Abstract
One promising approach for the gene therapy of Duchenne muscular dystrophy (DMD) is exon skipping. When thinking of possible intervention on human, it is very crucial to identify the most appropriate antisense sequences able to provide the highest possible skipping efficiency. In this article, we compared the exon 51 skipping activity of 10 different antisense molecules, raised against splice junctions and/or exonic splicing enhancers (ESEs), expressed as part of the U1 small nuclear RNA (snRNA). The effectiveness of each construct was tested in human DMD myoblasts carrying the deletion of exons 48–50, which can be treated with skipping of exon 51. Our results show that the highest skipping activity and dystrophin rescue is achieved upon expression of a U1 snRNA-derived antisense molecule targeting exon 51 splice sites in combination with an internal exon sequence. The efficacy of this molecule was further proven on an exon 45–50 deletion background, utilizing patient's fibroblasts transdifferentiated into myoblasts. In this system, we showed that the selected antisense was able to produce 50% skipping of exon 51.
Introduction
Duchenne muscular dystrophy (DMD) is one of the most severe neuromuscular diseases, affecting 1:3,500 live males. DMD is a monogenic disorder caused by mutations in the largest gene of higher eukaryotes, the gene encoding for the dystrophin protein (DMD).1,2 About 98% of the 2.4-Mb DMD gene accounts for intron sequences, whereas the remainder constitutes of 79 exons and seven different promoters, which direct the expression of tissue-specific isoforms. Additional isoforms also arise from alternative splicing or polyadenylation.3
In skeletal muscle tissues, dystrophin is localized on the inner face of sarcolemma, the muscle fiber plasma membrane, where it interacts with the cytoskeletal actin by its N-terminal domain, and with a complex of proteins localized on the sarcolemma, named dystrophin-associated protein complex, through its C-terminal. These interactions allow muscle force transduction and are essential for fiber integrity.4 Several other crucial aspects of the muscle fiber physiology, like calcium homeostasis,5 and epigenetic control of gene expression, have recently emerged as dystrophin-dependent (ref. 6; Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M et al., in preparation), providing an explanation for the dramatic consequence observed on the muscle tissue of Duchenne patients where dystrophin is absent.
Due to the well-defined genetic alteration, many studies have concentrated on finding possible therapeutic approaches for the treatment of DMD.7,8,9 With a few exceptions, all DMD mutations (deletions and duplications) lead to frameshifts, resulting in the formation of a premature termination codon that impairs dystrophin translation.10 On the contrary, when mutations do not perturb the correct reading frame, as in the case of Becker muscular dystrophy, the translation of a partially functional protein is still occurring. Notably, Becker muscular dystrophy patients carrying deletions of about half of the DMD gene show very mild symptoms.11,12 These observations suggested a new therapeutic strategy based on the possibility of manipulating the splicing of the dystrophin precursor mRNA in order to induce the skipping of specific exons and rescuing a correct reading frame. This can be obtained by antisense molecules that, by pairing with splice junctions or exonic splicing enhancers (ESEs), prevent exon recognition by the splicing machinery.
Different groups developed chemically modified antisense oligonucleotides (AONs) directed against splice junctions or ESEs, and found that this strategy could effectively rescue dystrophin both in vitro13 and in vivo.14,15,16 Safety and local efficacy of the intramuscular AON injections were tested in phase I clinical trials and systemic trials recently started.17,18
Small nuclear RNAs (snRNA) have also been utilized as vectors for stable antisense expression in order to circumvent the major limitation of the AON approach that, given the limited oligo stability, requires reiterative administrations. U1 and U7 snRNA-derived antisense molecules have been shown to be effective in inducing exon skipping and dystrophin rescue both in human DMD myoblasts19,20,21 and in the mdx mouse.22,23,24 Furthermore, long-term and body-wide effectiveness of the exon-skipping therapy was obtained by systemic injection of adeno-associated viral (AAV) vector expressing the U1-derived antisense molecules into mdx mice.25
In this article, we tested different U1 snRNA-antisense sequences for the ability of inducing exon 51 skipping by targeting different combinations of ESEs and splice junctions. Ten different antisense molecules were realized and the most effective construct was selected by a triple screening in DMD myoblasts, in DMD fibroblasts transdifferentiated into myoblasts and on luciferase reporter construct.
Results
Design and expression analysis of antisense molecules against exon 51 of the DMD gene
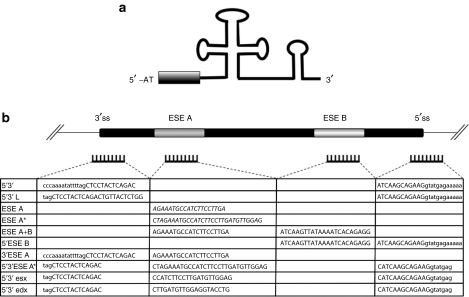
The U1 snRNA was utilized as carrier to express 10 different antisense molecules for exon 51 skipping. Nucleotides from position 3–10 at the 5′-end of U1 snRNA, required for the recognition of the 5′ splice site, were substituted with antisense sequences complementary to different portions of exon 51 and its splice sites (Figure 1a). Because we previously observed that both splice sites have to be targeted in order to induce efficient exon skipping,19 the first constructs produced contained antisense sequences against both splice junctions (5′3′ and 5′3′ L constructs; Figure 1b). Moreover, because ESEs have been shown to represent effective target substrates for efficient exon skipping26,27 we also produced chimeric constructs containing antisense sequences against putative ESE elements,28 alone or in combination with splice junctions (Figure 1b).
Figure 1.
Designing and cloning of the U1-antisense-RNA molecules. (a) Schematic representation of the chimeric U1-antisense snRNA. The grey box indicates the location of the antisense sequences. (b) Table summarizing the 10 different constructs produced together with the corresponding target regions on exon 51 and flanking intron sequences (uppercase – exonic regions; lowercase – intron sequences). Costructs ESE A and ESE A* contains the sequences (indicated in italics) currently utilized in the Prosensa17 and AVI Biopharma18 clinical trials, respectively.
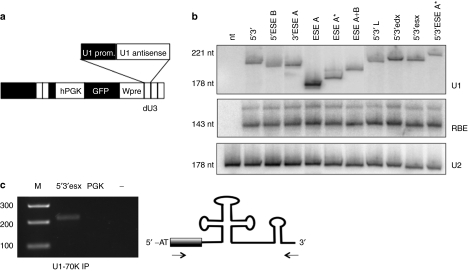
The strong polymerase II–dependent U1 snRNA gene promoter and termination sequences were used to derive antisense expression cassettes, which were cloned in the dU3 portion of the 3′ long terminal repeat region of the pRRLSIN.cPPT.PGK/GFP.WPRE lentiviral vector29 (Figure 2a). Because the different constructs considerably extend the length of the U1 snRNA (in some cases up to 58 nucleotides), we first checked their expression and stability in HeLa transfection experiments. The relative expression activity was tested by co-transfection with the U16-RBE plasmid30 and normalized for the endogenous U2 snRNA. Northern blot analysis indicated that all the chimeric molecules accumulated at fairly similar levels (Figure 2b). Moreover, immunoprecipitations of nuclear extracts with U1-70K antibodies followed by reverse transcriptase (RT)-PCR (Figure 2c) indicated that the U1-chimeric snRNAs are still able to form snRNPs.31
Figure 2.
U1-antisense expression in HeLa cells. (a) Schematic representation of pRRL/U1-antisense constructs. The U1-antisense expression cassettes were cloned in the dU3 region of the downstream LTR of the pRRLSIN.cPPT.PGK/GFP.WPRE lentiviral backbone. (b) HeLa cells were transfected with the different antisense constructs together with the U16-RBE plasmid (expressing a 143-nt long modified U16 snoRNA). Northern blot analysis was performed with probes against: U1 snRNA (panel U1), U16-RBE (panel RBE), and U2snRNA (panel U2). The two latter hybridizations are used to normalize for transfection efficiency and as loading control, respectively. The filter was cut so as to exclude the endogenous U1 snRNA hybridization. On the side, the molecular sizes are reported. (c) Nuclear extracts were prepared from the 5′3′esx sample from the experiment in a and immunoprecipitated with anti U1-70K antibodies. The RNA, extracted from the Ippt pellet, was analyzed for the presence of the 5′3′esx transcript by reverse transcriptase-PCR with primers shown on the right panel. GFP, green fluorescent protein complementary DNA; hPGK, human phosphoglycerate kinase promoter; nt, nucleotide.
Study of exon-skipping activity in human DMD myoblasts
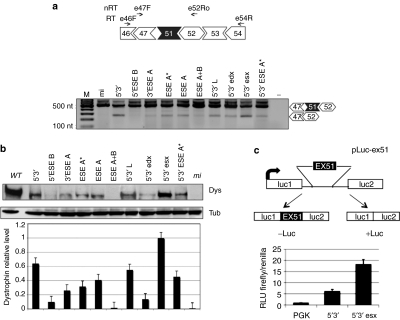
Lentiviral particles for each of the constructs were produced in 293T cells and used to infect DMD myoblasts (Δ48–50) carrying the deletion of exons 48–50 (provided by the Telethon Neuromuscular Biobank). In this case, skipping of exon 51 allows rescue of the correct reading frame and the production of a dystrophin protein 210 amino acids shorter than the wild type. Δ48–50 cells were infected with comparable amounts of the different recombinant lentiviruses. After infection, they were induced to differentiate and samples for RNA and protein analysis were collected after 7 days. Exon 51 skipping was evaluated by nested RT-PCR analysis performed on 200 ng of total RNA (Figure 3a), while western blot was performed on 50 µg of total proteins (Figure 3b). Dystrophin levels, measured in three independent experiments, were normalized against the tubulin signal; the relative values are reported in the histogram of Figure 3b. With the exception of 5′ESE B and ESE A+B constructs, all the samples revealed skipping activity and, among these, 5′3′ and 5′3′esx were the most effective. Notably, the skipping activity paralleled very well the efficiency of dystrophin rescue (Figure 3b), with constructs 5′3′ and 5′3′ esx displaying the highest activity.
Figure 3.
Exon 51 skipping and dystrophin rescue in DMD Δ48–50 primary myoblasts and selection of the best performing construct with the Luciferase-based splicing-reporter assay. (a) Top: schematic representations of the Δ48–50 dystrophin complementary DNA between exons 46 and 54 together with the positions of reverse transcriptase-PCR oligos (E46F and E54R—RT), and nested PCR oligos (E47F and E52Ro—nRT). Bottom: nested RT-PCR on RNA extracted from Δ48–50 cells infected with the antisense-expressing lentiviruses. Unskipped and skipped products are indicated on the right. Molecular sizes are indicated on the left. (b) Representative western blot of proteins (50 µg) extracted from infected Δ48–50 cells, probed with anti-dystrophin (Dys) and anti-tubulin (Tub) antibodies (WT: 2 µg of proteins from wild-type skeletal muscle cells; mi: 50 µg of proteins from mi cells). The histogram at the bottom indicates dystrophin levels normalized on tubulin signals. Values are obtained from three independent experiments; error bars: means ± SD. (c) Top: schematic representation of the pLuc-ex51construct. Exon 51 and part of its flanking introns were cloned between the Firefly luciferase exons. From this construct, luciferase expression is obtained only upon exon 51 skipping. Bottom: histogram showing the relative luminescence units (RLU) from C27 myoblast cells transfected with pLuc-ex51 and pTK (Renilla luciferase expressing plasmid), together with either 5′–3′ or 5′–3′esx constructs. An empty lentiviral vector is used as control (PGK). DMD, Duchenne muscular dystrophy; mi, mock infected; −: negative control without RNA.
Comparison between the 5′3′ and 5′3′esx constructs was further tested using a luciferase-based splicing-reporter assay32 in which luciferase is produced only when skipping of the inserted exon 51 is occurring (Figure 3c, upper panel—construct pLuc-ex51). The 5′3′ and 5′3′esx expressing plasmids were co-transfected with the pLuc-ex51 and with a Renilla reporter. Luciferase activity assays (Figure 3c, lower panel) indicated that 5′3′esx is inducing a higher level of exon skipping with respect to 5′3′, confirming the data obtained in DMD cells. These results indicated that the ESE target region present in the 5′3′esx construct plays an important role in exon 51 splicing and that it is able to improve skipping efficiency when combined to antisense sequences against splice junctions.
Exon skipping in DMD fibroblasts transdifferentiated into myoblasts
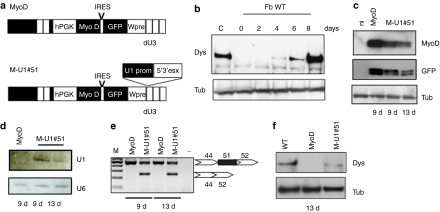
In order to have a simplified screening procedure for testing the activity of antisense constructs on patients' samples, we set up a protocol for fibroblasts transdifferentiation into myoblasts. Infection of WT fibroblasts with the MyoD lentivirus (Figure 4a) produced efficient conversion into myoblasts and myotubes as shown by dystrophin synthesis (Figure 4b) and morphological analysis (not shown). Dystrophin starts to be produced at 4 days after shifting to differentiation medium and its accumulation increases with the progression of the myogenic program (8 days). We then applied the transdifferentiation protocol to DMD fibroblasts in order to test the skipping activity of the 5′3′esx construct. A skin biopsy with a different genetic background (deletion of exons 45–50 − Δ45–50), also treatable with exon 51 skipping, was infected with the M-U1#51 lentivirus carrying the MyoD and 5′3′esx expression cassettes (Figure 4a). Muscle differentiation was induced 2 days after infection and at day 9 and 13, samples were collected for RNA and protein analysis. Figure 4c shows the expression of MyoD and GFP, which are encoded on the same vector. Figure 4d shows that the expression of the antisense-RNA persisted at 9 and 13 days and it corresponded to a very high efficiency of skipping (almost 50%, Figure 4e) and rescue of dystrophin synthesis (Figure 4f).
Figure 4.
Transdifferentation of DMD Δ45–50 fibroblasts to myoblasts and dystrophin rescue upon exon 51 skipping. (a) Schematic representation of MyoD and M-U1#51 constructs. The U1 cassette expressing the 5′3′esx construct was cloned into the dU3 region. (b) Western blot analysis with anti-dystrophin (Dys) and anti-tubulin antibodies (Tub) on proteins from MyoD-infected wild-type fibroblasts (Fb WT) collected at days 0, 2, 4, 6, and 8 after induction of differentiation (C: proteins from human skeletal muscle cells). Δ45–50 fibroblasts were infected with MyoD or M-U1#51 and collected at 9 and 13 days of differentiation. Samples were analyzed by: (c) western blot, for MyoD and GFP expression, (d) northern blot, for antisense-RNA expression, (e) nested RT-PCR for exon skipping, and (f) western blot for dystrophin rescue. Western blot with anti-tubulin (Tub) antibodies was used as a loading control. cDNA, complementary DNA; DMD, Duchenne muscular dystrophy; GFP, green fluorescent protein cDNA; hPGK, human phosphoglycerate kinase promoter; IRES, internal ribosome entry site; MyoD, MyoD cDNA.
Discussion
Exon skipping is one of the most promising gene therapy approaches for the treatment of DMD. Two phase I clinical trials have been concluded17,18 using different kinds of modified-AONs delivered to the patients by local intramuscular injections. In both studies, the analysis performed 3–4 weeks after treatment revealed a high percentage of dystrophin positive fibers and proved to be safe and well tolerated.
A parallel strategy to the use of synthetic oligonucleotides is represented by a gene therapy approach in which the antisense molecule is continuously expressed as part of a cellular RNA. Several groups have indeed proven the feasibility of this approach through the use of expression cassettes for snRNA-based antisense molecules delivered to mdx mice muscle cells by AAV vectors.22,23,24 Further studies in the mdx model have also shown that a single systemic injection into young (6 week old) animals of AAV1-U1 antisense-RNA constructs was effective in maintaining the physiological and molecular benefits for the entire lifespan of the animals.25 These results encouraged the idea that one single treatment could be effective in providing a long-term benefit and provided the basis for designing protocols with potential applications of AAV-mediated gene therapy in human.
The objective of this work was the selection of the most effective antisense-RNA for the skipping of exon 51 of the human DMD gene. Previous work in our lab indicated that both splice junctions had to be targeted by the antisense molecules in order to produce efficient skipping.19 However, more recent evidences indicated the very crucial function of ESEs in dictating splicing efficiency26 and as powerful targets for antisense strategies involving either exon skipping or inclusion.33 Furthermore ESEs seem to play a critical role in the context of the dystrophin pre-mRNA where the recognition of the correct exon sequences could be hampered by the huge intron size.34 Therefore, in analogy to what has been done for synthetic oligos, it was mandatory to design and test a collection of antisense molecules against different combinations of such target sequences in order to select the winner construct for further preclinical and clinical studies. Moreover, due to the utilization of an snRNA carrier, it was important to assess the activity of the different antisense combinations when part of the U1 snRNA ribonucleoprotein.
Ten different U1 snRNA-based constructs combining antisense sequences directed against splice junctions and ESEs were designed, cloned and tested for in vivo stability and exon 51 skipping activity. All of them were shown to stably accumulate in the cell despite the fact that the U1 snRNA size was sometime considerably modified by the addition of the antisense sequences. Notably, these molecules were still able to bind the U1-70K protein, which represents an hallmark for the assembly of stable U1 snRNP particles, thus explaining the reason for their stable accumulation in the cell.
The exon 51 skipping behavior of each antisense molecule was tested by infecting myoblasts derived from patient biopsies with the deletion of exons 48–50. This allowed us to select the two best performing molecules (5′3′ and 5′3′esx), differing only for an additional antisense element against a putative ESE in the 5′3′esx construct. Subsequently, their activity was further compared using a luciferase-based splicing-reporter assay that confirmed the higher activity of the 5′3′esx construct. These data indicated that the ESE target region present in the 5′3′esx construct plays an important role in exon 51 splicing and that, in the U1 snRNA context, is able to improve skipping efficiency when associated to antisense sequences against splice junctions. These results, together with the fact that the U1 constructs containing only anti-ESE antisense have very poor activities, allowed us to conclude that U1 snRNA works efficiently, as expected, primarily through recognition of splice junctions. This appears to be relevant difference with the U7 snRNA vector that works efficiently in the recognition of ESE sequences independently from the targeting of splice sites20,21 and from synthetic oligos, which act independently of any carrier RNA.17,18
The activity of the winner construct was finally tested on a different genetic background (deletion 45–50) utilizing transdifferentiated patients' fibroblasts. The possibility of testing exon-skipping activity in fibroblasts derived from skin biopsies is very important, considering that muscle biopsies are invasive surgical interventions and DMD patients are basically children, already weakened by the course of their pathology.
In conclusion, the results of these studies indicated that the 5′3′esx construct is very active in inducing skipping of exon 51 in two different DMD mutant backgrounds and in two different cellular systems and therefore, it can be considered the best candidate to be selected for future preclinical and clinical studies.
Previous work in mice indicated that the delivery of AAV viruses carrying U1-antisense expression cassettes provided a very good system for body-wide rescue of dystrophin synthesis. Moreover, the observation that the beneficial effect of this treatment lasted the entire lifespan of the animal without any secondary/toxic effect, together with the persistence of expression from AAV vectors in muscle of nonhuman primates for >6 years,35 suggested that the same strategy could be applied for a long-term treatment of human DMD patients. Translating this approach to large animals or humans is not so obvious due to the high doses of vector required and the immune response to capsid proteins if subsequent injections would be required. However, recent articles demonstrated that the formation of anti-AAV1 antibodies can be successfully abrogated by an immunomodulation treatment, providing a protocol with potential applications to AAV-mediated therapies in humans.36,37 Further studies and accurate preclinical tests will be required in order to provide the appropriate answers to these open issues.
Materials and Methods
Antisense clones construction
U1#51 5′3′: This first clone was PCR-derived from the previously described U1-5′ (ref. 19) and double antisense cassette was inserted in the U1 cassette already used for U1#23.
Clones U1#51 were obtained by inverse PCR on the construct pRRL-5′3′.
U1#51 ESE A:
U1#51AON1R
(5′-AGAAATGCCATCTTCCTTGAATGAGATCTTGGGCCTCTGC-3′);
U1F2 (5′-GGCAGGGGAGATACCATGATC-3′).
U1#51ESEA*:
U1#51ESEA*F
(5′-AGATGGCATTTCTAGGGCAGGGGAGATACCATGATC-3′);
U1#51ESEA*R
(5′-TCCTTGATGTTGGAGATGAGATCTTGGGCCTCTGC-3′)
U1#51ESEA+B:
U1#51AON1F
(5′-TCAAGGAAGATGGCATTTCTGGCAGGGGAGATACCATGATC-3′)
U1#51AON2R
(5′-ATCAAGTTATAAAATCACAGAGGATGAGATCTTGGGCCTCTGC-3′).
U1#513′ESEA:
U1#51-3′F
(5′-GTCTGAGTAGGAGCTAAAATATTTTGGGGGCAGGGGAGATACCATGATC-3′);
U1#51 AON1R
(5′-AGAAATGCCATCTTCCTTGAATGAGATCTTGGGCCTCTGC-3′)
U1#515′ESEB:
U1#51ESEBF
(5′-CCTCTGTGATTTTATAACTTGATGGCAGGGGAGATACCATGATC-3′);
U1#515R
(5′-ATCAAGCAGAAGGTATGAGAAAAAATGAGATCTTGGGCCTCTGC-3′)
U1#515′3′ESEA*:
U1#515′3′ESEA*F
(5′-GGAAGATGGCATTTCTAGGTCTGAGTAGGAGCTGGCAGGGGAGATACCATGATC-3′);
U1#515′3′ESEA*F
(5′-TTGATGTTGGAGCATCAAGCAGAAGGTATGAGATGAGATCTTGGGCCTCTGC-3′)
U1#515′3′Esx:
U1#515′3′sxF
(5′-TCAAGGAAGATGGGTCTGAGTAGGAGCTGGCAGGGGAGATACCATGATC-3′);
U1#515′3′sxR
(5′-TGTTGGAGCATCAAGCAGAAGGTATGAGATGAGATCTTGGGCCTCTGC-3′)
U1#515′3′Edx:
U1#515′3′dxF
(5′-TCCAACATCAAGGTCTGAGTAGGAGCTGGCAGGGGAGATACCATGATC-3′);
U1#515′3′dxR
(5′-GGTACCTGCATCAAGCAGAAGGTATGAGATGAGATCTTGGGCCTCTGC-3′)
U1#515′3′L:
U1#515′3′LF
(5′-CAGAGTAACAGTCTGAGTAGGAGCTAGGCAGGGGAGATACCATGATC-3′);
U1#515′3′LR
(5′-GATCAAGCAGAAGGTATGAGAAAAAATGAGATCTTGGGCCTCTGC-3′).
The PCR fragments were digested with NheI and inserted into the NheI-digested pRRLSIN.cPPT.PGK/GFP.WPRE lentiviral backbone.29 The clone pCCL-MyoD/5′3′esx was derived from pRRL-5′3′ESX with a digestion reaction using endonucleases SalI and ScaI, and cloning the resulting excised insert into the SalI–ScaI-digested pCCL-MyoD (provided by Maurizia Caruso).
Cell cultures. Cultures of primary myoblasts were first preplated in order to separate fibroblasts from the primary line, seeded in Human Skeletal Muscle Growth Medium (PromoCell, Heidelberg, Germany) and grown in a humidified incubator, at 5% CO2 and 37 °C.
Cultures of primary fibroblasts were established through out-growth from both human DMD and healthy skin biopsies in RPMI with 15% fetal calf serum, 1% penicillin–streptomycin and 1% glutamine (GIBCO/BRL Life Technologies, Grand Island, NY). Fibroblasts were then maintained as described above.
Virus preparation and cell transduction. The 293T cells were plated on 25-cm diameter plates (three plates/virus) and co-transfected with lentiviral constructs and packaging plasmids Plp1, Plp2, and PlpV/SVG (Invitrogen, Carlsbad, CA) according to the four-plasmid transient transfection method previously described.29
Supernatant was collected for the following 2 days after the transfection and then centrifuged in a Beckmann Ultracentrifuge at 20,000g for 2 hours. Pellets were resuspended in Hank's balanced salt solution buffer (Invitrogen).
Transduction efficiency was tested infecting HeLa cells: cells were then collected for RNA and protein extraction, in order to test antisense and GFP expression, respectively.
The day before transduction, myoblasts or fibroblasts were seeded in growth medium, on 60-mm diameter plates (at least two for each different virus), at a density of 5 × 105 cells per plate.
The next 2 days they were infected twice with lentiviruses and polybrene (4 mg/ml).
The second day from the last infection, cells were induced to differentiate with Human Skeletal Muscle Differentiation Medium (PromoCell). After 7 days (for myoblasts) or 9 and 13 days (for fibroblasts) of differentiation, cells were washed twice with complete phosphate-buffered saline (PromoCell) and collected with the help of a gum scraper with 300 µl of protein buffer [100 mmol/l Tris–HCl pH 7.4, 1 mmol/l EDTA, 2% sodium dodecyl sulfate, 1× Complete EDTA-free Protease Inhibitor Cocktail (Roche, Applied Science, Mannheim, Germany)] for protein extraction, or with 1 ml of TRIZOL reagent (Invitrogen) for RNA extraction.
Proteins extraction and analysis. Proteins were extracted from cell plates with 100 mmol/l Tris–HCl (pH 7.4), 1 mmol/l EDTA, 2% sodium dodecyl sulfate (SDS), and a protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). Samples were placed on a rotary shaker at 4 °C for 30 minutes, centrifuged at 13,200 r.p.m. for 20 minutes at 4 °C and supernatant containing protein extracts was collected. Concentration was assessed with the BCA assay (PIERCE; Thermo Fisher Scientific, Rockford, IL), according to the manufacturer's instructions.
Western blot. Fifty micrograms of protein extracts (for dystrophin and tubulin) were loaded onto a NuPAGE Tris–Acetate Minigel 3–8% 1 mm (Invitrogen); 15 µg (for GFP and MyoD) were run on a NuPAGE Bis–Tris minigel 10% 1 mm (Invitrogen).
Running and blotting were performed in an XCell SureLock Minicell (Invitrogen) according to the manufacturer's instructions and proteins were transferred to a nitrocellulose transfer membranes (Protran; Schleicher & Schuell BioScience, Keene, NH).
Membranes were blocked with 10% nonfat dry milk (Difco Skim Milk; Becton & Dickinson, Le Pont de Claix, France), incubated for 1 hour with primary antibody [for dystrophin: NCL-DYS1 (Novocastra, Newcastle upon Tyne, UK) diluted 1:40 in 3% milk; for tubulin: anti-tubulin AA12 (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:200 in Tris-buffered saline with Tween 20; for GFP: anti-GFP ab290 (AbCAM, Cambridge, UK) diluted 1:2,500 in Tris-buffered saline with Tween 20; for MyoD: anti-MyoD 5.8 A (Santa Cruz Biotechnology), diluted 1:500 in 2% milk], washed in Tris-buffered saline with Tween 20 (Sigma Aldrich, St Louis, MO), and incubated with goat anti-mouse IgG (H+L) horseradish peroxidase–conjugated secondary antibody (diluted 1:5,000 in 3% milk) for dystrophin, tubulin, and MyoD, or ImmunoPure Goat Anti-Rabbit IgG Peroxidase Conjugated (PIERCE; Thermo Fisher Scientific) for GFP for 1 hour. Protein detection was carried out with SuperSignal chemiluminescent substrate (PIERCE; Thermo Fisher Scientific).
RNA preparation and analysis. Cells were harvested with 1 ml of Trizol (Invitrogen) and RNA was extracted according to the manufacturer's instructions; concentration was assessed with Nanodrop ND-1000 Spectrophotometer (CELBIO, Pero, Milan, Italy).
RT-PCR. Dystrophin mRNA was analyzed by RT-PCR on 200 ng of total RNA with oligos E46F (5′-GCTAGAAGAACAAAAGAATAT-3′) for deletion 48–50 or oligo E43F (5′-CTACAACAAAGCTCAGGTCG-3′) for deletion 45–50 and E54R (5′-CTTTTATGAATGCTTCTCCAAG- 3′) for 40 cycles with the Access RT-PCR system (Promega, Madison, WI). Four microliters of the RT-PCR products were then used as template for the nested reaction performed with oligo E47F (5′-TTACTGGTGGAAGAGTTGCC-3′) and E52Ro (5′-TTCGATCCGTAATGATTGTTCTAGCC-3′) for 30 cycles.
A 10 µl of the reactions were run on a 2% agarose–ethidium bromide gel and signals were revealed on a UV transilluminator.
Northern blot. Northern analysis was performed as already described.38 Briefly, 10 µg of total RNA were loaded onto a 6% polyacrylamide gel, run at 17 mA, and transferred into a Hybond–N+ nitrocellulose membrane (Amersham, GE Healthcare Life sciences, Buckinghamshire, UK) for 16 hours at 10 volts and 4 °C. Membranes were hybridized with probe α-U1 (5′-CAGGGGAAAGCGCGAACGCAGTCCCCCA-3′) and revealed using a Typhoon TRIO Variable Mode Imager system (Amersham, GE Healthcare Life sciences), with ImageQuant TL.I program.
pLuc-ex51 construction. The insert ex51 was obtained by PCR from genomic DNA using the oligos:
5′-AAGGAAAAAAGCGGCCGCATGAGAATGAGCAAAATCGT-3′ and
5′-CCTTAATTAAGAGACAACTATTCTTGTAAG-3′.
The italicized bases are NotI and PacI restriction sites.
Ex51 is made of the whole exon 51 flanked by 268 bp of intron 50 and 263 bp of intron 51. The PCR fragments were digested with NotI and PacI enzymes and inserted into the NotI and PacI-digested pcDNA3.1-Luc vector.32
C27 transfections. C2.7 myoblasts39 are a mouse myogenic cell line derived from the C2 cells, isolated from mouse-activated satellite cells.40 C27 cells were plated on 35-mm diameter plates; they were co-transfected with 2 µg of pLuc-ex51, 2 µg of the lentiviral vector carrying the antisense expression cassette and 50 ng of a Renilla Luciferase expressing construct, used as a transfection efficiency control. The transfection was performed according to the Lipofectamine 2000 protocol (Invitrogen). Cells were grown in Dulbecco's modified Eagle's medium 10% fetal bovine serum for 36 hours.
Luciferase assay. C27 cells were collected using 250 µL of passive lysis buffer and the assay was performed according to Dual-Luciferase Reporter Assay System protocol (Promega).
Acknowledgments
We thank Marina Mora and Telethon Neuromuscular Biobank (Istituto Carlo Besta, Milan, Italy) for providing 8962 Δ48–50 cells, Carlo Manzoni (Università Cattolica del Sacro Cuore, Rome, Italy) for providing skin biopsies, Akio Masuda (Division of Neurogenetics Center for Neurological Diseases and Cancer, Nagoya University, Japan) for providing pcDNA3.1-Luc vector, and Maurizia Caruso (CNR Istituto Neurobiologia e Medicina Molecolare, Rome, Italy) for providing pCCL-MyoD; we also thank Marcella Marchioni (CNR, Istituto di Biologia e Patologia Molecolari, Rome, Italy) for technical help. This work was partially supported by grants from: Telethon (GGP07049), Parent Project Italia, EU project SIROCCO (LSHG-CT-2006-037900), ESF project “NuRNASu” and PRIN. Authors declare no conflict of interest.
REFERENCES
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C., and, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM., and, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Blake DJ, Weir A, Newey SE., and, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Gailly P. New aspects of calcium signaling in skeletal muscle cells: implications in Duchenne muscular dystrophy. Biochim Biophys Acta. 2002;1600:38–44. doi: 10.1016/s1570-9639(02)00442-9. [DOI] [PubMed] [Google Scholar]
- Colussi C, Mozzetta C, Gurtner A, Illi B, Rosati J, Straino S, et al. HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc Natl Acad Sci USA. 2008;105:19183–19187. doi: 10.1073/pnas.0805514105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir LA., and, Chamberlain JS. Emerging strategies for cell and gene therapy of the muscular dystrophies. Expert Rev Mol Med. 2009;11:e18. doi: 10.1017/S1462399409001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trollet C, Athanasopoulos T, Popplewell L, Malerba A., and, Dickson G. Gene therapy for muscular dystrophy: current progress and future prospects. Expert Opin Biol Ther. 2009;9:849–866. doi: 10.1517/14712590903029164. [DOI] [PubMed] [Google Scholar]
- Manzur AY., and, Muntoni F. Diagnosis and new treatments in muscular dystrophies. Postgrad Med J. 2009;85:622–630. doi: 10.1136/jnnp.2008.158329. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H., and, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, et al. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- Love DR, Flint TJ, Genet SA, Middleton-Price HR., and, Davies KE. Becker muscular dystrophy patient with a large intragenic dystrophin deletion: implications for functional minigenes and gene therapy. J Med Genet. 1991;28:860–864. doi: 10.1136/jmg.28.12.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Bremmer-Bout M, Aartsma-Rus A, de Meijer EJ, Kaman WE, Janson AA, Vossen RH, et al. Targeted exon skipping in transgenic hDMD mice: a model for direct preclinical screening of human-specific antisense oligonucleotides. Mol Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD., and, Wilton SD. Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J Gene Med. 2006;8:207–216. doi: 10.1002/jgm.838. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, et al. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Δ48–50 DMD cells. Proc Natl Acad Sci USA. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C, Suter D, Pauli C, Dunant P, Lochmüller H, Burgunder JM, et al. U7 snRNAs induce correction of mutated dystrophin pre-mRNA by exon skipping. Cell Mol Life Sci. 2003;60:557–566. doi: 10.1007/s000180300047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchaouir R, Meregalli M, Farini A, D'Antona G, Belicchi M, Goyenvalle A, et al. Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice. Cell Stem Cell. 2007;1:646–657. doi: 10.1016/j.stem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C, et al. Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model. Proc Natl Acad Sci USA. 2006;103:3758–3763. doi: 10.1073/pnas.0508917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C, et al. Chimeric adeno-associated virus/antisense U1 small nuclear RNA effectively rescues dystrophin synthesis and muscle function by local treatment of mdx mice. Hum Gene Ther. 2006;17:565–574. doi: 10.1089/hum.2006.17.565. [DOI] [PubMed] [Google Scholar]
- Denti MA, Incitti T, Sthandier O, Nicoletti C, De Angelis FG, Rizzuto E, et al. Long-term benefit of adeno-associated virus/antisense-mediated exon skipping in dystrophic mice. Hum Gene Ther. 2008;19:601–608. doi: 10.1089/hum.2008.012. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, De Winter CL, Janson AA, Kaman WE, Van Ommen GJ, Den Dunnen JT, et al. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Heemskerk JA, De Winter CL, Van Ommen GJ., and, Van Deutekom JC. Therapeutic modulation of DMD splicing by blocking exonic splicing enhancer sites with antisense oligonucleotides. Ann N Y Acad Sci. 2006;1082:74–76. doi: 10.1196/annals.1348.058. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ., and, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D, Cittadini A, Latronico MV, Borello U, Aycock JK, Drusco A, et al. ‘Advanced' generation lentiviruses as efficient vectors for cardiomyocyte gene transduction in vitro and in vivo. Gene Ther. 2003;10:630–636. doi: 10.1038/sj.gt.3301936. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Michienzi A, De Angelis FG., and, Bozzoni I. The Rev protein is able to transport to the cytoplasm small nucleolar RNAs containing a Rev binding element. RNA. 1999;5:993–1002. doi: 10.1017/s1355838299990064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR., and, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Masuda A, Matsuura T, Ito M, Okushin K, Engel AG, et al. Tannic acid facilitates expression of the polypyrimidine tract binding protein and alleviates deleterious inclusion of CHRNA1 exon P3A due to an hnRNP H-disrupting mutation in congenital myasthenic syndrome. Hum Mol Genet. 2009;18:1229–1237. doi: 10.1093/hmg/ddp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., and, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL., and, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Gao GP, Grant RL, Schnell MA, Zoltick PW, Rozamus LW, et al. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood. 2005;105:1424–1430. doi: 10.1182/blood-2004-06-2501. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS, et al. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. 2007;15:1160–1166. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- Lorain S, Gross DA, Goyenvalle A, Danos O, Davoust J., and, Garcia L. Transient immunomodulation allows repeated injections of AAV1 and correction of muscular dystrophy in multiple muscles. Mol Ther. 2008;16:541–547. doi: 10.1038/sj.mt.6300377. [DOI] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B., and, Bozzoni I. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinset C, Montarras D, Chenevert J, Minty A, Barton P, Laurent C, et al. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation. 1988;38:28–34. doi: 10.1111/j.1432-0436.1988.tb00588.x. [DOI] [PubMed] [Google Scholar]
- Yaffe D., and, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]