Abstract
Thioxolone acts as a prodrug in the presence of carbonic anhydrase II (CA II), whereby the molecule is cleaved by thioester hydrolysis to the carbonic anhydrase inhibitor, 4-mercaptobenzene-1,3-diol (TH0). Thioxolone was soaked into the proton transfer mutant H64A of CA II in an effort to capture a reaction intermediate via X-ray crystallography. Structure determination of the 1.2 Å resolution data revealed the TH0 had been modified to a 4,4′-disulfanediyldibenzene-1,3-diol, a product of crystallization conditions, and a zinc ligated 2,4-dihydroxybenzenesulfenic acid, most likely induced by radiation damage. Neither ligand was likely a result of an enzymatic mechanism. 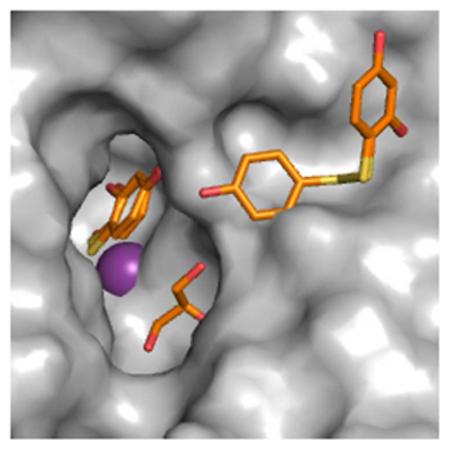
Keywords: Carbonic anhydrase, free radical damage, synchrotron radiation, thioxolone, sulfenic acid
Carbonic anhydrase (CA) is a zinc metalloenzyme that catalyzes the reversible hydration of CO2 to HCO3−.1 There are 14 CA isozymes expressed in humans, which serve a variety of functions including respiration, pH homeostasis, and fluid retention.1 Carbonic anhydrase inhibitors (CAIs) have been used to treat a variety of disorders including glaucoma, epilepsy, hypertension, and altitude sickeness.2 In addition to carbon dioxide hydration, CA isozyme II (CA II) has a promiscuous esterase activity that is capable of cleaving a wide variety of substrates.3 Recently the anti-psoriatic medication thioxolone (6-hydroxy-1,3-benzenozathiol-2-one) was shown to have inhibitory activity on several CA isozymes, including CA II.4,5 Thioxolone was shown to behave like a prodrug, being cleaved into 4-mercaptobenzene-1,3-diol (TH0) via thioester hydrolysis in CA II.6
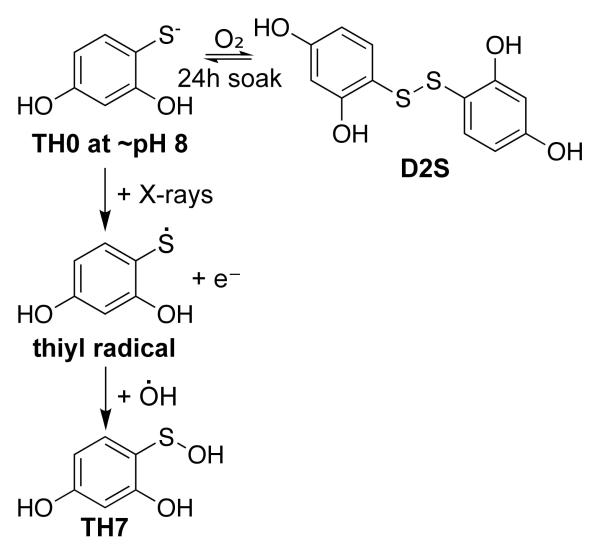
In an effort to capture a reaction intermediate, thioxolone was soaked into crystals of an H64A mutant of CA II (CA II H64A) which has reduced proton transfer.7 X-ray structure determination of synchrotron collected data revealed, not only the conversion of the thiol in TH0 to 2,4-dihydroxybenzenesulfenic acid (TH7), stabilized by the active site zinc (Figure 1A), but also a disulfide bridged version of TH0, 4,4′-disulfanediyldibenzene-1,3-diol (D2S) (Figure 1B). Attempts to reconcile these data with an enzymatic mechanism failed. However when one considers the drug soak conditions and a direct ionization event by X-ray irradiation, a hypothesis can be formulated to account for the observed final products (Scheme 1).
Figure 1. CA II H64A-Ligand Complex.
(A) Surface rendering of CA II (grey) oriented down the active site cleft. Ligands are shown as sticks. All figures are colored as follows: ligand carbons are orange; Protein carbons, grey, oxygens, red; nitrogens, blue; sulfurs, yellow. Zn2+ is a purple sphere. Waters are red spheres. Symmetry related protein carbons are colored cyan. GOL denotes glycerol. (B) A stereo image of the ligands and their interacting residues. 2|Fo-Fc| averaged kick map of (C) TH0/TH7 and (D) D2S/TH0, respectively, contoured to 1.2σ. Figure generated in Pymol 8.
Scheme 1.
Proposed Model of Ligand Generation.a
aAbbreviations: 4-mercaptobenzene-1,3-diol (TH0), 2,4-dihydroxybenzenesulfenic acid (TH7), and 4,4′-disulfanediyldibenzene-1,3-diol (D2S).
A crystal of CA II H64A was soaked with thioxolone then cryo-cooled to 100 K and subjected to high energy X-rays (λ=0.938 Å) at the Cornell High Energy Synchrotron Source (CHESS) beamline F1. Diffraction data was collected to 1.2 Å resolution. For a full account of the structure determination refer to Experimental Methods. The structure was refined to an Rcryst of 11.6% and an Rfree of 13.7% with root mean squared deviations of 0.01 Å for bond lengths and 1.5° for bond angles (PDB ID: 3iqk, Table 1).
Table 1.
Data Processing and Model Statistics.
| Data Statistics | |
| Space Group | P21 |
| Cell Dimensions |
a=43.4 Å, b=41.4 Å, c=72.1 Å, β=104.1° |
| Resolution (Å) | 15.6-1.2 (1.24-1.2) a |
| Rsym(%)b | 5.5 (34.2) |
| I/σ(I) | 12.6 (2.7) |
| Completeness (%) | 92.4 (78.2) |
| Redundancy | 2.4 (2.1) |
| Refinement | |
| No. Reflections | 67156 (5117) |
| Rcrystc/Rfreed/Ralldata (%) | 11.6 / 13.7/ 11.7 |
| No. Atoms | |
| protein | 2345 |
| TH0 (2) / TH7 / D2S | 18 / 10 / 18 |
| zinc/ glycerol/ sulfate/ water |
1 / 6 / 5 / 313 |
| B-factors (Å2) | |
| Protein (main/side) | 10.4 / 14.3 |
| TH0/TH7/D2S | 15.0 / 12.9 / 19.2 |
| zinc/ glycerol/ sulfate/ water |
5.9/11.4/15.5/25.8 |
| Ramachandran Preferred/Allowed (%) | 93.2 / 6.8 |
| RMSDe length (Å) / angle (°) |
0.012 / 1.51 |
Values in parenthesis are for highest resolution shell.
Rsym=(Σ|I-<I>|/Σ<I>)×100.
Rcryst=(Σ|Fobs|-|Fc|/Σ|Fobs|)×100.
Rfree is calculated the same as Rwork, except it uses 5% of reflection data omitted from refinement.
The root mean squared deviation from ideal values.
Initially TH0 was modeled into the two positions previously identified in the structure of wild-type CA II soaked in thioxolone.6 However several regions of positive Fo-Fc density remained implying that the TH0s were insufficient to satisfy the electron density. These observations included of a single 15σ peak located 1.4 Å from the sulfur of the zinc-bound TH0 and 2.2 Å from the zinc itself indicating an atom (modeled as an oxygen) covalently bonded to the sulfur and coordinating to the zinc. The observed zinc-bound ligand(s) was therefore reinterpreted as both a TH0 and a TH7 with occupancies of 0.33 and 0.67, respectively, which fully accounted for the previously observed residual density (Figure 1A).
The newly modeled TH7 had several unique features. First, the C3-S-O4 angle was 92.9°, as compared to the predicted 100° and seen in the 1-hydroxysulfanyl-4-mercapto-butane-2,3-diol complexed with RNA 3′-terminal phosphate cyclase (PDB ID 1qmh).9,10 Secondly, a short non-bonded distance of 2.15 Å is observed between O1 and O4. It has been previously reported that in a high energy environment the O of a hydroxysulfanyl group might favor a short hydrogen bond to an adjacent electronegative atom rather than selecting a geometrically favorable position.9
The second ligand binding region is aligned distally from the zinc-bound ligand(s) (TOC Figure and Figure 1B). Positive Fo-Fc density matching two TH0 molecules connected by disulfide bond was observed at the mouth of the active site cleft, designated D2S. The torsion angle of the disulfide conformed to ideal geometry and bond distances.11-13 There was significantly less density and higher temperature factors around the half of the molecule adjacent to TH7/TH0. This region has fewer interactions than the rest of the molecule, however it is also less solvent exposed. As a result, D2S was modeled as having partial occupancy of 0.71 with the remainder being modeled as TH0 and a water.
Neither of the TH7 or D2S could have been formed by the hydrolysis of the thioester bond of the thioxolone molecule. The zinc-bound hydroxide, the initiating nucleophile, would preferentially attack the sulfur over the oxygen of the thioester leading to the cleavage of the sulfur-oxygen bond.6 However the end product of the reaction, the TH0 molecule, has an exposed thiol group, which would form a thiolate anion at the pH of crystallization (approximately pH 8). Given the mildly oxidizing condition a population of D2S and TH0 in equilibrium is likely to occur (Scheme 1). This was confirmed by the presence of the D2S molecule in data collected on an in-house diffractometer indicating that its presence is not radiation dependent (data not shown).
The appearance of TH7 is not so easily explained. Sulfenic acid does not form spontaneously in solution. It is generated via irradiation, though it can be accomplished by exposure to chlorite.9,14,15 Sulfur radicals are particularly vulnerable to hydrolysis by hydroxyl radicals to a sulfenic acid, but typically it is an intermediate state and spectroscopic studies show its presence is transitory.16 The zinc ion likely stabilizes the transition state allowing it to be captured by X-ray crystallography. The synchrotron beam generates the necessary radicals for this chemistry. The estimated radiation dose absorbed by this crystal is approximately 3×106 to 3×107 Gy.17 Additionally, it has also been estimated that each absorbed 12 keV (approx. 1.0 Å wavelength) photon generates up to 500 ionization and excitation events in a protein crystal.18 The ionization energy of a thiol is 9.46eV or 5×10−10 Gy, assuming the crystal has a density similar to water, so it seems very likely that the molecule would form a thiyl radical (RS.−).19,20 The CA II active site is heavily solvated which, upon radiation exposure, would become hydroxyl radicals, reacting with the thiyl radical to become a sulfenic acid (Scheme 1). Hydroxyl radicals may become mobile 110 K, a temperature that could easily be achieved by the distance of the crystal from the cryo-stream, cryo-stream positional error, and crystal size (~1 mm × 0.5 mm × 0.2 mm).17,21,22 This could account for the relatively small movements of a hydroxyl towards the thiyl radical. It is also plausible that the zinc contributes to this reaction, perhaps through reduction by secondary ionization events or by coordinating the two ions placing them in proximity to one another, but its role in the mechanism is not fully understood. The H64A mutation itself appears to play no part, as it is over 8 Å away from the TH0/TH7, 10 Å away from D2S, and blocked by a glycerol, which was used as a cryo-protectant during data collection. Curiously, during the low radiation control experiments the occupancy of the zinc-bound ligand proved too low to be modeled (data not shown).
It is important to note that these molecules are not seen in mass spectrometric (MS) analysis of wild type CA II cleaved thioxolone.6 The absence of D2S may be attributed to the MS sample processing conditions (pH 7.5 and significantly lower sulfate concentration) that would favor the TH0 rather than D2S side of equilibrium. The lack of TH7 could be due to molecular instability in solution. It is the unique environment of the CA II active site that stabilizes this molecule allowing it to be visualized via crystallography.
Growing numbers of higher flux synchrotron radiation sources have opened doors in macromolecular crystallography allowing the collection of ever higher resolution data. With the use of these higher flux sources there is the possibility of radiation-induced degradation of the sample. It was the high resolution data collected from a synchrotron that both created the TH7 ligand and allowed it to be identified. There is a large body of literature about the effects of X-rays on cryo-cooled protein crystals.13,21,23,24 However the adduct formation reported here has not been previously described and serves as a reminder of the unpredictable nature of free radical chemistry.
Experimental Methods
Expression and purification
A plasmid containing human CA II with His 64 mutated to Ala (CA II H64A) was prepared as described previously.25,26 The plasmid was transformed in BL21(DE3)pLysS cells, expressed following protocols described elsewhere.27 The protein was purified by affinity chromatography using paminomethylbenzenesulfonamide-agarose (pAMBS, Sigma) as described previously.27 The CA II fractions were dialyzed into 50 mM Tris-Cl, pH 7.8, then concentrated to ~15 mg mL−1 with an Amicon Centrifugal Filter Device (Millipore). Purity was checked using SDS-PAGE and concentration was calculated by measuring the absorption of the protein solution at 280 nm and assuming an extinction coefficient of 5.5 × 104 M−1 cm−1.
Crystallization and diffraction data collection
The hanging-drop method utilizing vapor diffusion was used to grow mutant H64A crystals at room temperature.28 Crystallization drops were prepared by mixing 5 l protein (~15 mg ml−1 in 50 mM Tris-HCl pH 7.8) with 5 l precipitant solution (50 mM Tris-HCl pH 8.0, 2.6 M ammonium sulfate) and equilibrating the drop against 1 ml precipitant solution. Crystals used in the drug soak appeared within 5 days.
CA II H64A crystals were soaked in 2 ml of thioxolone (100 mM in 50% DMSO) at room temperature with 10 ml of stabilizing solution (50 mM Tris-HCl pH 8.0, 2.6 M ammonium sulfate) for ~24 h. Prior to data collection, crystals were also soaked in a cryoprotectant of 30% glycerol and 2.6 M sodium sulfate in 50 mM Tris pH 7.8, then flash-cooled to 100 K.
Data were collected at the Cornell High Energy Synchrotron Source (CHESS) beamline F1 using a wavelength of 0.938 Å, images were collected on a Quantum 4 CCD detector system. A total of 120 images were collected to 1.2 Å resolution, using 1° oscillation with 30 s exposure time and a crystal-to-detector distance of 90 mm. The data was merged using DENZO and scaled with SCALEPACK from the HKL suite.29 Details of data collection and processing are given in Table 1.
Structure determination and model refinement
The complex structure was determined by molecular replacement using the previously solved H64A HCAII structure with solvents removed (PDB code 1g0f) as the search model. Initially, the structure was refined isotropically in SHELXL.30 Difference Fourier maps were calculated and three large distinct electron densities were identified in the |Fo|-|Fc| map of the active site region. Two of these correlated with the locations of the ligands in the structure of thioxolone soaked wild-type CA II (PDB ID 2osf)6 and the third one was matched to the glycerol binding site of CA II (PDB ID 3sh4)31. The coordinates for two TH0 molecules and one glycerol were modeled into the density and restraints were generated using SHELXPRO.30 Subsequent refinement revealed the thiol from one TH0 molecule appeared to be disulfide bonded to a third, less ordered TH0, making a 4,4′-disulfanediyldibenzene-1,3-diol (D2S). Waters were picked, all full occupancy molecules were refined anisotropically, and riding hydrogens were added to all residues except the NHs of His and the OH of Tyr, Thr, and Ser. Inspection of the remaining difference density revealed a ~15σ positive peak 1.8 Å adjacent to the thiol of the remaining TH0 and seemingly coordinated to the zinc. The TH0 was remodeled as 4-hydroxysulfanylbenzene-1,3-diol (TH7). Additional discrepancies in the |Fo|-|Fc| density prompted a switch in the refinement program. Hydrogens were added and occupancies were refined in PHENIX as well as the calculation of both 2|Fo|-|Fc| and |Fo|-|Fc| kick maps.32,33 The resulting maps prompted a dual occupancy refinement of TH7 overlaid with a TH0 molecule and the D2S molecule overlaid with a TH0. Final model statistics are listed in Table 1.
Acknowledgements
The authors would like to thank the staff at CHESS for their help and support at the F1 station during X-ray data collection and Dr. Arthur Robbins for technical insight and encouragement during the refinement stages. This work was supported in part by a National Institutes of Health grant (GM25154) and a Maren Foundation grant.
Reference
- 1.Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding. Chem Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Supuran CT, Scozzafava A, Conway J. Carbonic anhydrase : its inhibitors and activators. CRC Press; Boca Raton: 2004. [Google Scholar]
- 3.Whitney PL. Inhibition and modification of human carbonic anhydrase B with bromoacetate and iodoacetamide. Eur J Biochem. 1970;16:126–135. doi: 10.1111/j.1432-1033.1970.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 4.Iyer R, Barrese AA, 3rd, Parakh S, Parker CN, Tripp BC. Inhibition profiling of human carbonic anhydrase II by high-throughput screening of structurally diverse, biologically active compounds. J Biomol Screen. 2006;11:782–791. doi: 10.1177/1087057106289403. [DOI] [PubMed] [Google Scholar]
- 5.Innocenti A, Maresca A, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors: thioxolone versus sulfonamides for obtaining isozyme-selective inhibitors? Bioorg Med Chem Lett. 2008;18:3938–3941. doi: 10.1016/j.bmcl.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Barrese AA, 3rd, Genis C, Fisher SZ, Orwenyo JN, Kumara MT, Dutta SK, Phillips E, Kiddle JJ, Tu C, Silverman DN, et al. Inhibition of carbonic anhydrase II by thioxolone: a mechanistic and structural study. Biochemistry. 2008;47:3174–3184. doi: 10.1021/bi702385k. [DOI] [PubMed] [Google Scholar]
- 7.Duda D, Tu C, Qian M, Laipis P, Agbandje-McKenna M, Silverman DN, McKenna R. Structural and kinetic analysis of the chemical rescue of the proton transfer function of carbonic anhydrase II. Biochemistry. 2001;40:1741–1748. doi: 10.1021/bi002295z. [DOI] [PubMed] [Google Scholar]
- 8.DeLano WL. Delano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 9.Lapinski L, Gerega A, Sobolewski AL, Nowak MJ. Thioperoxy derivative generated by UV-induced transformation of N-hydroxypyridine-2(1H)-thione isolated in low-temperature matrixes. J Phys Chem A. 2008;112:238–248. doi: 10.1021/jp077365r. [DOI] [PubMed] [Google Scholar]
- 10.Palm GJ, Billy E, Filipowicz W, Wlodawer A. Crystal structure of RNA 3′-terminal phosphate cyclase, a ubiquitous enzyme with unusual topology. Structure. 2000;8:13–23. doi: 10.1016/s0969-2126(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 11.Weik M, Berges J, Raves ML, Gros P, McSweeney S, Silman I, Sussman JL, Houee-Levin C, Ravelli RB. Evidence for the formation of disulfide radicals in protein crystals upon X-ray irradiation. J Synchrotron Radiat. 2002;9:342–346. doi: 10.1107/s0909049502014589. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 13.Ravelli RB, McSweeney SM. The ‘fingerprint’ that X-rays can leave on structures. Structure. 2000;8:315–328. doi: 10.1016/s0969-2126(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 14.Bonifacic M, Schafer K, Mockel H, Asmus KD. Primary Steps in Reactions of Organic Disulfides with Hydroxyl Radicals in Aqueous-Solution. Journal of Physical Chemistry. 1975;79:1496–1502. [Google Scholar]
- 15.Darkwa J, Olojo R, Chikwana E, Simoyi RH. Antioxidant chemistry: Oxidation of L-cysteine and its metabolites by chlorite and chlorine dioxide. Journal of Physical Chemistry A. 2004;108:5576–5587. [Google Scholar]
- 16.Varmenot N, Berges J, Abedinzadeh Z, Scemama A, Strzelczak G, Bobrowski K. Spectral, kinetic, and theoretical studies of sulfur-centered reactive intermediates derived from thioethers containing an acetyl group. Journal of Physical Chemistry A. 2004;108:6331–6346. [Google Scholar]
- 17.Gruner SM. Cornell High Energy Synchrotron Source. Ithaca, NY: 2010. Personal Communication. [Google Scholar]
- 18.O'Neill P, Stevens DL, Garman EF. Physical and chemical considerations of damage induced in protein crystals by synchrotron radiation: a radiation chemical perspective. J Synchrotron Radiat. 2002;9:329–332. doi: 10.1107/s0909049502014553. [DOI] [PubMed] [Google Scholar]
- 19.Cheng BM, Chew EP, Hung WC, Eberhard J, Lee YP. Photoionization studies of sulfur radicals and products of their reactions. J Synchrotron Radiat. 1998;5:1041–1043. doi: 10.1107/S0909049597016075. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong DA, Sun Q, Schuler RH. Reduction potentials and kinetics of electron transfer reactions of phenylthiyl radicals: Comparisons with phenoxyl radicals. Journal of Physical Chemistry. 1996;100:9892–9899. [Google Scholar]
- 21.Garman EF. Radiation damage in macromolecular crystallography: what is it and why should we care? Acta Crystallogr D Biol Crystallogr. 2010;66:339–351. doi: 10.1107/S0907444910008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkire RW, Duke NEC, Rotella FJ. Is your cold-stream working for you or against you? An in-depth look at temperature and sample motion. J Appl Crystallogr. 2008;41:1122–1133. [Google Scholar]
- 23.Weik M, Ravelli RB, Kryger G, McSweeney S, Raves ML, Harel M, Gros P, Silman I, Kroon J, Sussman JL. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colletier JP, Bourgeois D, Sanson B, Fournier D, Sussman JL, Silman I, Weik M. Shoot-and-Trap: use of specific x-ray damage to study structural protein dynamics by temperature-controlled cryo-crystallography. Proc Natl Acad Sci U S A. 2008;105:11742–11747. doi: 10.1073/pnas.0804828105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsman C, Behravan G, Jonsson BH, Liang ZW, Lindskog S, Ren XL, Sandstrom J, Wallgren K. Histidine 64 is not required for high CO2 hydration activity of human carbonic anhydrase II. FEBS Lett. 1988;229:360–362. doi: 10.1016/0014-5793(88)81156-6. [DOI] [PubMed] [Google Scholar]
- 26.Forsman C, Behravan G, Osterman A, Jonsson BH. Production of active human carbonic anhydrase II in E. coli. Acta Chem Scand B. 1988;42:314–318. doi: 10.3891/acta.chem.scand.42b-0314. [DOI] [PubMed] [Google Scholar]
- 27.Khalifah RG. Histidine-200 alters inhibitor binding in human carbonic anhydrase B. A carbon-13 nuclear magnetic resonance identification. Biochemistry. 1977;16:2236–2240. doi: 10.1021/bi00629a030. [DOI] [PubMed] [Google Scholar]
- 28.McPherson A. Preparation and Analysis of Protein Crystals. John Wiley and Sons; New York: 1982. [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Vol. 276. Yale University; New Haven, CT, USA: 1997. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick GM. A short history of SHELX. Acta Cryst. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 31.Sippel KH, Robbins AH, Domsic J, Genis C, Agbandje-McKenna M, McKenna R. High-resolution structure of human carbonic anhydrase II complexed with acetazolamide reveals insights into inhibitor drug design. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:992–995. doi: 10.1107/S1744309109036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Praaenikar J, Afonine PV, Guncar G, Adams PD, Turk D. Averaged kick maps: less noise, more signal… and probably less bias. Acta Crystallogr D Biol Crystallogr. 2009;65:921–931. doi: 10.1107/S0907444909021933. [DOI] [PMC free article] [PubMed] [Google Scholar]