Abstract
The microRNA (miRNA) “sponge” method was introduced three years ago as a means to create continuous miRNA loss of function in cell lines and transgenic organisms. Sponge RNAs contain complementary binding sites to a miRNA of interest, and are produced from transgenes within cells. As with most miRNA target genes, a sponge's binding sites are specific to the miRNA seed region, which allows them to block a whole family of related miRNAs. This transgenic approach has proven to be a useful tool to probe miRNA functions in a variety of experimental systems. Here we will discuss the ways sponge and related constructs can be optimized and review recent applications of this method with particular emphasis on stable expression in cancer studies and in transgenic animals.
Keywords: microRNA, sponge, antisense, inhibitor, transgenic, decoy
INTRODUCTION
The widespread involvement of microRNAs (miRNAs) in regulating developmental processes, physiological responses, and pathological conditions in animals has been amply demonstrated (He and Hannon 2004; Bushati and Cohen 2007; Bartel 2009). Nonetheless, the specific functions of each miRNA in the various contexts in which it is expressed are only beginning to be discovered. The typical miRNA is computationally predicted to regulate hundreds of target genes (Friedman et al. 2009), and while there has been progress in compiling sets of predicted targets into pathways (Tsang et al. 2010), every prediction still needs to be experimentally validated. The best experimental approaches create a loss of function in the miRNA of interest. Loss-of-function approaches are superior because they reveal functions that depend on physiological miRNA levels; by contrast, adding exogenous miRNA to the system can result in repression of nonphysiological target mRNAs since miRNA–target interaction is strongly concentration-dependent (S Mukherji, MS Ebert, GZ Zheng, JS Tsang, PA Sharp, A van Oudenaarden, in prep.).
There are three general methods for miRNA loss-of-function studies: genetic knockouts, antisense oligonucleotide inhibitors (Meister et al. 2004; Krützfeldt et al. 2005; Ørom et al. 2006) and sponges (Ebert et al. 2007). The sponge mRNA, which contains multiple target sites complementary to a miRNA of interest, is a dominant negative method (see Fig. 1). When the sponge is expressed at high levels, it specifically inhibits the activity of a family of miRNAs sharing a common seed (miRNA nucleotides 2–7, the major specificity determinant for target recognition [Lewis et al. 2003]). While deleting the gene encoding a miRNA is the only way to guarantee complete loss of its activity, the sponge method offers several advantages. First is the convenience of making dominant negative transgenics over knockouts, and the applicability to a broader range of model organisms and cell lines. Second, many miRNAs have seed family members encoded at multiple distant loci; due to this functional redundancy, these miRNAs would have to be knocked out individually and the animals bred to generate the complete knockout strain. Furthermore, some miRNA precursors are transcribed in clusters; the proximity of the miRNAs within a cluster may make it difficult to cleanly delete one miRNA without affecting the processing of the others. Since sponges interact with the mature miRNA, their effectiveness is unaffected by the clustering of miRNA precursors.
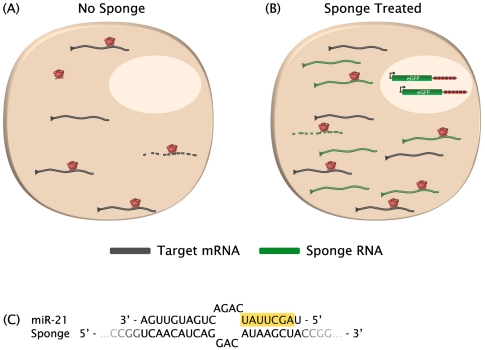
FIGURE 1.
(A) In the absence of sponge treatment, target mRNAs (gray) for a particular miRNA seed family (red complexes) are repressed. Dashed lines indicate mRNA decapping and degradation. (B) After introduction of the sponge transgene, sponge mRNAs (green) are expressed at a high level and sequester the miRNA complexes, rescuing the expression of the endogenous targets. Sponge-treated cells can be identified by their eGFP reporter expression. (C) Pairing of a miRNA with a bulged sponge site shows mismatches opposite miRNA nucleotides 9–12. The miRNA seed region is highlighted.
Sponges also offer advantages over chemically modified antisense oligonucleotide inhibitors for many research applications. First, these antisense inhibitors appear to be specific for one miRNA as they depend upon extensive sequence complementarity beyond the seed region (Davis et al. 2006; Esau 2008). Thus, to neutralize a family of miRNAs may require the delivery of a mixture of perfectly complementary oligonucleotides. In addition, many cells both in vitro and in vivo are resistant to the uptake of oligonucleotides. By contrast, for difficult-to-transfect cell lines or cells in vivo, the sponge transgene can be delivered by a viral vector. Inclusion of an open reading frame for a selectable marker or reporter gene in the vector allows for selection or screening, fluorescence-activated cell sorting, or even laser capture microdissection of cells strongly expressing the sponge. This makes it possible to isolate a fraction of cells in which the family of miRNAs is strongly inhibited, which can reveal even subtle changes in target gene expression. In principle, one could include regulatory elements in the sponge promoter to make it drug-inducible or tissue-specific for the tissue of choice. By contrast, the cholesterol-modified “antagomir” oligonucleotides that can be injected into the mouse cannot access all tissues, and mostly accumulate in the liver (Krützfeldt et al. 2005). Finally, antagomirs require repeated administration in large doses to inhibit a miRNA over long durations, whereas one could generate germline transgenic sponge-expressing animals to continuously inhibit the miRNA of interest for the lifetime of the animal.
Although sponge technology has advantages in more biological experiments, antisense type technology is more promising from the perspective of therapeutically inhibiting miRNAs. This promise depends on continued development of oligonucleotide chemistries and improvement in the delivery of antisense oligonucleotides to cells and tissues.
SPONGE DESIGN
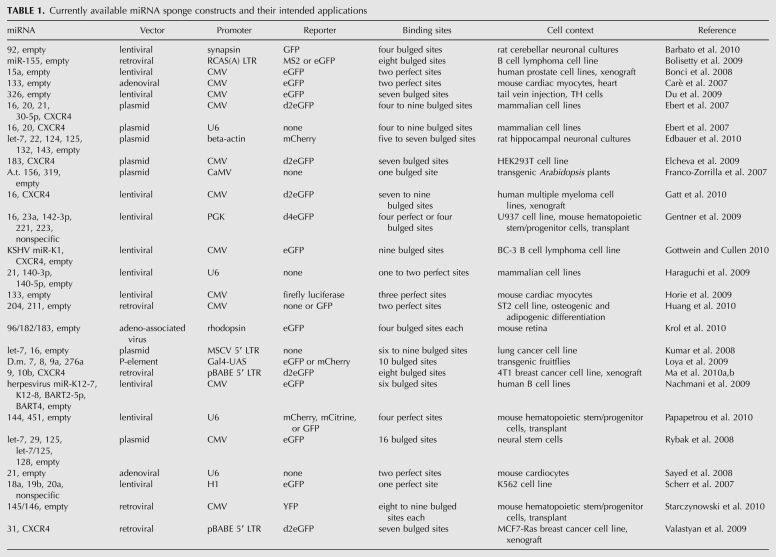
Variations of miRNA sponge type constructs have been described as target mimics (Franco-Zorrilla et al. 2007), decoys (Carè et al. 2007), miRNA target (miRT) sequences (Gentner et al. 2009), miRNA erasers (Sayed et al. 2008), and lentivirus-mediated antagomirs (Scherr et al. 2007; for currently available miRNA sponge constructs and their intended applications, see Table 1). The miRNA binding sites in these constructs are either perfectly antisense or contain mismatches in the middle positions, which if perfectly base-paired would be vulnerable to Ago2-mediated endonucleolytic cleavage. Sponges with sites perfectly complementary to the miRNA show some inhibitory activity (Carè et al. 2007; Ebert et al. 2007; Scherr et al. 2007; Bonci et al. 2008; Sayed et al. 2008; Gentner et al. 2009; Haraguchi et al. 2009; Horie et al. 2009; Huang et al. 2010; Papapetrou et al. 2010), perhaps because miRNAs complexed with the catalytically inactive Argonautes 1, 3, and 4 can still be titrated by these sites without cleavage of the sponge RNA. More effective are sponges containing bulged sites that are mispaired opposite miRNA positions 9–12 (Ebert et al. 2007, Gentner et al. 2009), presumably because they form a more stable interaction with the miRNA, including miRNA complexed with Ago2.
TABLE 1.
Currently available miRNA sponge constructs and their intended applications
Typical sponge constructs contain four to 10 binding sites separated by a few nucleotides each. Increasing the number of binding sites may have diminishing marginal utility, as each site increases the probability of sponge RNA degradation. Variations in the bulged mismatches and the spacers can be introduced to reduce the risk of recombination during cloning and to reduce the risk of introducing unintended binding motifs for other regulatory factors. Sites are normally placed in an unstructured, noncoding region of the RNA. For PolIII-generated sponge RNAs, which lack a 5′ cap and 3′ poly(A) tail, terminal stem–loops can be included as stabilizing elements (Ebert et al. 2007). Another type of transgenic antisense inhibitor, TuD (“tough decoy”) RNAs, place the miRNA binding site or sites in the single-stranded regions of short stem–loops, precisely presenting them for binding to miRNA complexes (Haraguchi et al. 2009).
The efficacy of a miRNA sponge depends not just on the affinity and avidity of binding sites, but also on the concentration of sponge RNAs relative to the concentration of the miRNA. To maximize sponge expression, the strongest available promoter for the cell type of interest should be used, e.g., a CMV promoter in many mammalian cell lines. For transient assays, plasmid transfection can deliver the highest dose of the sponge transgene. For viral delivery of sponges, transduction with high multiplicity of infection should be performed. Since random integration of the sponge transgene may disrupt an endogenous gene, it is advisable to generate multiple clonal lines or make polyclonal lines. The choice of viral vector can contribute to DNA copy number and to cell-type specificity. Sponges delivered in vivo can also make use of tissue-specific promoters in cases where the miRNA of interest is expressed in multiple cell types that could confound an observed loss-of-function phenotype. In principle, stably propagated episomal vectors (Kimchi 1999) should be an effective alternative to chromosomally inserted sponge transgenes.
For inclusion of a reporter in the sponge construct, any protein-coding gene that can be tolerated at high expression levels is allowable. Preferably it is placed directly upstream of the miRNA binding sites (as opposed to being in a separate cistron in the same vector), such that the protein expression directly represents sponge RNA expression. Drug resistance markers allow for stringent selection of high sponge-expressing clones. Fluorescent reporters enable quantitative analysis and sorting of individual live cells, and can be diversified with different colors representing different miRNA sponges.
LIMITATIONS OF THE SPONGE METHOD
It should be noted that optimized sponges may still exhibit different degrees of inhibition in different contexts. Where miRNA concentration is very high, complete titration demands a very high and possibly unachievable dose of sponge RNA. On the other hand, in cells expressing a large pool of endogenous targets for the miRNA family of interest, there should be less free miRNA available (Arvey et al. 2010), so a lower dose of sponge RNA should suffice to give strong inhibition. High expression levels of sponge transgenes such as those that express a GFP reporter are not known to create any off-target effects, but such effects should be ruled out by comparing phenotypes and gene expression profiles not only between the miRNA sponge and an empty or nonspecific control vector treatment, but also between the control vector treatment and no treatment.
Determining whether a sponge treatment is successfully inhibiting the miRNA of interest is more challenging than validating the success of genetic miRNA deletion, which results in a clean loss of the mature miRNA. The efficacy of sponge constructs can be validated in cell culture by reporter assay or assays for the expression of known target genes. Typically a luciferase reporter fused to miRNA binding sites or a confirmed target 3′ UTR is measured in the presence of the miRNA sponge or a negative control sponge containing no binding sites or nonspecific sites. In the presence of the miRNA of interest, the luciferase reporter should be significantly derepressed by the miRNA sponge. In this case it is also difficult to assess whether inhibition of different seed family members occurs to the same degree, as a given target reporter is regulated by all of the expressed members of the seed family.
TRANSIENT APPLICATIONS FOR miRNA SPONGES
The immediate application of miRNA sponges as first described was transient treatment and assay in cell culture models. A number of reports demonstrate the flexibility of the method with respect to cell type, promoter, vector, reporter gene, and type of miRNA targeted (see Table 1). Sponges were transfected or transduced into human, mouse, and rat cell lines such as nonsmall cell lung cancer (Kumar et al. 2008), B cell lymphoma (Bolisetty et al. 2009), embryonic neural stem cells (Rybak et al. 2008), and dissociated hippocampal neurons (Edbauer et al. 2010). Sponge RNAs were transcribed from strong promoters such as CMV (Rybak et al. 2008; Elcheva et al. 2009), U6 (Sayed et al. 2008), and viral LTRs (Kumar et al. 2008). The most commonly used vectors were plasmids (Kumar et al. 2008; Rybak et al. 2008; Elcheva et al. 2009; Edbauer et al. 2010), but some used retroviruses (Bolisetty et al. 2009), lentiviruses (Horie et al. 2009; Nachmani et al. 2009) or adenovirus (Sayed et al. 2008). Individual miRNAs, e.g., miR-155 (Bolisetty et al. 2009), or large seed families, e.g., let-7 (Kumar et al. 2008), were successfully targeted. The most common reporter gene was eGFP (Rybak et al. 2008; Bolisetty et al. 2009; Elcheva et al. 2009; Nachmani et al. 2009), but mCherry (Edbauer et al. 2010) and luciferase (Horie et al. 2009) were also used. Typically, cellular assays and target validation assays (visualization of derepressed target protein or 3′ UTR reporter expression) were performed 24–72 h after introduction of the sponge construct.
One fortuitous aspect of sponge treatment is that it may cause a significant and specific reduction in the miRNA level (Rybak et al. 2008; Sayed et al. 2008; Horie et al. 2009), in some cases even to an extent that the miRNA is undetectable by Northern blot (Sayed et al. 2008). This may indicate that miRNA–target interaction stimulates degradation of the miRNA. Target reporter sites with extensive complementarity to the 3′ end of the miRNA appear to accelerate exonucleolytic trimming of the miRNA in fruitflies and mammalian cells (Ameres et al. 2010). This phenomenon may occur at both centrally bulged and perfect sponge sites. Another positive outcome is the absence of any feedback response that would up-regulate the miRNA upon introduction of increased target sites in the form of the miRNA sponge. Even though early results with transiently introduced sponges were encouraging, it was not certain that sponge mRNAs would be able to accumulate to levels sufficient to inhibit miRNA in stable expression formats. Recent results indicate that this is possible.
STABLE miRNA SPONGE EXPRESSION
Continuous expression of the sponge inhibitor makes it possible to perform long-term miRNA loss-of-function studies in cell culture and in vivo assays, such as bone marrow reconstitution and cancer xenografts. Several groups have achieved stable miRNA sponge activity by expressing the transgene from chromosomal integrations (Scherr et al. 2007; Bonci et al. 2008; Gentner et al. 2009; Haraguchi et al. 2009; Huang et al. 2010; Valastyan et al. 2009; Barbato et al. 2010; Gatt et al. 2010; Gottwein and Cullen 2010; Ma et al. 2010a,b; Papapetrou et al. 2010; Starczynowski et al. 2010; see Table 1). The challenge for stable expression is to produce a sufficient dose of sponge mRNA given much lower transgene copy numbers compared to transient plasmid transfection. The good news from recent reports is that even partial miRNA inhibition can yield measurable and interesting phenotypes.
One of the applications of stable sponge expression is to probe the roles of miRNAs in differentiation pathways. Barbato et al. used a lentiviral sponge in post-mitotic primary cerebellar granule neurons to assay the effect of miR-92, which is down-regulated over the course of neuronal maturation. At 6 d in vitro, the sponge-expressing neurons showed derepression of the potassium chloride cotransporter KCC2 and electrophysiological changes in response to GABA treatment (Barbato et al. 2010). Similarly, Huang et al. used a stably expressed retroviral sponge in mesenchymal stem cells to assay the role of miR-204 in an in vitro differentiation time-course lasting 10 d. Continuous inhibition of miR-204 strongly reduced adipogenic differentiation while increasing markers of osteogenic differentiation (Huang et al. 2010). Papapetrou et al. sought to probe the role of the erythroid-specific, closely clustered miRNAs, miR-144 and miR-451, in blood cell development. To this end they used lentiviral sponges marked with a different color fluorescent reporter for each miRNA to dissect their relative contributions in erythropoiesis (Papapetrou et al. 2010). Bone marrow reconstitution was performed with a 1:1 mixture of a green control sponge with a red (miR-144) or a yellow (miR-451) sponge, or both. Three to four weeks after transplantation, the competitive repopulation of the chimeric blood was analyzed by flow cytometry. Both miRNAs were found to be required for normal progression through the first stage of erythroblast maturation, and their simultaneous inhibition showed that they act additively.
One of the most common applications of stably expressed sponges is to mimic the down-regulation of specific miRNAs that are aberrantly expressed in certain disease states. For example, by screening miRNA expression and metastatic potential of a panel of mammary cell lines, Valastyan et al. identified miR-31 as strongly down-regulated in aggressive metastatic cancer (Valastyan et al. 2009). They set up an experimental model wherein human nonmetastatic breast cancer cells transduced with retroviral eGFP sponges for miR-31 or an irrelevant sequence were orthotopically implanted in mouse mammary fat pads. Primary tumor size was not significantly affected by the inhibition of miR-31, but, while the control sponge tumors did not metastasize, miR-31 sponge tumors metastasized to the lungs, forming 10 times more lesions (easily identifiable by their GFP fluorescence). This result allowed the investigators to identify miR-31 as a suppressor of metastasis. A similar approach was taken to show that miR-10b (Ma et al. 2010a) and miR-9 (Ma et al. 2010b) promote breast cancer metastasis. The recent finding that reduction in the expression of a tumor suppressor by a mere 20% can promote the development of cancer (Alimonti et al. 2010) suggests that screens with sponges, which may alter target gene expression to a similar extent, could be generally informative.
A related experiment is the application of a sponge to mimic the genetic state of patients with a genomic deletion of a particular miRNA or miRNA cluster. For example, the miR-15a-16-1 cluster is located within a region of chromosome 13q14 that is frequently deleted in leukemia, prostate cancer, and other malignancies (Bottoni et al. 2005; Bonci et al. 2008; Bandi et al. 2009; Hanlon et al. 2009; Corthals et al. 2010; Gatt et al. 2010). Bonci et al. and Gatt et al. used lentiviral GFP sponges with sites for miR-15a and miR-16, respectively, and tested transduced human prostate cancer and multiple myeloma cell lines by xenograft assay. In both cases the miR-15/16-inhibited cancers developed larger, more invasive tumors than their negative controls; in the multiple myeloma study, the animals showed substantially decreased survival, from a median of 80 to 31 d. Analysis of the tumors implicated several signaling pathways in which the miR-15/16 family acts to suppress survival, proliferation, and invasiveness (Gatt et al. 2010).
Another instance of a disease-associated miRNA cluster deletion occurs in the 5q- subtype of myelodysplastic syndrome (MDS) (Starczynowski et al. 2010). In this case the miRNAs in the cluster, miR-145 and miR-146a, have different seeds. To model the partial loss of these two miRNAs in hematopoietic stem/progenitor cells, Starczynowski et al. used a combination sponge containing eight to nine bulged sites for each miRNA. Cells transduced with retroviral YFP sponges were transplanted into lethally irradiated recipient mice, and were mixed with wild-type cells to mimic the chimerism of human 5q- patients. Eight weeks post-transplantation, the animals' blood cells manifested most of the features of MDS. Observation over the long term proved the benefit of including a fluorescent reporter in the competition assay: Over the course of several months, YFP+ cells were depleted from the blood of the sponge-transduced (but not vector control) recipients, yet thrombocytosis was still evident, indicating a cell nonautonomous effect of miRNA depletion. This correlated with an increased serum IL-6 concentration attributable to the derepression of miR-146 target gene TRAF6. Sustained, systemic phenotypes may result from transient miRNA perturbation in a subset of cells if secreted cytokines operate in a positive feedback loop, as in the recently described inflammatory cascade driven by IL6, let-7 down-regulation, and NF-κB (Iliopoulos et al. 2009). As in the case of miR-15a-16-1 depletion in cancer, the ability of the stable sponge to partially knock down miRNA activity provides a good mimic for the partial loss of miRNA expression in patients with a heterozygous deletion. The miR-145-146a miRNA cluster was shown to be haploinsufficient in conferring protection against disease (Starczynowski et al. 2010).
IN VIVO APPLICATIONS FOR miRNA SPONGES
Delivery of sponge constructs to tissue in live mice is feasible with the use of viral vectors. Carè et al. used an adenoviral eGFP sponge to inhibit miR-133 in cardiac myocytes in vivo in a mouse model of cardiac hypertrophy (Carè et al. 2007). Du et al. used a lentiviral miR-326 sponge with tropism for CD4+ T-cells and delivered it to the blood system by a tail-vein injection of mice with experimental autoimmune encephalomyelitis (EAE), a model of multiple sclerosis. Sponge treatment reduced the development of autoimmune IL-17 secreting TH-17 cells and ameliorated the histological signs of EAE (Du et al. 2009). Adeno-associated virus (AAV) vectors are another delivery option suitable for long-term sponge treatment: They can infect nondividing cells and give continuous high expression from a nonrandom integration site (Kotin et al. 1990). Krol et al. used an AAV vector to deliver sponges to mice subretinally. In this case, the eGFP sponge was driven by the rhodopsin promoter to allow for specific expression in photoreceptor cells, and each animal received a combination sponge for three light-regulated miRNAs (miR-182, -96, and -183) in one eye and an empty control vector (lacking miRNA binding sites) in the other (Krol et al. 2010). Three weeks post-injection, retinas were isolated and dissected into retinal layers using laser capture microdissection for eGFP-expressing cells. Western blotting revealed strong derepression for the target glutamate transporter SLC1A1.
The first transgenic organisms made to express miRNA sponges were plants (Franco-Zorrilla et al. 2007). These incorporated a single bulged binding site for the miRNA of interest in the context of an overexpressed noncoding RNA, and successfully generated phenotypes opposite those of the corresponding miRNA-overexpressing plants.
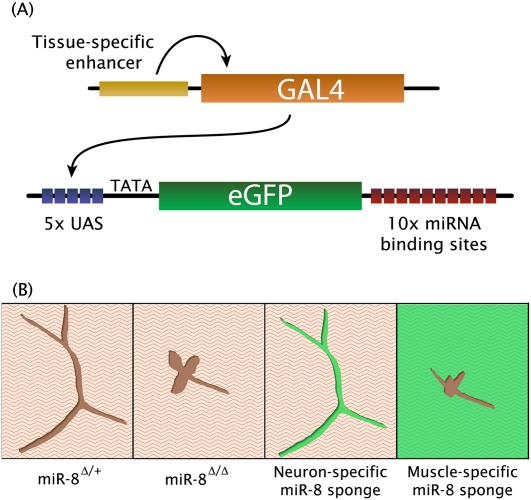
Stable, germline miRNA sponge expression in an animal model organism was first achieved in Drosophila using the Gal4-UAS (Upstream Activation Sequence) system (Loya et al. 2009; see Fig. 2). The sponge constructs consist of five UAS elements, a fluorescent reporter, and ten bulged miRNA binding sites in the 3′ UTR. Gal4 expressed from a tissue-specific promoter drives high expression of the sponge transgene. These inhibitors were able to completely suppress a neomorphic phenotype caused by an overexpressed miRNA in the eye, and to largely rescue expression of a target UTR reporter regulated by an endogenous miRNA in the wing imaginal disc. Hypomorphic phenotypes were enhanced by means of a sensitized background: the heterozygous miRNA deletion mutant, which has a reduced level of the miRNA but no detectable phenotype on its own. In this background, the sponge transgenics could phenocopy miRNA-null mutant flies. Varying the number of transgene copies also modulated the inhibitory effect, which could be used in combination with the miRNA genetic background to generate allelic series. The power of the Gal4 inducible system to dissect a null phenotype was shown by inhibiting a miRNA's activity in specific subtypes of cells. It is known that the miR-8 knockout has neuromuscular junction defects; activating the expression of a miR-8 sponge specifically in neurons or in muscle cells revealed the locally required activity (and regulation of the target gene Ena) in the post-synaptic muscle cell, even though miR-8 is present in both pre- and post-synaptic cells. The ability to probe the miRNA function in restricted subsets of cells could be critical, as there are cases of miRNA–target interactions restricted to one cell type; an extreme example is miR-273 repressing the transcription factor die-1 in the right chemosensory ASE neuron, and lsy-6 repressing cog-1 in the left chemosensory ASE neuron in Caenorhabditis elegans (Chang et al. 2004).
FIGURE 2.
(A) Tissue-specific expression of the Gal4 transcription factor was used to drive miRNA sponge expression under the control of upstream activating sequences (UAS) in transgenic fruit flies. (B) Dissection of a complex phenotype using tissue-specific sponges. A developmental defect in the axonal branching and synaptic boutons of neuromuscular junctions (NMJ) was observed in the miR-8 knockout (second panel) and in miR-8 heterozygous flies expressing a miR-8 sponge inhibitor specifically in muscle (fourth panel). Wild-type appearance of the NMJ is seen in the miR-8 heterozygote (first panel) and in miR-8 heterozygous flies expressing a miR-8 sponge specifically in neurons (third panel). Sponge expression is indicated by GFP fluorescence (shown in green).
OUTLOOK
Transgenic vertebrates expressing sponges are a work in progress. The recent development of the Tol2 transposon system and various Gal4 strains should facilitate the introduction of sponge transgenes for tissue-specific expression in zebrafish (Asakawa and Kawakami 2008). In the mouse, an inducible sponge could be created by means of the Cre-lox system (to remove a transcriptional stop cassette with tissue-specific recombinase expression) or with a tet-responsive element driving the sponge and tissue-specific reverse tet transactivator (rtTA) expression in combination with feeding the animal doxycycline. A sensitized background of DGCR8 and/or Dicer heterozygosity, which shows partially reduced levels for some miRNAs (Murchison et al. 2005; Wang et al. 2007), might enhance the loss of function. It should be noted, however, that the Dicer heterozygous state can accelerate the development of tumors in mouse models (Kumar et al. 2009). It remains to be shown whether in vivo sponge expression will provide a faithful alternative to genetic knockouts of miRNA families. For the miR-15/16 and miR-144/451 clusters, their respective roles as suppressors of tumor growth and promoters of erythropoiesis are supported by deletion experiments (Klein et al. 2010; Rasmussen et al. 2010). For miR-133 on the other hand, the knockout mouse does not develop cardiac hypertrophy (Liu et al. 2008) as is observed when a viral sponge construct or antagomirs are delivered to the heart (Carè et al. 2007). Further experiments will be necessary to clarify this discrepancy and to prove the utility of the sponge method in vivo.
ACKNOWLEDGMENTS
We thank Mary Lindstrom for help preparing the figures. This work was supported by United States Public Health Service grant R01-CA133404 from the National Institutes of Health (to P.A.S.) and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2414110.
REFERENCES
- Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, et al. 2010. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet 42: 454–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS 2010. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol 6: 363 doi: 10.1038/msb.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K 2008. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ 50: 391–399 [DOI] [PubMed] [Google Scholar]
- Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E 2009. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res 69: 5553–5559 [DOI] [PubMed] [Google Scholar]
- Barbato C, Ruberti F, Pieri M, Vilardo E, Costanzo M, Ciotti MT, Zona C, Cogoni C 2010. MicroRNA-92 modulates K(+) Cl(−) co-transporter KCC2 expression in cerebellar granule neurons. J Neurochem 113: 591–600 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolisetty MT, Dy G, Tam W, Beemon KL 2009. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol 83: 12009–12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. 2008. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14: 1271–1277 [DOI] [PubMed] [Google Scholar]
- Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC 2005. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol 204: 280–285 [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat Med 13: 613–618 [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ Jr, Frøkjaer-Jensen C, Lockery S, Hobert O 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789 [DOI] [PubMed] [Google Scholar]
- Corthals SL, Jongen-Lavrencic M, de Knegt Y, Peeters JK, Beverloo HB, Lokhorst HM, Sonneveld P 2010. Micro-RNA-15a and micro-RNA-16 expression and chromosome 13 deletions in multiple myeloma. Leuk Res 34: 677–681 [DOI] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C 2006. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 34: 2294–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G 2009. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol 10: 1252–1259 [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA 2007. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Goswami S, Noubissi FK, Spiegelman VS 2009. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell 35: 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau CC 2008. Inhibition of microRNA with antisense oligonucleotides. Methods 44: 55–60 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt ME, Zhao JJ, Ebert MS, Chu Z, Mani M, Gazit R, Carrasco DE, Dutta-Simmons J, Adamia S, Minvielle SB, et al. 2010. Blood (in press). [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L 2009. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 6: 63–66 [DOI] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol 84: 5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon K, Rudin CE, Harries LW 2009. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL). PLoS ONE 4: e7169 doi: 10.1371/journal.pone.0007169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Ozaki Y, Iba H 2009. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 37: e43 doi: 10.1093/nar/gkp040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Horie T, Ono K, Nishi H, Iwanaga Y, Nagao K, Kinoshita M, Kuwabara Y, Takanabe R, Hasegawa K, Kita T, et al. 2009. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun 389: 315–320 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao L, Xing L, Chen D 2010. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 28: 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K 2009. An epigenetic switch involving NK-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi A 1999. Functional approaches to gene isolation in mammalian cells. Science 285: 299 doi: 10.1126/science.285.5426.299a [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. 2010. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17: 28–40 [DOI] [PubMed] [Google Scholar]
- Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI 1990. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci 87: 2211–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, et al. 2010. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141: 618–631 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T 2008. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci 105: 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23: 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN 2008. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 22: 3242–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA 2009. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods 6: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA 2010a. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 28: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. 2010b. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5: 376–385 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Kauppinen S, Lund AH 2006. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 10: 137–141 [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, Korkola JE, Sadelain M 2010. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 28: 287–296 [DOI] [PubMed] [Google Scholar]
- Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O'Carroll D 2010. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M 2008. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell 19: 3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M 2007. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res 35: e149 doi: 10.1093/nar/gkm971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, et al. 2010. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med 16: 49–58 [DOI] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS, van Oudenaarden A 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell 38: 140–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA 2009. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137: 1032–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]