Abstract
Caenorhabditis elegans GLD-3 is a five K homology (KH) domain-containing protein involved in the translational control of germline-specific mRNAs during embryogenesis. GLD-3 interacts with the cytoplasmic poly(A)-polymerase GLD-2. The two proteins cooperate to recognize target mRNAs and convert them into a polyadenylated, translationally active state. We report the 2.8-Å-resolution crystal structure of a proteolytically stable fragment encompassing the KH2, KH3, KH4, and KH5 domains of C. elegans GLD-3. The structure reveals that the four tandem KH domains are organized into a globular structural unit. The domains are involved in extensive side-by-side interactions, similar to those observed in previous structures of dimeric KH domains, as well as head-to-toe interactions. Small-angle X-ray scattering reconstructions show that the N-terminal KH domain (KH1) forms a thumb-like protrusion on the KH2–KH5 unit. Although KH domains are putative RNA-binding modules, the KH region of GLD-3 is unable in isolation to cross-link RNA. Instead, the KH1 domain mediates the direct interaction with the poly(A)-polymerase GLD-2, pointing to a function of the KH region as a protein–protein interaction platform.
Keywords: post-translational control, germline development, RNA-binding protein, protein–protein interaction, SAXS
INTRODUCTION
The K homology (KH) domain is one of the most abundant domains in RNA-binding proteins. It was originally identified as a conserved region present in three copies in the heterogeneous nuclear ribonucleoprotein (hnRNP) K (Siomi et al. 1993) and has since been identified in proteins from all three domains of life (Valverde et al. 2008). KH domains span from 70 to 100 amino acids and fold into globular structures with a three-stranded antiparallel β-sheet flanked by three α-helices. The loop between the two N-terminal α-helices features the conserved GxxG sequence that is involved in nucleic acid interactions. Degenerate KH domains lacking the GxxG sequence have also been described and were shown to mediate protein–protein interactions (Buttner et al. 2005; Oddone et al. 2007) or have been implicated in modulating substrate-binding affinities (Musco et al. 1996).
KH domains are usually present in multiple copies in eukaryotic proteins. Besides remarkable cases like Vigilin, which contains 15 tandem KH domains, numerous proteins contain up to half a dozen (Valverde et al. 2008). The clustering of KH domains can in principle give rise to different structural and functional properties. Consecutive domains might be arranged as “beads on a string” or alternatively fold into higher-order structures that are either preformed as rigid platforms or induced upon ligand binding. The functional implication is that individual domains might either act independently or cooperatively, for example, to increase nucleic acid recognition (Lunde et al. 2007). Further clustering might also be achieved by the tendency of individual KH domains to self-associate (Du et al. 2007; Valverde et al. 2008). However, these homo-oligomerization properties have generally been extrapolated from biochemical and crystallographic studies of protein fragments containing either one or two KH domains. To date, it remains unclear whether the clustering of single or double KH domains observed in crystal structures is imposed by packing interactions or whether it might reflect a genuine physiological state.
We have addressed the structural organization of a five KH domain-containing protein, Caenorhabditis elegans GLD-3 (germline development defective 3). GLD-3 belongs to the Bicaudal-C (Bic-C) protein family. The founding member of this protein family was originally identified in Drosophila and shown to be involved in oogenesis and patterning of the embryo (Ashburner et al. 1983; Mohler and Wieschaus 1986; McKnight et al. 1992; Mahone et al. 1995). In C. elegans, GLD-3 is required for many aspects of germline development and embryogenesis (Eckmann et al. 2002, 2004). It operates at the level of post-translational control to regulate the timely expression of specific developmental proteins (Eckmann et al. 2002; Wang et al. 2002). One mechanism involves the extension of the poly(A) tail of the target mRNAs and requires a complex between GLD-3 and GLD-2, a noncanonical poly(A)-polymerase (PAP) (Wang et al. 2002). GLD-2 lacks the RNA-binding domain characteristic of nuclear PAPs and instead associates with the KH region of GLD-3 for presumed RNA targeting (Wang et al. 2002; Eckmann et al. 2004). The GLD-3 KH region has also been implicated in binding another developmental regulator protein, GLS-1 (Rybarska et al. 2009). It is unclear how the five KH domains of GLD-3 are arranged to support these macromolecular interactions. Using a combination of crystallography, small-angle X-ray scattering and biochemical studies, we show that GLD-3 contains KH domains that function independently to bind GLD-2 (KH1) and cooperatively to form a higher-order structure (KH2, KH3, KH4, KH5).
RESULTS AND DISCUSSION
Four tandem KH domains of GLD-3 form a proteolytically stable unit
We expressed and purified the KH region of C. elegans GLD-3 (KH1–KH5, residues 1–460) (Eckmann et al. 2002; Rybarska et al. 2009) and assessed its stability upon protease treatment. Limited proteolysis is often used to assess the presence of globular regions and flexible portions of a given polypeptide and is particularly useful to probe the conformation of multidomain proteins such as GLD-3. We incubated GLD-3 KH1–KH5 with a set of proteases with different specificities. Addition of GluC, trypsin, or chymotrypsin all resulted in a stable fragment of ∼43 kDa (Supplemental Fig. 1). N-terminal Edman sequencing and mass spectrometry analysis identified the fragments as spanning residues 88–460 (GluC cleavage) and 95–460 (trypsin cleavage). Based on sequence similarity with eukaryotic KH proteins of known structure, the cleavage sites are predicted to reside in loop regions of KH domain 1 (KH1) (Fig. 1A). The 43-kDa fragment including KH2 to KH5 appeared stable even at higher protease concentrations. This indicated that neither the short linkers predicted to connect KH2–KH3 and KH4–KH5 nor the relatively long linker predicted to connect KH3–KH4 is susceptible to protease cleavage. The stability of KH2–KH5 in the presence of proteases suggests that it forms a compact structural unit.
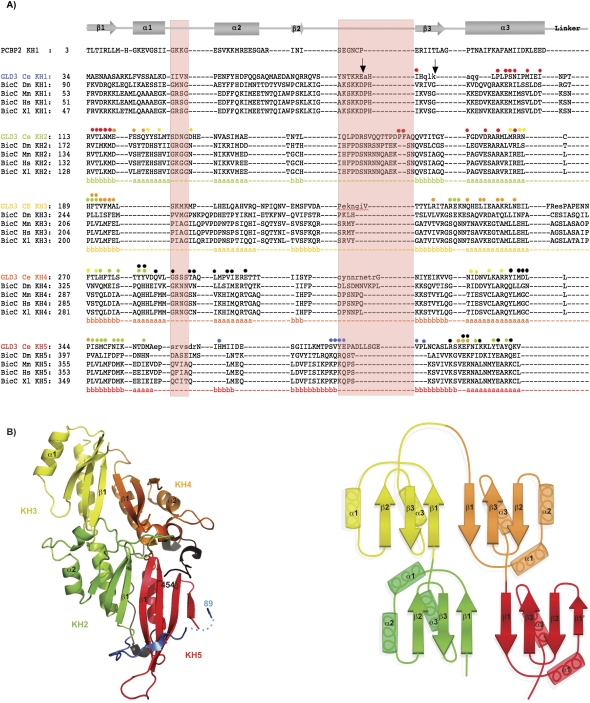
FIGURE 1.
The KH2, KH3, KH4, and KH5 domains of GLD-3 form a globular structure. (A) Structure-based sequence alignment of the C. elegans GLD-3 KH domains. The sequences of the KH domains of C. elegans (Ce) GLD-3 are aligned with the KH1 domain of human Poly(C) binding protein 2 (PCBP2), shown on top together with a schematic of the secondary structure elements: (arrows) β-strand; (rectangles) α-helices. The alignment of KH2 to KH5 is based on the GLD-3 structure, while for KH1 the alignment is based on sequence similarity and secondary structure predictions. Included are the sequences of the related Bic-C proteins from D. melanogaster (Dm), Mus musculus (Mm), Homo sapiens (Hs), and Xenopus laevis (Xl) aligned to C. elegans GLD-3 using T-coffee (Notredame and Suhre 2004). The annotated secondary structure of GLD-3 is shown in colored letters below the alignment (a, α-helix; b, β-strand); (green) KH2; (yellow) KH3; (orange) KH4; (red) KH5. Small letters represent disordered residues not seen in the crystal structure (molecule A). (Light red shading) The invariable (GxxG) and variable loop regions of canonical KH domains. GLD-3 Glu-C and Trypsin protease cleavage sites are marked with black arrows (at Glu 87 and Lys 94, respectively). Colored dots above GLD-3 KH domains highlight residues involved in intramolecular interactions with another KH domain (colors as above) or with the N-terminal and C-terminal segments present in the construct used (blue and black dots, respectively). The interacting residues were identified with the program AquaProt (Reichmann et al. 2007). The C-terminal segment present in the structure downstream from KH5 is not shown in the alignment. (B, left) Cartoon representation of the crystal structure of GLD-3 KH2–KH5. The four KH domains are colored: (green) KH2, (yellow) KH3, (orange) KH4, (red) KH5. The molecule is viewed at the β-sheet side of the KH domains (“front” side). (Blue) The segment N-terminal to KH2 (dots represent a disordered region); (black) the segment C-terminal to KH5 that is present in the construct crystallized. The N terminus and C terminus are labeled (residues 89 and 454). This and all other structure figures were generated with PyMOL (Delano Scientific). (B, right) A schematic representation of the KH2–KH5 architecture of GLD-3. The secondary structure elements are labeled. The head-to-toe dimers (KH2–KH3 and KH4–KH5) form two augmented β-sheets.
Crystal structure of the KH2–KH3–KH4–KH5 region of GLD-3
Guided by the proteolysis results, we expressed and crystallized a fragment of GLD-3 consisting of residues 88–460. The crystal structure was solved by a two-wavelength anomalous dispersion experiment using selenomethionine as an anomalous scatterer. The final model is refined at 2.8 Å resolution, with free-R and working-R factors of 27.6% and 25.8%, respectively, and good stereochemistry (Table 1). The crystal asymmetric unit contains two molecules (termed A and B) that are virtually identical (root mean square deviation [RMSD] of 0.59 Å over 313 Cα atoms). The final model encompasses residues 89 to 454, with the exception of disordered residues mostly present in loop regions, particularly in the so-called variable loop (Fig. 1A; Table 1).
TABLE 1.
X-ray data collection and refinement statistics
GLD-3 KH2 is formed by residues 113–187, followed by residues 189–259 for KH3, 270–342 for KH4, and 344–419 for KH5 (Fig. 1A). The individual domains adopt the β1-α1-α2-β2-β3-α3 sequel of secondary structure elements characteristic of type I KH motifs (Fig. 1B). A deviation from this pattern is found in KH3, where helix α2 is missing. Another deviation is found in KH5, with a β-strand (β1′) at the position where helix α2 would normally be expected (residues 367–371 in Fig. 1A). However, the β-strand conformation of these residues is likely due to crystal lattice contacts, as it is stabilized by interactions with a symmetry-related molecule and with the N-terminal residues 89–91 of the construct used (Supplemental Fig. 2A). The segment N-terminal to KH2 (light blue in Fig. 1B) stretches with residues 100–110 onto a hydrophobic patch of the neighboring KH5 domain. This interaction is likely an artifact of truncating the KH1 domain (Fig. 1A). The segment C-terminal to KH5 (residues 420–454; black in Fig. 1B) wraps around KH5 in an extended conformation, sealing the interaction with KH4 (see below). Although the C-terminal segment does not belong to the canonical sequence of KH motifs, it contributes to the structural organization of the KH2–KH5 region, explaining its stability in the proteolysis experiment. In this study, we refer to GLD-3 88–460 as KH2–KH5l (where the subscript l stands for long) and to the shorter GLD-3 110–460 construct as KH2–KH5s (where the N terminus of KH2 is defined based on the structural analysis).
The KH2–KH3–KH4–KH5 domains of GLD-3 assemble into a higher-order structure
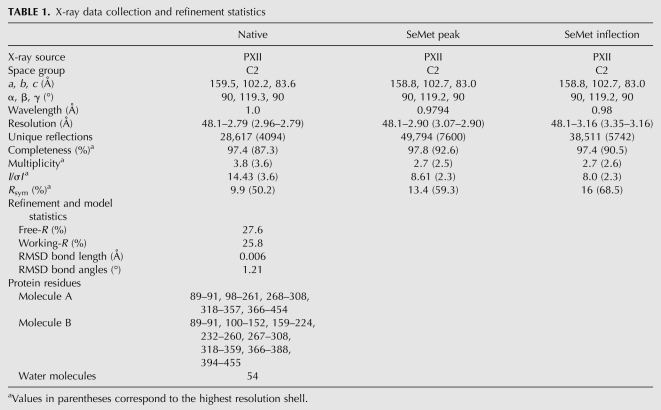
GLD-3 KH2–KH5l folds into a globular structure with approximate dimensions of 85 × 45 × 40 Å that is formed by the compact arrangement of the four individual KH domains (Fig. 1B). KH2 interacts head-to-toe with KH3 and side-by-side with KH5. Analogously, KH4 interacts head-to-toe with KH5 and side-by-side with KH3. The linkers connecting the head-to-toe dimers consist of a single amino-acid residue (Cys 188 for KH2–KH3 and Leu 343 for KH4–KH5), while the side-by-side positioning of the consecutive KH3–KH4 domains is mediated by a 10-residue-long linker (residues 260–269) that is disordered in the structure. The four KH domains adopt the same orientation, presenting the β-sheets on the one side of the molecule (front) and the α-helices on the other (back) (Fig. 1B). The side-by-side dimers result in two augmented β-sheets along KH2–KH5 and along KH3–KH4. The extended β-sheets are formed by the interactions of the respective β1-strands, which pack against each other in an antiparallel manner with extensive hydrophobic contacts. The side-by-side dimers also interact via polar contacts between the respective α3 helices (Fig. 2, left panel). The intramolecular interactions between KH2–KH5 and between KH3–KH4 are extensive (2495 Å2 and 1418 Å2 of buried surface area, respectively). Instead, the head-to-toe arrangements of KH2–KH3 and of KH4–KH5 result in a smaller interaction surface (1000 Å2 and 727 Å2 of the buried surface area, respectively). The KH2–KH3 interactions are mediated by hydrophilic contacts of the loop regions (Fig. 2, right panel), while the corresponding regions of KH4–KH5 are strengthened by hydrophobic contacts involving the segment C-terminal to KH5 (residues 434–454) (Supplemental Fig. 2B). This C-terminal segment also provides additional interactions to the side-by-side dimers (packing with residues 423–434 against the α3-helices of KH2 and KH5). The relative position and intramolecular interactions of the four KH domains are identical in the two molecules of the crystal asymmetric unit. Since the two molecules are involved in different crystal lattice contacts, it is unlikely that this arrangement is due to crystal packing effects.
FIGURE 2.
The clustering of KH domains in GLD-3 is mediated by conserved interactions. The left panel shows the interactions between side-by-side dimers. The molecule is viewed at the α-helical side of the KH domains (“back” side, related by a 180° rotation around a vertical axis in respect to the view in Fig. 1B). The close-up view highlights the contacts between KH2 and KH5 (conserved residues labeled in bold; see also Supplemental Fig. 3). The right panel shows the interactions between head-to-toe dimers. The close-up view highlights the contacts between KH4 and KH5. The N-terminal residues 89–112 and the C-terminal residues 420–454 are not shown for clarity.
The mode of interaction of the head-to-toe dimers has no similarity to that of KH proteins of known structure. The formation of the augmented β-sheet of the KH2–KH5 and KH3–KH4 dimers is instead similar to that reported for the KH1–KH2 regions of the poly(C) binding protein (PCBP2, PDB code 2JZX) and of Nova-1 (PDB code 2ANN) (Supplemental Fig. 2C, left panel). It is also similar to that observed in the crystals of isolated KH domains, for example, between symmetry-related Nova-1 KH3 domains (Lewis et al. 1999) or symmetry-related PCBP2 KH1 domains (Supplemental Fig. 2C, right panel; Du et al. 2005, 2008). The KH2–KH5 architecture of GLD-3 does not show the other type of interface reported from crystals of isolated KH domains, namely, a helix–helix packing between symmetry-related α3-helices as seen in Nova-2 KH3 (Lewis et al. 1999; Valverde et al. 2008). Analysis of the GLD-3 KH2–KH5l structure shows that the two molecules present in the asymmetric unit pack via a helix–helix contact of the α1–α2-helices of their KH2–KH3 modules (Supplemental Fig. 2D). However, this is a crystal-packing effect, since static light-scattering experiments show that GLD-3 88–460 behaves mostly as a monomer in solution, with little multimerization/aggregation (data not shown).
The presence of five consecutive KH domains is a conserved feature in the Bic-C family of proteins (Eckmann et al. 2002). Although C. elegans GLD-3 shows only weak overall sequence similarity to Drosophila melanogaster Bic-C (23% identity), several interface residues mediating the side-by-side dimer contacts between the α3-helices of KH2–KH5 and KH3–KH4 are conserved (Fig. 1; Supplemental Fig. 3). In addition, differences are predicted in the length of the variable loop regions, but not in the length of the linkers connecting the KH domains (Fig. 1). Thus, it is possible that other Bic-C family members might share a similar higher-order structure.
RNA-binding properties of the GLD-3 KH region
Drosophila Bic-C has been shown to bind to poly(G) and poly(U) RNAs in pull-down experiments in vitro (Saffman et al. 1998; Braddock et al. 2002) and a similar preference in RNA binding has been reported for mouse Bic-C (Bouvrette et al. 2008). While Bic-C proteins contain several GxxG motifs that are involved in nucleic acid binding in canonical KH domains (Braddock et al. 2002), the KH domains of GLD-3 are degenerate in that none features the GxxG sequence (Fig. 1). In addition, no significant patches of positively charged residues can be identified on the surface of the GLD-3 KH2–KH5l structure (data not shown).
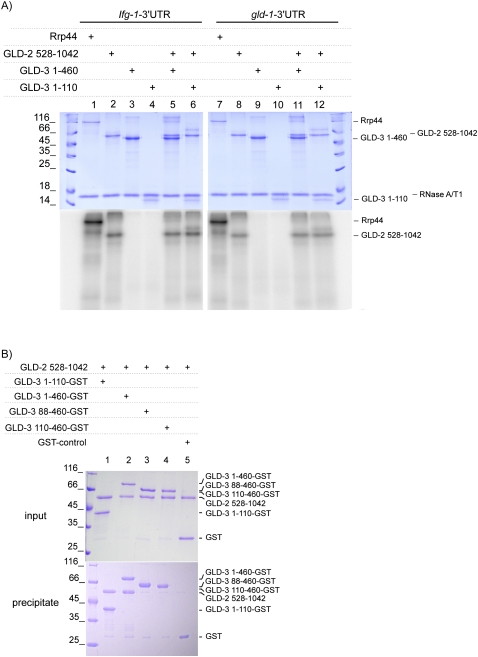
To test RNA-binding, we first used fluorescence anisotropy with a fluorescein-labeled poly(U) RNA oligo to measure the binding affinities with GLD-3, but no accurate measure could be obtained for either the KH1–KH5 or the KH2–KH5 proteins (Kds higher than 300 μM and 1 mM, respectively) (data not shown). We next investigated whether binding of the GLD-3 KH region could be detected with a physiologically more relevant RNA substrate. We incubated GLD-3 with a body-labeled RNA corresponding to the 3′ untranslated region (UTR) of the gld-1 mRNA, an in vivo target of GLD-2–GLD-3 polyadenylation (Suh et al. 2006). After ultraviolet (UV) irradiation at 254 nm, we observed no cross-linking with KH1–KH5 (Fig. 3A, lane 9). In contrast, the conserved poly(A)-polymerase region of GLD-2 (residues 528–1042) was able to cross-link RNA (Fig. 3A, lane 8). The extent of cross-linking by GLD-2 was similar whether in the absence or in the presence of GLD-3 (which is able to form a binary complex with the poly(A)-polymerase; see below) (Fig. 3A, lanes 11,12). A similar cross-linking pattern was obtained with the 3′ UTR of Ifg-1 (Fig. 3A, lanes 3,5,6), an unrelated RNA sequence that is not known as an in vivo target of GLD-2–GLD-3 activity. These data indicate that the GLD-3 KH domains contribute little to the RNA-binding properties of our recombinant polyadenylating complex, which appears to act in vitro in a sequence-independent manner.
FIGURE 3.
RNA-binding and protein-binding capabilities of the GLD-3 KH region. (A) RNA cross-linking experiment with GLD-3 in the absence and presence of GLD-2. Proteins and [α-32P]-body-labeled RNA corresponding to the gld-1 and Ifg-1 3′ UTR were cross-linked under UV light and separated on a 15 % SDS-PAGE. The gel was stained with Coomassie (upper gel) and analyzed by PhosphorImaging (lower gel). (B) Binding of the GLD-3 KH1 region to GLD-2. Stoichiometric complexes were obtained by pull-down experiments with GST-tagged GLD-3 KH1 and GLD-3 KH1-KH5 (lanes 1,2) but not with GLD-3 KH2-KH5l and KH2-KH5s (lanes 3,4). The Coomassie-stained SDS-PAGE gels show the input (upper gel) and the eluate (lower gel).
Protein-binding properties of the GLD-3 KH region
It has been previously shown by yeast two-hybrid experiments that GLD-3 interacts with GLD-2 via KH1 (Eckmann et al. 2004). To assay for direct protein–protein interactions, we carried out GST-pull-down experiments with recombinant proteins. The poly(A)-polymerase region of GLD-2 (residues 528–1042) forms a 1:1 stoichiometric complex with GLD-3 KH1 (residues 1–110) and GLD-3 KH1–KH5 (1–460) (Fig. 3B, lanes 1,2). No interaction could be detected with the KH2–KH5 region of GLD-3 (Fig. 3B, lanes 3,4) or GST alone (lane 5). Thus, KH1 is necessary and sufficient for binding GLD-2.
The KH region of GLD-3 has also been shown by yeast two-hybrid experiments to be an interacting domain for GLS-1, another factor involved in C. elegans germline development (Rybarska et al. 2009). It has been reported that a temperature-sensitive point mutation in KH4 (Gly327 to Arg) specifically impairs the interaction with GLS-1 and not with GLD-2 (Rybarska et al. 2009). Analysis of the structure suggests that this mutation is likely to severely perturb the structure of KH4, which would, in turn, have ripple destabilizing effects on the neighboring KH domains. Indeed, recombinant expression of GLD-3 1–460 with the G327R mutation results in insoluble protein (data not shown).
Structural characterization of the GLD-3 KH region in solution by SAXS
Given that the KH1 domain of GLD-3 can function independently of the KH2–KH5 region to bind GLD-2, we investigated how the first KH domain contributes to the overall structural organization using small-angle X-ray scattering (SAXS). We measured SAXS data with GLD-3 KH1–KH5 and GLD-3 KH2–KH5s. The samples were loaded onto a size exclusion chromatography column coupled to a quasi-elastic multiangle laser light scattering (MALS) detector to separate potential aggregates and to measure the absolute molecular weight and hydrodynamic radius simultaneously (Putnam et al. 2007). The SAXS measurements showed no radiation damage and no aggregation (Fig. 4A), a necessary prerequisite for the shape analysis.
FIGURE 4.
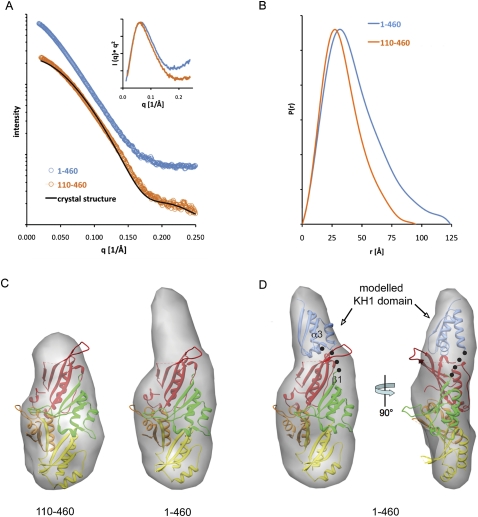
Solution structure of the GLD-3 KH1–KH5 region. (A) Experimental scattering profiles of GLD-3 1–460 (blue) and GLD-3 110–460 (orange). The theoretical scattering curve from the crystal structure of GLD-3 88–460 is shown as a black line. The inlet shows the calculated Kratky plots, suggesting a more flexible solution structure for the 1–460 polypeptide from the increase at high angles. (B) Distance distribution functions P(r) of the GLD-3 constructs computed from experimental SAXS data shown in the same colors as in A. The P(r) functions are normalized to unity at their maxima. (C) The average SAXS envelopes of GLD-3 110–460 (left) and 1–460 (right) calculated with GASBOR, displayed in transparent, volumetric representations and superposed with the crystal structure of GLD-3 88–460 (colored according to Fig. 1B). (D) Transparent, volumetric representation of GLD-3 1–460 in front view (left) and rotated 90° (right), showing the possible location of the KH1 domain of GLD-3 1–460 (blue, representing residues 35–110; black dots highlight a possible connection between KH1 α3 and KH2 β1).
The radius of gyration (RG) and maximum particle dimension (Dmax) that were computed directly from the scattering curve of GLD-3 110–460 (Fig. 4B) are consistent with those observed in the crystal structure (RG of 26.4 Å; Dmax of 95 Å from the SAXS data and 85 Å from the crystal structure). The small difference between the experimental and calculated scattering curves might be due to the flexible loops missing in the crystal structure (Fig. 4A). The absolute molecular weight (MW) of GLD-3 110–460 determined by MALS measurements matches the expected value (40 kDa) and confirms the monomeric nature of the KH2-KH5 fragment in solution. SAXS measurements on GLD-3 1–460 indicate that the KH1–KH5 region is also monomeric (molecular mass of 55 kDa derived from the MALS experiment) and is significantly more elongated than the KH2–KH5 fragment (RG of 34.6 Å; Dmax of 124 Å) (Fig. 4B). The Kratky plots suggest that the 1–460 polypeptide might exhibit increased conformational flexibility as compared to the globular 110–460 fragment (Fig. 4A).
Ab initio methods were employed to reconstruct low-resolution shapes. Because these methods do not provide unique three-dimensional models, several independent reconstructions were carried out starting from random initial approximations to compute the averaged model. The 10 individual bead models of GLD-3 110–460 displayed similar compact shapes (normalized spatial discrepancy [NSD] values between 0.98 and 1.02) (Supplemental Fig. 4). The averaged model, representing the common structural features of all individual reconstructions at lower resolution, fits well with the crystal structure of the KH2–KH5 region (Fig. 4C, left panel). Thus, the SAXS data are not consistent with a flexible KH2–KH5 protein with a different KH arrangement over time in solution. Rather, they support the presence of the same arrangement of the KH2–KH5 domains that we observe in the two molecules of the crystal asymmetric unit.
In the case of GLD-3 1–460 (KH1–KH5), the 10 independent reconstructions show more variability (NSD values between 1.25 and 1.53) (Supplemental Fig. 4), consistently with the increased flexibility detected in the Kratky plots (Fig. 4A). The averaged SAXS envelope shows additional density at one end of the molecule as compared to the shape of the 110–460 construct (Fig. 4C, right panel). This additional density has a size compatible to that of another KH domain, which can be fitted together with the KH2–KH5 crystal structure roughly in a head-to-toe orientation with respect to KH2 (Fig. 4D). Although this model is consistent with the presence of a short linker connecting KH1 and KH2 as would be expected from a head-to-toe arrangement, it is not possible to precisely define the orientation and position of KH1 in the low-resolution SAXS reconstruction. In addition, other structural features might come into play, in particular, the flexibility of the KH1 attachment suggested by the access to proteases (Supplemental Fig. 1) and by the variation of the individual reconstructions (Supplemental Fig. 4). Nevertheless, the SAXS data of the KH1–KH5 protein unambiguously indicate a protruding region on the KH2–KH5 globular fold that corresponds to the additional KH domain.
Conclusions
The five tandem KH domains of GLD-3 are organized into two structural and functional units. The crystallographic analysis we report here shows that KH2, KH3, KH4, and KH5 form a higher-order structure. The KH2–KH5 globular organization is preformed rather than induced by ligand binding and appears to be a relatively rigid platform held together by extensive intramolecular interactions. KH1 is separated upon limited proteolysis, suggesting that it is not engaged in strong intramolecular contacts with the other KH domains. SAXS reconstructions show that KH1 forms a thumb-like extension protruding from KH2–KH5. KH1 also acts as an individual structural unit in that it mediates the direct interaction with the poly(A)-polymerase GLD-2 independently from KH2–KH5. Notably, none of the KH domains of GLD-3 contains the GxxG loop characteristic of nucleic-acid-binding KH domains. Indeed, we find that the KH1–KH5 region of GLD-3 is unable to cross-link RNA in vitro and does not significantly increase the RNA-cross-linking properties of GLD-2. Different scenarios can be envisioned for how GLD-3 contributes to the poly(A)-polymerase activity of GLD-2: KH1 could contribute parts of a composite catalytic site, for example, or it could induce a conformational change in the poly(A)-polymerase. Understanding the mode of action of GLD-3 KH1 in the poly(A)-polymerase complex and how the KH2-KH5 region might interact with other C. elegans germline specific proteins awaits future structural studies.
MATERIALS AND METHODS
Cloning, expression, and purification
GLD-3 (88–460, 1–460, 1–110, and 110–460) and GLD2 528–1042 from C. elegans were amplified by polymerase chain reaction (PCR) using NcoI/NotI restriction sites. The final constructs were expressed as GST-tagged TEV-cleavable proteins using a pET-derived expression vector in Escherichia coli BL21 Gold pLyS cells (Stratagene) and B834 (DE3) cells (Novagen) in the case of selenomethionine-derivatized protein.
For protein expression, bacterial cells were grown in Terrific Broth (TB) at 18°C and induced with 1 mM IPTG for 16 h. Expression of selenomethionine-derivatized protein was carried out in minimal medium in the presence of 50 mg/L selenomethionine. Native and selenomethionine-derivatized GLD-3 88–460 was purified using a GSH-Sepharose affinity step (GE Healthcare) followed by TEV-cleavage and ion-exchange chromatography (HiTrap Q; GE Healthcare). The final purification step was carried out by size-exclusion chromatography (Superdex S200; GE Healthcare) in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 % glycerol, and 2 mM DTT. Purification of GLD-3 1–460, GLD-3 1–110, GLD-3 110–460, and GLD-2 528–1042 was performed with the same protocol.
In vitro cross-linking and GST-pull-down experiments
3′-UTR RNAs were generated by in vitro transcription in the presence of [α-32P]UTP (Perkin-Elmer) with the MEGAshortscript transcription kit (Ambion), followed by denaturing gel purification. For cross-linking, proteins (50 pmol of GLD-2 and/or 60 pmol of GLD-3) were mixed with 1 pmol of 32P-body-labeled RNA to a final 20-μL reaction volume in 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM magnesium diacetate, 10% (w/v) glycerol, 0.1% (w/v) NP-40, and 1 mM DTT. After incubating for 1 h at 4°C, reaction mixtures were irradiated with UV light (λ = 254 nm) for 30 min on ice before denaturation with 0.1% SDS and digestion with RNase A/T1 for 1 h at 37°C. Proteins cross-linked to RNA fragments were separated on a 15% (w/v) SDS-PAGE and visualized by Coomassie staining and PhosphorImaging (GE Healthcare).
For GST-pull-down experiments, GST-tagged GLD-3 proteins or GST alone (0.5 mg/mL) were incubated with stoichiometric amounts of untagged GLD-2 on GSH-Sepharose (GE Healthcare) for 2 h at 4°C in 20 mM Tris (pH 7.5), 150 mM NaCl, 20% glycerol, and 2 mM DTT. Beads were washed three times with an appropriate amount of binding buffer. Samples taken before incubation and beads were analyzed on a 15% SDS-PAGE, and proteins were stained with Coomassie Blue.
Crystallization and structure determination
Both native and selenomethionine-derivatized crystals were obtained using the sitting-drop vapor diffusion method at room temperature after mixing 0.2 μL of protein with 0.2 μL of reservoir solution containing 1.5 M (NH4)2SO4 and 0.1 M HEPES (pH 7.0). The crystals were cryoprotected in mother liquor supplemented with 20% glycerol, mounted in nylon loops, and flash-frozen in liquid nitrogen for data collection at 100 K. All X-ray diffraction data were collected at the PXII beamline of the Swiss Light Source (SLS) synchrotron in Villigen (Switzerland). The data sets were processed and scaled with XDS (Kabsch 2010). The GLD-3 88–460 crystals belong to the monoclinic space group C2 and contain two molecules in the asymmetric unit. A multi-wavelength anomalous diffraction (MAD) experiment with a selenomethionine-substituted crystal was performed by collecting a 2.9-Å-resolution data set at the peak wavelength of 0.9794 Å and a 3.1-Å-resolution data set at the inflection wavelength of 0.98 Å. The data processing statistics are summarized in Table 1.
The program autoSHARP/SHARP was used to find 17 selenium atom sites and to calculate phases to 3.5 Å (Vonrhein et al. 2007). Phase improvement was carried out with SOLOMON and resulted in an interpretable electron density map that could be used for manual model building with COOT (Emsley and Cowtan 2004). Refinement of the model was performed with CNS (Brunger et al. 1998) and included bulk solvent corrections, and individual B-factor and rigid body refinement. For cross-validation, 5% of the original reflections were omitted from refinement and used to calculate the free R-factor. After cyclic rounds of manual model building and refinement, the GLD-3 88–460 selenomethionine model was used to phase a 2.8-Å native data set by molecular replacement with PHASER (McCoy et al. 2007). The structure was refined using CNS. The stereochemical quality of the final model was assessed with MolProbity (Davis et al. 2007), showing that 98% and 0.64% of the residues are in the favored and disallowed regions of the Ramachandran plot, respectively. The model statistics are summarized in Table 1.
Small-angle X-ray scattering (SAXS)
SAXS experiments were performed at the SIBYLS beamline 12.3.1 at the Advanced Light Source (ALS) synchrotron in Berkeley, California. Size-exclusion chromatography was performed prior to SAXS measurements with a 24-mL Superdex 75 gel filtration column. One hundred microliters of protein (GLD-3 1–460 and GLD-3 110–460) was applied to the column in 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 2 mM DTT. MALS measurements were performed with a DynaPro quasi-elastic light scattering detector (Wyatt Technology). Simultaneous concentration measurements were made with an Optilab rEX refractive index detector connected in tandem to the MALS detector. The system was calibrated with BSA at 10 mg/mL using a refractive index increment of 0.172.
A dilution series of proteins was used for each condition, starting at a protein concentration of ∼5 mg/mL. Samples and buffer were loaded into a 96-well plate (Nunc) and covered with protective film. Samples were automatically loaded into the cuvette with a Hamilton syringe robot as described (Hura et al. 2009). SAXS measurements were made at three X-ray exposures of 1, 10, and 1 sec at room temperature. Overlaying the two short exposures assessed sample radiation damage. Interparticle interference during SAXS data collection was detected by comparing the lowest scattering angles for the concentration series. The data free of interparticle interference were merged and used for subsequent analyses. Fits to the Guinier region were made using autoRg (Robert Rambo, Lawrence Berkeley National Lab, Berkeley, CA). SAXS-based ab initio modeling was performed with GASBOR (Svergun et al. 2001). Ten independent computational bead models were derived from each SAXS data set and averaged using DAMAVER (Volkov and Svergun 2003)
Data deposition
The coordinates and structure factors have been deposited in the Protein Data Bank with PDB code 3N89.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the staff of the PX beamlines at the Swiss Light Source (Villigen, Switzerland) for assistance during data collection; C. Basquin for fluorescence anisotropy and static light scattering experiments; the MPI-Biochemistry Core Facility for mass spectrometry and Edman sequencing; and K. Valer-Saldana, S. Pleyer, and J. Basquin at the MPI-Biochemistry crystallization facility. We also thank members of our laboratories for comments and critical reading of the manuscript and Monika Krause for the drawing in Figure 1B. This study was supported by the Max Planck Gesellschaft (E.C., C.E.), the Sonderforschungsbereich SFB646, and the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (DFG) (E.C.), a FOR855 (DFG) grant (C.R.E.), and a DFG grant (K.N.). (former Büttner).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2315010.
REFERENCES
- Ashburner M, Detwiler C, Tsubota S, Woodruff RC 1983. The genetics of a small autosomal region of Drosophila melanogaster containing the structural gene for alcohol dehydrogenase. VI. Induced revertants of scutoid. Genetics 104: 405–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvrette DJ, Price SJ, Bryda EC 2008. K homology domains of the mouse polycystic kidney disease-related protein, Bicaudal-C (Bicc1), mediate RNA binding in vitro. Nephron Exp Nephrol 108: e27–e34 [DOI] [PubMed] [Google Scholar]
- Braddock DT, Louis JM, Baber JL, Levens D, Clore GM 2002. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature 415: 1051–1056 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP 2005. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, et al. 2007. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Lee JK, Tjhen R, Li S, Pan H, Stroud RM, James TL 2005. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 Å. J Biol Chem 280: 38823–38830 [DOI] [PubMed] [Google Scholar]
- Du Z, Lee JK, Fenn S, Tjhen R, Stroud RM, James TL 2007. X-ray crystallographic and NMR studies of protein–protein and protein–nucleic acid interactions involving the KH domains from human poly(C)-binding protein-2. RNA 13: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fenn S, Tjhen R, James TL 2008. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J Biol Chem 283: 28757–28766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell 3: 697–710 [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Crittenden SL, Suh N, Kimble J 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL II, Tsutakawa SE, Jenney FE Jr, Classen S, Frankel KA, Hopkins RC, et al. 2009. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6: 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK 1999. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure 7: 191–203 [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G 2007. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahone M, Saffman EE, Lasko PF 1995. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J 14: 2043–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight GL, Reasoner J, Gilbert T, Sundquist KO, Hokland B, McKernan PA, Champagne J, Johnson CJ, Bailey MC, Holly R, et al. 1992. Cloning and expression of a cellular high density lipoprotein-binding protein that is up-regulated by cholesterol loading of cells. J Biol Chem 267: 12131–12141 [PubMed] [Google Scholar]
- Mohler J, Wieschaus EF 1986. Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics 112: 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musco G, Stier G, Joseph C, Castiglione Morelli MA, Nilges M, Gibson TJ, Pastore A 1996. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 85: 237–245 [DOI] [PubMed] [Google Scholar]
- Notredame C, Suhre K 2004. Computing multiple sequence/structure alignments with the T-coffee package. Curr Protoc Bioinformatics 3: 3.8.1–3.8.28 [DOI] [PubMed] [Google Scholar]
- Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M 2007. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep 8: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA 2007. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Reichmann D, Cohen M, Abramovich R, Dym O, Lim D, Strynadka NC, Schreiber G 2007. Binding hot spots in the TEM1-BLIP interface in light of its modular architecture. J Mol Biol 365: 663–679 [DOI] [PubMed] [Google Scholar]
- Rybarska A, Harterink M, Jedamzik B, Kupinski AP, Schmid M, Eckmann CR 2009. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet 5: e1000494 doi: 10.1371/journal.pgen.1000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman EE, Styhler S, Rother K, Li W, Richard S, Lasko P 1998. Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol 18: 4855–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21: 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci 103: 15108–15112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH 2001. Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L 2008. Structure and function of KH domains. FEBS J 275: 2712–2726 [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI 2003. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G 2007. Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316 [DOI] [PubMed] [Google Scholar]