Abstract
The biomimetic design of silk/silica fusion proteins was carried out, combining the self assembling domains of spider dragline silk (Nephila clavipes) and silaffin derived R5 peptide of Cylindrotheca fusiformis that is responsible for silica mineralization. Genetic engineering was used to generate the protein-based biomaterials incorporating the physical properties of both components. With genetic control over the nanodomain sizes and chemistry, as well as modification of synthetic conditions for silica formation, controlled mineralized silk films with different silica morphologies and distributions were successfully generated; generating 3D porous networks, clustered silica nanoparticles (SNPs), or single SNPs. Silk serves as the organic scaffolding to control the material stability and multiprocessing makes silk/silica biomaterials suitable for different tissue regenerative applications. The influence of these new silk-silica composite systems on osteogenesis was evaluated with human mesenchymal stem cells (hMSCs) subjected to osteogenic differentiation. hMSCs adhered, proliferated, and differentiated towards osteogenic lineages on the silk/silica films. The presence of the silica in the silk films influenced osteogenic gene expression, with the upregulation of alkaline phosphatase (ALP), bone sialoprotein (BSP), and collagen type 1 (Col 1) markers. Evidence for early bone formation as calcium deposits was observed on silk films with silica. These results indicate the potential utility of these new silk/silica systems towards bone regeneration.
Introduction
Biomaterials with tunable nanoscale features can offer improved performance for in vivo applications.1 Silica is widely used in industry, medicine, and nanotechnology where structural control is necessary but also challenging.2,3 Here we describe a new procedure using ambient conditions, for the unusual and detailed control of silica morphology and distribution on the surface of silk films utilizing genetically engineered chimeric proteins. A genetic combination of spider dragline silk sequence (Nephila clavipes) and the silaffin derived R5 peptide of the diatom (Cylindrotheca fusiformis) led to the bioinspired synthesis of 3D porous silica networks, clustered silica nanoparticles (SNPs), or single/isolated SNPs. We anticipate that these silica-based biomaterials will have applications in tissue regeneration and drug delivery due to the ability to regulate the location and morphological features of the silica. The silk component serves as an organic scaffold that controls material stability and allows multiple modes of processing. Silica serves as an important osteoinductive element with potential control of remodeling rate and tissue regeneration outcomes in vivo. Additionally, Si-derived nanostructures with strong morphological and spatial control are attractive for electronics,4 biosensors,5 microfluidic devices,6 and DNA microarray technology.7 The novelty in material design also allows for applications in biodopants8 and protein-silica nanocomposites.9
Silica biomineralization has attracted much attention due to the remarkable morphological control of silica nanopatterns synthesized in vivo that exceed current synthetic and technological capabilities in vitro. Examples of controlled silica biosynthesis are the skeletal architectures from diatoms formed under physiological conditions (aqueous environment, neutral pH, low temperatures) and driven by biomolecules,10,11 in contrast with harsh industrial synthesis conditions. Studies to date show widespread bioapplications of porous silica nanostructures in drug delivery,12 targeted therapies,13 and cell labeling, where internalization shows no cytotoxic effects.14 Silica can also be easily derivatized to immobilize antibodies or proteins, and once supported on a stable scaffold carrier make excellent implant material. Silica nanoparticles enhanced and stabilized the fluorescence of encapsulated organic dyes15 and when bioconjugated find applications in imaging, sensing, targeting, and detection of single cells. Also, inorganic-organic silica hybrids have potential use in optoelectronics,16 molecular machines,17 tunable lasers,18 or electrodes for biofuel cells.19
Silica nanostructures of different morphologies and distributions synthesized on silk films have not been previously fabricated. Other bioinspired silica particle formation20 as well as silica structures with different morphologies (arch shapes, fibers)21 have appeared. The present work represents a new type of biomaterial with many benefits. First, silk/silica binding chimeric proteins precipitate silica in situ,22 at room temperature with commercially available reagents, without the need for specialized equipment. Second, silks self assemble into highly stable β-sheet structures23 with remarkable mechanical properties that improve material properties and stability against organic solvents, water and strong acids and bases. Third, the biodegradability of silk24 and the aqueous processing conditions for silica mineralization make it suitable for use in living tissues. Further, since the sizes and distributions of the silica component can be controlled in the bioengineering process, new levels of control of degradation lifetime can be tailored into the materials. Fourth, silks can be processed into fibers,25 films,26 porous matrices27 or hydrogels.28 Finally, surfactant free silica nanostructures offer silanol groups that can be utilized for a diverse range of applications.
The objective of the current study was to design, clone and express a new family of silk-silica fusion proteins, wherein silica morphology and distribution could be controlled to a high degree, and osteogenesis would be promoted from stem cells.
Experimental Section
Chemicals
All chemicals were purchased from Sigma-Aldrich (MO, USA) or Fluka (WI, USA) and used without further purification. Cell medium ingredients were purchased from Invitrogen (CA, USA) and Sigma-Aldrich (MO, USA).
Preparation of silk/silica chimeric proteins
Silk/silica chimeric proteins were genetically engineered and expressed using our previous procedure.29 Expressed proteins were purified using Ni-NTA resin (Qiagen, CA, USA) and dialyzed (membrane MWCO 2,000 g mol–1; Pierce, Woburn, MA, USA) in 100 mM phosphate buffer at pH 5.5 for 1 day changing buffer 2 times and against distilled water for 3 days changing the water 3 times a day. The silk solutions were freeze dried to obtain the protein powder.
Preparation of silk and silk/silica films
Hexafluoro-2-propanol (HFIP) based silk solutions were prepared at a concentration of 2.5% and 5% wt./vol. and stirred overnight to ensure complete protein dissolution. Next, 50 μL of the silk solutions were cast on the bottom of polystyrene boat used subsequently for SEM imaging and twenty-four-well tissue culture plate (TCP) used for cell culture. Films were left to dry for 24 hours. Next, 2 mL of 90% methanol solution in water was added to each boat and incubated for 10 minutes to induce β-sheet formation in the silk films. Then, methanol was removed and films were left to dry overnight in a fume hood. This step was used to generate silk films that were stable in an aqueous environment and suitable for cell culture. Films were sterilized with 70% ethanol, left to dry, and placed under UV light for 5 minutes in a laminar flow hood. TCP only was used as a control. Prior to cell seeding, 1 mL of cell culture medium was added to each well, soaked for 30 min, and then aspirated.
Human mesenchymal stem cells (hMSCs)
Bone marrow aspirate from a young healthy donor was obtained from Lonza (Walkersville, MD). Frozen low passage (2 or 3) hMSC stocks were thawed and suspended in growth medium containing high glucose Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1% antibiotic/antimycotic, 1% non-essential amino acids, and 10 ng basic fibroblast growth factor (bFGF). The cells were plated onto the silk and silk/silica films at a density of 5,000 cells/well in a 24-well plate and kept in a humidified incubator at 37°C and 5% CO2. The cells were cultured in hMSC media until 85% confluency and then the medium was changed to an osteogenic medium containing high glucose Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1% antibiotic/antimycotic, 1% non-essential amino acids, 100 nM dexamethasone, 10 mM β-glycerolphosphate, and 0.05 mM L-ascorbic acid 2-phosphate. The medium was changed every 3–4 days. Cell growth and shape were monitored using a phase contrast light microscope (Carl Zeiss, Jena, Germany) equipped with a Sony Exwave HAD 3CCD (Sony Electronics, Inc., USA) color video camera.
Gene expression using real–time RT-PCR analysis
After osteogenic culture, the cells from three separate wells of each type were lysed in 0.35 mL Buffer RLT (Qiagen #79216, CA, USA) containing 10% mercaptoethanol, followed by shredding in a QIAshedder (Qiagen #79656, CA, USA). RNA was isolated from the cells using an RNeasy Mini Kit (Qiagen #74106, CA, USA). From this RNA, cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems #4368814, MA, USA) following the manufacturer's instructions. The cDNA samples were analyzed for expression of alkaline phosphatase, collagen type I, and bone sialoprotein relative to the GAPDH housekeeping gene using Assay-on-Demand™ Gene Expression kits with TaqMan® Universal PCR Master Mix (ABI #4364340) (Applied Biosystems AoD probes, MA, USA). The data were analyzed using the ABI Prism 7000 Sequence Detection Systems software.
SEM Characterization. Parameters
After 2 weeks of osteogenic culture, the basal parts of the tissue culture dishes were removed and fixed in 2.5% vol/vol glutaraldehyde solution in PBS for 30 minutes. Next, the samples were washed in DI water and dehydrated by 20 min incubation in a series of ethanol solutions in water (10%, 30%, 60%, 80%, 90%, 100% vol/vol. Surface morphology down to the nanometer levels was evaluated using a Carl Zeiss (Carl Zeiss SMT, Germany) Ultra 55 Field Emission Scanning Electron Microscope (FESEM) operating at an accelerating voltage of 10 kV and using an in-lens ion annular secondary electron detector. The bottom of the tissue culture plate (TCP) well with cast film was cut and coated with a carbon layer (Baltec Carbon Coater, model MED-020, Liechtenstein) to ensure sample conductivity necessary for imaging. Elemental composition of the silk film surface was evaluated using an Energy Dispersive X-ray Spectrometer (EDAX) connected to the SEM instrument.
Results and Discussion
Scheme 1 shows the strategy for biomaterial formation and mineralization method that led to three types of silk/silica surfaces. Five percent and 2.5% wt./vol. silk films produce two different morphologies of silica deposits, a 3D dense network and aggregates of SNPs distributed over the silk surface, respectively. Silica was deposited on the silk film surface presumably through electrostatic interactions of negatively charged silica species and the R5 peptide.29 The additional positive charge on the silk surface was induced via charged imidazole groups of the N-terminal histidine tag which enhanced silica deposition.30,31 Also, 2.5% silk films produced dispersed SNPs on the silk film surface when glycerol was present in the mineralizing solution. The addition of glycerol aids in dispersion of SNPs on the silk film surface likely through stabilization of SNPs thus preventing aggregation. Figure 1 compares silica morphologies and distribution on the surface of silk films formed under the different conditions. Frames A-B of Figure 1 show scanning electron microscopy (SEM) images of a 5% silk film after the silicification reaction. The surface exhibits silica nanostructures arranged in dense 3D networks, consistent with the model in Scheme 1. The abundance of positive charges expected for 5% silk films induces a strong attraction for silica species to the surface of silk film, causing the immediate precipitation of silica that uniformly covers the whole silk film surface. The high microporosity of the silica deposits increases the amount of surface hydroxyl groups that can be functionally modified, forming new inorganic-organic hybrids. This type of material can become compatible in various solutions and is currently being examined for its potential as a drug delivery vehicle. Frames C-D of Figure 1 show SEM images of SNPs precipitated on a 2.5% silk film surface. The surface contains a lower overall density of silica nanostructures (2.4/μm2), as compared to 100% coverage discussed above, and more importantly the silica morphology changed drastically with predominant clusters of SNPs compared to the widespread porous network observed for the 5% silk films. The average diameter of SNPs is fairly uniform (400 nm ± 90 nm). The decreased concentration of silk lowers the overall content of R5 peptide and histidine moieties on the film surface that leads to localized distribution of positive charges and local precipitation of SNPs. The addition of glycerol to the silicification reaction on 2.5% silk films changes the SNP distribution on the surface of silk films as shown in Figure 1, frames E-F. The surface is covered with uniformly spread isolated SNPs in the density of SNPs 1.6/μm2. The nanometer size range and separation of SNPs over large areas offers unique properties of this biomaterial with respect to applications in tissue engineering. The surfactant free SNPs surfaces can be easily modified for targeted therapies or for imaging.
Scheme 1.

Method to control silica distribution on silk films.
Figure 1.

SEM images of (A-B) 5%, (C-D) 2.5%, and (E-F) 2.5% (glycerol) wt./vol. silk films with silica nanostructures.
The effect of methanol treatment on silk films was evaluated to induce a structural transition from random coil to secondary β-sheet structure;32 the stable conformation in aqueous environment. The tendency of the films to form β-sheets can be evaluated using FTIR spectroscopy by analyzing the amide I (1600-1700 cm-1) and amide II (1500-1600 cm-1) regions of the silk film spectrum as shown in Figure 2A. The characteristic peaks at 1627 cm-1 and 1533 cm-1 correspond to the N-H stretch and N-H bend respectively and indicate hydrogen bond and β-sheet formation within the film, after solvent treatment.
Figure 2.
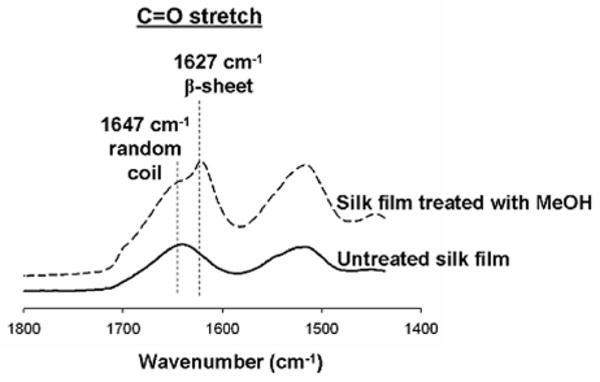
Comparison of FTIR spectra of silk film before and after methanol treatment. Methanol induces β-sheet formation within the silk film as shown as a shift in the FTIR spectrum.
The silica formation on silk/silica films was confirmed by EDAX analysis and a representative spectrum is shown in Figure 2B. The presence of silicon and oxygen peaks at 1.74 keV and 0.623 keV respectively indicate that the nanostructures observed in SEM images (Figure 1) originate from silica. The oxygen content is partially attributed to the underlying silk film as well as carbon (also coating) and nitrogen.
Enhancement of new bone formation with silica containing bone implants has already been established. Nevertheless, use of nanostructural silica in a supporting matrix is often desired to enable material biodegradation within appropriate time frames matching the rates of new bone tissue formation. It was also shown that primary human osteoblasts adhere better to roughened mineral surfaces.33 We tested our silk/silica films to support the human mesenchymal stem cells (hMSCs) growth and osteogenic differentiation. Control samples were silk film only without silica mineralization and TCP. Cells showed good adhesion to all silk/silica films and proliferation rates reaching 85% confluence on day 5, the starting point of osteogenic differentiation. Cells grown on plastic reached 95% confluence on day 5, showing similar growth rates. Cell morphology was examined by light microscopy at two time points, 5 days after cell seeding and 2 days after differentiation into osteogenic lineage. Before differentiation similar cell densities and morphologies were observed on all silk films, showing spindle-like shapes characteristic of undifferentiated fibroblastic hMSCs, with example morphology shown in Figure 4A. After 2 days of osteogenic stimulation cells spread evenly on silk film surfaces forming a thin coating with closely packed flattened cells, as can be observed on a representative image in Figure 4B. No differences in cell morphology were observed between the silk/silica samples and the control samples, although after 2 weeks of osteogenic culture some detachment and cell clustering was observed for cells grown on TCP and silk only but not on silk/silica films.
Figure 4.

Optical microscope images of hMSCs on 5% wt/vol silk films mineralized with silica before (A) and after (B) differentiation into osteogenic lineages.
The ability of silica dissolution ions to enhance new bone formation has already been established with bioactive glasses commonly used as bone implants.34,35 We further evaluated silk/silica films for the ability to modulate hMSCs differentiation. Expression of bone-specific markers was quantified using real-time RT-PCR after 2 weeks of osteogenic culture. Figure 5A shows that alkaline phosphatase was upregulated on all silk/silica films with respect to the control samples (silk only and TCP). Figure 5B shows that bone sialoprotein (BSP) gene expression was upregulated when the cells were cultured on 5% silk films mineralized with silica, with respect to the control samples. Expression of collagen I was as well elevated on 5% silk films mineralized with silica in osteogenic culture when compared to the controls. The upregulation of ALP on all silk/silica films, BSP and Col 1 on 5% silk/silica film in osteogenic culture suggests the influence of silica presence on gene expression towards upregulation of osteogenesis. Also, since BSP upregulation is an indication of early apatite nucleation36 we further tested 5% silk/silica film with EDAX mapping to evaluate calcium presence. After 2 weeks in osteogenic cell culture we observed calcium mineral deposits on 5% silk/silica film surface (Figure 6), the main component of early apatite. The silicon signal was also observed indicating residual silica minerals.
Figure 5.

Osteogenic gene expression of cells grown on 5% wt./vol. silk films with silica or silk only (Silk) and tissue culture plastic (TCP) after exposure to osteogenic stimulants for 2 weeks as compared. Each column represents the mean and standard deviation of N = 3 independent cultures. P<0.05.
Figure 6.

SEM image and EDAX mapping of 5% wt./vol. silk film with silica after 2 weeks of hMSCs osteogenic culture.
Conclusions
A method to control silica deposition on the surface of stable silk films was demonstrated. Different mineral phase loading and morphologies can be tailored synthetically thus altering the biomaterial properties. The size scale, the all-aqueous conditions, and room temperature processing offer in situ reactions in tissue compatible environments to replace currently used bulk silica or bioactive glasses, as well as benefits for imaging and delivery.37 Bioengineering offers fine control over the protein design in terms of nanodomain size, chemistry, and morphology to regulate the biomaterial properties. This approach can be extended to introduce alternative fusions of inorganic phases for other applications. Preliminary studies with hMSCs show support of silk/silica films towards cell attachment and upregulation of osteogenic gene markers, the latter pronounced highly on 5% silk film with porous silica network. These results indicate that silk/silica system enhances osteogenesis.
Figure 3.

EDAX spectrum of silk/silica film exhibiting strong peaks for elemental silicon and oxygen which confirm that nanostructures observed on the surface of silk film originate from silica.
Acknowledgments
This research was supported by NIH (DE017207) and AFOSR (FA 9550-07-1-0079). This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF award no. ECS-0335765. CNS is part of the Faculty of Arts and Sciences at Harvard University.
References
- 1.Sakiyama-Elbert SE, Hubbell JA. Annu Rev Mater Res. 2001;31:183–201. [Google Scholar]
- 2.Tan W, W K, He X, Zhao XJ, Drake T, Wang L, Bagwe RP. Med Res Rev. 2004;24:621–638. doi: 10.1002/med.20003. [DOI] [PubMed] [Google Scholar]
- 3.Coradin T, Livage J. Acc Chem Res. 2007;40:819–826. doi: 10.1021/ar068129m. [DOI] [PubMed] [Google Scholar]
- 4.Neaton JB, M DA, Ashcroft NW. Phys Rev Lett. 2000;85:1298–1301. doi: 10.1103/PhysRevLett.85.1298. [DOI] [PubMed] [Google Scholar]
- 5.Kilian KA, B T, Gooding J. J Chem Commun. 2009;6:630–640. doi: 10.1039/b815449j. [DOI] [PubMed] [Google Scholar]
- 6.Yeung ES. Annu Rev Phys Chem. 2004;55:97–126. doi: 10.1146/annurev.physchem.54.011002.103820. [DOI] [PubMed] [Google Scholar]
- 7.Murthy BR, N JKK, Selamat ES, Balasubramanian N, Liu WT. Biosensors and Bioelectronics. 2008;24:723–728. doi: 10.1016/j.bios.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Silver J, W R, Ireland TG, Fern GR, Zhang S. Nanotechnology. 2008;19:095302/1–095302/7. doi: 10.1088/0957-4484/19/9/095302. [DOI] [PubMed] [Google Scholar]
- 9.Ramanathan M, L HR, Sarsenova A, Wild JR, Ramanculov EK, Olsen EV, Simonian AL. Colloids and Surfaces B:Biointerfaces. 2009;73:58–64. doi: 10.1016/j.colsurfb.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Sumper M, Brunner E. ChemBioChem. 2008;9:1187–1194. doi: 10.1002/cbic.200700764. [DOI] [PubMed] [Google Scholar]
- 11.Perry CC, Keeling-Tucker T. J Biol Inorg Chem. 2000;5:537–550. doi: 10.1007/s007750000130. [DOI] [PubMed] [Google Scholar]
- 12.Radu DR, Lai CY, Jeftinija S, Lin VSY. J Am Chem Soc. 2004;126:13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, L M, Sherman S, Xia T, Kovochich M, Nel AE, Zink JI, Tomanoi F. Nanobiotechnol. 2007;3:89–95. doi: 10.1007/s12030-008-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, Tsai CP, Huang HY, Kuo CT, Hung Y, Huang DM, Chen YC, Mou CY. Chem Mater. 2005;17:4570–4573. [Google Scholar]
- 15.MacCraith BD, McDonagh C. J Fluor. 2002;12:333–342. [Google Scholar]
- 16.Innocenzi P, Lebeau B. J Mater Chem. 2005;15:3821–3831. [Google Scholar]
- 17.Angelos S, Johansson E, Stoddart JF, Zink JI. Adv Funct Mater. 2007;17:2261–2271. [Google Scholar]
- 18.Reisfeld R, Weiss A, Saraidarov T, Yariv E, Ishchenko AA. Polym Adv Technol. 2004;15:291–301. [Google Scholar]
- 19.Lim J, Malati P, Bonet F, Dunn B. J Electrochem Soc. 2007;154:A140–A145. [Google Scholar]
- 20.Steinmetz NF, S SN, Barclay JE, Rallapalli G, Lomonossoff GP, Evans DJ. Small. 2009;5:813–816. doi: 10.1002/smll.200801348. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez F, Glawe DD, Naik RR, Hallinana KP, Stone MO. Biomacromolecules. 2004;5:261–265. doi: 10.1021/bm034232c. [DOI] [PubMed] [Google Scholar]
- 22.Wong Po Foo CP, P SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, Naik RR, Perry CC, Kaplan DL. PNAS. 2006;103:9428–9433. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto A, C J, Collette AL, Kim UJ, Altman GH, Cebe P, Kaplan DL. J Phys Chem B. 2006;110:21630–21638. doi: 10.1021/jp056350v. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Ogiso M, Minoura N. Biomaterials. 2003;24:357–365. doi: 10.1016/s0142-9612(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 25.Jin HJ, Fridrikh SV, Rutlege GC, Kaplan DL. Biomacromolecules. 2002;3:1233–1239. doi: 10.1021/bm025581u. [DOI] [PubMed] [Google Scholar]
- 26.Jin HJ, P J, Karageorgiou V, Kim UJ, Valluzzi R, Cebe P, Kaplan DL. Adv Func Mat. 2005;15:1241–1247. [Google Scholar]
- 27.Kim HJ, Kim UJ, Vujnak-Novakovic G, Min BH, Kaplan DL. Biomaterials. 2005;26:4442–4452. doi: 10.1016/j.biomaterials.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Kim UJ, Park JY, Li CM, Jin HJ, Valluzzi R, Kaplan DL. Biomacromolecules. 2004;5:786–792. doi: 10.1021/bm0345460. [DOI] [PubMed] [Google Scholar]
- 29.Wong Fo Poo C, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, Naik RR, Perry CC, Kaplan DL. PNAS. 2006;103:9428–9433. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belton DJ, Patwardhan SV, Perry CC. Chem Commun. 2005;27:3475–3477. doi: 10.1039/b504310g. [DOI] [PubMed] [Google Scholar]
- 31.Liang MK, Patwardhan SV, Danilovtseva EN, Annekov VV, Perry CC. J Mat Res. 2009;24:1700–1708. [Google Scholar]
- 32.Murphy AR, St P John, Kaplan DL. Biomaterials. 2008;29:2829–2838. doi: 10.1016/j.biomaterials.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gough JE, Notingher I, Hench LL. J Biomed Mater Res A. 2004;68:640–650. doi: 10.1002/jbm.a.20075. [DOI] [PubMed] [Google Scholar]
- 34.Christodoulou I, Buttery LDK, Tai G, Hench LL, Polak JM. J Biomed Mater Res Part B: Appl Biomater. 2006;77B:431–446. doi: 10.1002/jbm.b.30455. [DOI] [PubMed] [Google Scholar]
- 35.Bombonato-Prado KF, Bellesini LS, Junta CM, Marques MM, Passos GA, Rosa AL. J Biomed Mat Res Part A. 2008;88:401–408. doi: 10.1002/jbm.a.31701. [DOI] [PubMed] [Google Scholar]
- 36.Gordon JA, Tye CE, Sampaio AV, Underhill TM, Hunter GK, G HA. Bone. 2007;41:462–473. doi: 10.1016/j.bone.2007.04.191. [DOI] [PubMed] [Google Scholar]
- 37.Boccaccini AR, Notingher I, Maquet V, Jerome R. J Mater Sci Mater Med. 2003;14:443–450. doi: 10.1023/a:1023266902662. [DOI] [PubMed] [Google Scholar]


