;q26) translocation, and from pooled CFU-GM using the RNAeasy Mini Kit (Qiagen) per manufacturer's instructions. RNA conversion to complementary DNA and real-time PCR were performed using the Taqman One-Step RT-PCR kit (Applied Biosystems, Branchburg, NJ). Primers and probe were as follows: cEVI1-Forward 5′-ACC CAC TCC TTT CTT TAT GGA CC-3′, cEVI1-Reverse 5′-TGA TCA GGC AGT TGG AAT TGT G-3′, and cEVI1-Probe 5′-TGA GGC CTT CTC CAG GAT TCT TGT TTC AC-3′. Quantitative real-time PCR was performed as described above.
Characterization of retroviral integration sites by LAM-PCR. LAM-PCR and shotgun cloning were performed to identify proviral genomic insertion sites adjacent to the 5′ long terminal repeat as previously described.7,35 The January 2006 draft assembly of the rhesus macaque genome was used for site localization and validation using parameters as described.7,36
Ex vivo expansion of T cells. The mononuclear cell fraction of blood was isolated by gradient centrifugation on Lymphocyte Separation Medium (MP Biomedicals, Solon, OH) and T cells were then purified according to the manufacturer's instructions using nonhuman primate immunomagnetic anti-CD3 MACS beads (Miltenyi, Auburn, CA). Purified T cells (≥99% pure) were then expanded by coculturing with irradiated (75 Gy) human peripheral blood mononuclear cell feeder cells at a ratio of 1:10 in BioWhittaker X-VIVO 20 medium (Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone; ThermoScientific, Logan, UT) and 500 U/ml recombinant human interleukin 2 (Tecin; Hoffmann-LaRoche, Nutley, NJ). Initial cell density of cells was 1.4 × 106/ml in a 37 °C, 5% CO2 incubator. After 4-day culture, half the medium in the flask was replaced with fresh media and on day 5 of expansion; the cells were counted by Trypan blue exclusion on a hemacytometer and cell density was adjusted to 8 × 105/ml. T cells were counted every 2 days and kept at a density of 8 × 105/ml for 2 weeks. Cells were harvested for phenotypic and molecular analyses.
SUPPLEMENTARY MATERIAL Table S1. Master table of insertions.
Acknowledgments
These studies were supported by the intramural research program of the National Heart, Lung, and Blood Institute. We thank the staff of the 5 Research Court primate center for excellent animal care. T.J.G.: collection and analysis of data, wrote manuscript; S.S.: collection and analysis of data; R.L.: collection and analysis of data; A.L.: data analysis and interpretation; R.A.: collection and analysis of data; A.K.: collection and analysis of data; R.E.D.: data analysis and interpretation; R.W.C.: data analysis and interpretation; C.E.D.: conception, wrote, and approved the final version of the manuscript.
Supplementary Material
Master table of insertions.
REFERENCES
- Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Davé UP, Jenkins NA., and, Copeland NG. Gene therapy insertional mutagenesis insights. Science. 2004;303:333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, et al. Recurrent retroviral vector integration at the MDS1/EVI1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D., and, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary murine bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Düllmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XB, Beard BC, Trobridge GD, Wood BL, Sale GE, Sud R, et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métais JY., and, Dunbar CE. The MDS1-EVI1 gene complex as a retrovirus integration site: impact on behavior of hematopoietic cells and implications for gene therapy. Mol Ther. 2008;16:439–449. doi: 10.1038/sj.mt.6300372. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Gerhardt TM, Schmahl GE, Flotho C, Rath AV., and, Niemeyer CM. Expression of the Evi-1 gene in haemopoietic cells of children with juvenile myelomonocytic leukaemia and normal donors. Br J Haematol. 1997;99:882–887. doi: 10.1046/j.1365-2141.1997.4983304.x. [DOI] [PubMed] [Google Scholar]
- Abkowitz JL, Catlin SN, McCallie MT., and, Guttorp P. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood. 2002;100:2665–2667. doi: 10.1182/blood-2002-03-0822. [DOI] [PubMed] [Google Scholar]
- Shepherd BE, Kiem HP, Lansdorp PM, Dunbar CE, Aubert G, LaRochelle A, et al. Hematopoietic stem-cell behavior in nonhuman primates. Blood. 2007;110:1806–1813. doi: 10.1182/blood-2007-02-075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbey A, Stephens V., and, Bartholomew C. Loss of cell cycle control by deregulation of cyclin-dependent kinase 2 kinase activity in Evi-1 transformed fibroblasts. Cell Growth Differ. 1999;10:601–610. [PubMed] [Google Scholar]
- Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, et al. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, et al. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 2005;24:1976–1987. doi: 10.1038/sj.emboj.7600679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–845. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- Loré K, Seggewiss R, Guenaga FJ, Pittaluga S, Donahue RE, Krouse A, et al. In vitro culture during retroviral transduction improves thymic repopulation and output after total body irradiation and autologous peripheral blood progenitor cell transplantation in rhesus macaques. Stem Cells. 2006;24:1539–1548. doi: 10.1634/stemcells.2005-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatoku M, Sellers S, Agricola BA, Metzger ME, Kato I, Donahue RE, et al. Avoidance of stimulation improves engraftment of cultured and retrovirally transduced hematopoietic cells in primates. J Clin Invest. 2001;108:447–455. doi: 10.1172/JCI12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale JF, Hanazono Y, Sellers SE, Agricola BA, Metzger ME, Donahue RE, et al. Ex vivo expansion of genetically marked rhesus peripheral blood progenitor cells results in diminished long-term repopulating ability. Blood. 1998;92:1131–1141. [PubMed] [Google Scholar]
- Holyoake TL, Alcorn MJ, Richmond L, Farrell E, Pearson C, Green R, et al. CD34 positive PBPC expanded ex vivo may not provide durable engraftment following myeloablative chemoradiotherapy regimens. Bone Marrow Transplant. 1997;19:1095–1101. doi: 10.1038/sj.bmt.1700799. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Kirby MR, Metzger ME, Agricola BA, Sellers SE., and, Cullis HM. Peripheral blood CD34+ cells differ from bone marrow CD34+ cells in Thy-1 expression and cell cycle status in nonhuman primates mobilized or not mobilized with granulocyte colony-stimulating factor and/or stem cell factor. Blood. 1996;87:1644–1653. [PubMed] [Google Scholar]
- Wu T, Kim HJ, Sellers SE, Meade KE, Agricola BA, Metzger ME, et al. Prolonged high-level detection of retrovirally marked hematopoietic cells in nonhuman primates after transduction of CD34+ progenitors using clinically feasible methods. Mol Ther. 2000;1:285–293. doi: 10.1006/mthe.2000.0034. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Kuramoto K., and, Dunbar CE.2005Large animal models for stem and progenitor cell analysis. In: Coligan, JE (ed.). Current Protocols in Immunology. Wiley: Hoboken, NJ. pp. 22A.1.1–22A.1.29; [DOI] [PubMed] [Google Scholar]
- Larochelle A, Krouse A, Metzger M, Orlic D, Donahue RE, Fricker S, et al. AMD3100 mobilizes hematopoietic stem cells with long-term repopulating capacity in nonhuman primates. Blood. 2006;107:3772–3778. doi: 10.1182/blood-2005-09-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Zickler P, Hoffmann G, Haas S, Wissler M, Muessig A, et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002;100:2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- Hu J, Renaud G, Gomes TJ, Golmes T, Ferris A, Hendrie PC, et al. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol Ther. 2008;16:1617–1623. doi: 10.1038/mt.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Master table of insertions.