Abstract
Along the ribosome assembly pathway, various ribosomal RNA processing and modification reactions take place. Stem–loop 69 in the large subunit of Escherichia coli ribosomes plays a substantial role in ribosome functioning. It contains three highly conserved pseudouridines synthesized by pseudouridine synthase RluD. One of the pseudouridines is further methylated by RlmH. In this paper we show that RlmH has unique substrate specificity among rRNA modification enzymes. It preferentially methylates pseudouridine and less efficiently uridine. Furthermore, RlmH is the only known modification enzyme that is specific to 70S ribosomes. Kinetic parameters determined for RlmH are the following: The apparent KM for substrate 70S ribosomes is 0.51 ± 0.06 μM, and for cofactor S-adenosyl-L-methionine 27 ± 3 μM; the kcat values are 4.95 ± 1.10 min−1 and 6.4 ± 1.3 min−1, respectively. Knowledge of the substrate specificity and the kinetic parameters of RlmH made it possible to determine the kinetic parameters for RluD as well. The KM value for substrate 50S subunits is 0.98 ± 0.18 μM and the kcat value is 1.97 ± 0.46 min−1. RluD is the first rRNA pseudouridine synthase to be kinetically characterized. The determined rates of RluD- and RlmH-directed modifications of 23S rRNA are compatible with the rate of 50S assembly in vivo. The fact that RlmH requires 30S subunits demonstrates the dependence of 50S subunit maturation on the simultaneous presence of 30S subunits.
Keywords: post-transcriptional modification, ribosome assembly, 23S rRNA, RluD, RlmH, catalytic properties
INTRODUCTION
Ribosome biosynthesis is a complex, dynamic, highly coordinated, and energetically costly process composed of synthesis, processing, folding, modification, and assembly of all its numerous components. Nonetheless, synthesis of new ribosome subunits in prokaryotic cells is carried out fast and efficiently (Lindahl 1975; Bremer and Dennis 1996).
Ribosome assembly starts when ribosomal RNA (rRNA) is still being transcribed. Ribosomal proteins start to bind as soon as their binding sites emerge (Lewicki et al. 1993). Likewise, the processing and in all probability the enzymatic modification of the rRNA start before transcription is completed (Srivastava and Schlessinger 1989; Kaczanowska and Rydén-Aulin 2007). In vitro studies have shown that, while a number of modifications are synthesized on naked rRNA, synthesis of other modifications requires partially or even fully assembled subunits (Ofengand and Del Campo 2004; Kaczanowska and Rydén-Aulin 2007).
Ribosomal subunit assembly proceeds via assembly intermediate particles (Gegenheimer et al. 1977; Nierhaus 1991). Subunits acquire their final sedimentation value in less than a minute after the transcription of rRNA (Lindahl 1975). Freshly formed ribosome subunits are “immature” and enter the translating ribosome pool after an additional 1–2 min have passed (Lindahl 1975). Therefore, the rate-limiting step of ribosome assembly is the final maturation of subunits after the majority, if not all, of the ribosomal proteins have already bound to rRNA (Peil et al. 2008; Al Refaii and Alix 2009). Conformational rearrangements and a subset of rRNA processing and modification events are likely to take place during that time period (Holmes and Culver 2004; Kaczanowska and Rydén-Aulin 2007). In case of the large subunit, only a few modifications (Ψ1911, Ψ1915, Ψ1917, and Um2552) are shown to be introduced at the level of the 50S subunit (Bügl et al. 2000; Vaidyanathan et al. 2007). Incidentally, the corresponding modification enzymes RluD and RlmE have been implicated in ribosome assembly (Bügl et al. 2000; Caldas et al. 2000b; Gutgsell et al. 2005). However, the exact set and the rate of reactions taking place during this rate-limiting step of ribosome biosynthesis remains to be determined.
E. coli 23S rRNA stem–loop 69 is a universally conserved region which forms a distinct structure at the interface side of the 50S subunit and is a part of the intersubunit bridge B2a (Mitchell et al. 1992; Gabashvili et al. 2000; Yusupov et al. 2001; Schuwirth et al. 2005). Stem–loop 69 participates in several ribosome functions: In addition to 30S, it contacts the A-site tRNA and translation factors; it is involved in translation accuracy, initiation, termination, and ribosome recycling (O'Connor and Dahlberg 1995; Agrawal et al. 2004; Ali et al. 2006; Hirabayashi et al. 2006; Kipper et al. 2009).
Stem–loop 69 contains three highly conserved pseudouridines (Ψ) at positions 1911, 1915, and 1917 synthesized by the pseudouridine synthase RluD (Huang et al. 1998; Raychaudhuri et al. 1998; Ofengand et al. 2001; Ofengand 2002). Deletion of the rluD gene leads to a slow growth phenotype and massive defects in ribosome assembly (Ofengand et al. 2001; Gutgsell et al. 2005). Pseudouridines in stem–loop 69 have been shown to be necessary for efficient RF-2–directed translation termination in vivo (Ejby et al. 2007). RluD-directed pseudouridine isomerization takes place at the level of 50S subunits and is therefore one of the reactions occurring during the late steps of subunit maturation (Leppik et al. 2007; Vaidyanathan et al. 2007).
One of the pseudouridines in stem–loop 69 (position 1915) is further methylated by RlmH (Kowalak et al. 1996; Ero et al. 2008; Purta et al. 2008). RlmH is the only pseudouridine-specific methyltransferase identified to date (Ero et al. 2008). Cells lacking the rlmH gene have a clear growth disadvantage when competing with wild-type cells (Purta et al. 2008). RlmH prefers 70S ribosomes as substrate (Ero et al. 2008), which is an unprecedented case among ribosome modification enzymes. Following RluD in action and using 70S ribosomes as substrate implicate RlmH to take part in the final steps of ribosome biosynthesis (Ero et al. 2008; Purta et al. 2008).
Little is known about the quantitative aspects of ribosome biogenesis. The enzymes participating in ribosome maturation, while largely identified, are for the most part still poorly characterized. To describe the events of the large subunit maturation, the substrate specificity of RlmH was analyzed both in vivo and in vitro. We also determined the rate of rRNA pseudouridylation by RluD and methylation by RlmH and compared it with the previously published timeline of ribosome biogenesis (Lindahl 1975; Peil et al. 2008).
RESULTS
Nucleotide specificity of RlmH
RlmH catalyzes the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the nucleotide at position 1915 of E. coli 23S rRNA (Ero et al. 2008; Purta et al. 2008). Our previous results suggest that RlmH prefers pseudouridine over uridine as the methylation target in vitro (Ero et al. 2008). To determine the substrate nucleotide specificity of RlmH in vivo, we analyzed the nucleotide composition of 23S rRNA stem–loop 69 region isolated from various E. coli strains.
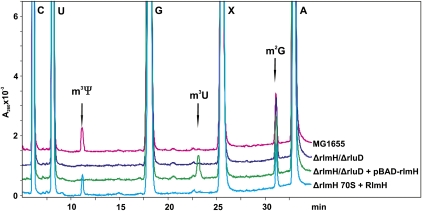
70S ribosomes were isolated from the E. coli wild-type strain MG1655 (wt), rluD knockout strain (ΔrluD), and rlmH/rluD double knockout strain (ΔrlmH/ΔrluD) with or without complementing plasmid-encoded (pBAD-rlmH) RlmH protein. Phenol-extracted RNA was used for the oligonucleotide-directed RNase H excision, generating an RNA fragment comprising nucleotides 1777–1922 of 23S rRNA. Nucleoside composition of the aforementioned RNA fragment was analyzed by RP-HPLC as described in Materials and Methods. Retention times of the modified nucleosides in 23S rRNA fragment under the conditions used are 4.9 min. (Ψ), 11.2 min. (m3Ψ), 23.2 min. (m3U), and 32.2 min. (m2G) according to Gehrke and Kuo (1989). Nucleoside absorbance profiles were recorded at 260 nm and peak areas were integrated. Obtained amounts of nucleosides are presented in relation to the 100% value of corresponding nucleosides of wild-type ribosomes. As an m3U-modified nucleoside was not detected in the wild-type probe, a fraction of m3U was calculated with respect to the maximum theoretical amount of m3U in the RNA fragment (uridine at position 1915 is completely methylated, assuming that molar absorptivity is the same for both U and m3U).
Nucleoside composition analysis (Fig. 1; Table 1) revealed that in comparison to the wild-type strain, ΔrluD and ΔrlmH/ΔrluD strains lack both pseudouridines and m3Ψ in their stem–loop 69 region, in agreement with the lack of the rluD gene. A small fraction (<10%) of m3U was detected in the ΔrluD strain but not in the ΔrlmH/ΔrluD strain that in addition lacks the rlmH gene. This finding indicates that in the absence of pseudouridine at position 1915, endogenous RlmH is able to methylate U1915, albeit inefficiently. Expression of RlmH protein from a plasmid (pBAD-rlmH) in the ΔrluD and ΔrlmH/ΔrluD strains causes a more significant amount (∼50%–60%) of m3U formation that can be attributed to the high level of protein overexpression from the arabinose-inducible pBAD plasmid construct (data not shown). Evidently, pseudouridine rather than uridine is the substrate preferred by RlmH in vivo.
FIGURE 1.
Modified nucleosides m3Ψ and m3U formed by RlmH in vivo and in vitro. HPLC analysis of nucleoside composition of a 23S rRNA fragment comprising nucleotides 1777–1922. The 23S rRNA fragments were isolated by RNase H treatment and gel electrophoresis from 70S ribosomes of E. coli wild-type strain (MG1655), isogenic strain lacking rlmH (ΔrlmH), or both rlmH and rluD genes (ΔrlmH/ΔrluD). For in vivo experiments, strains were complemented with a plasmid (pBAD-rlmH) overexpressing RlmH protein and for in vitro experiments, purified 70S ribosomes were treated with RlmH protein in vitro. Peaks corresponding to four standard nucleosides (C, U, G, and A) and three modified nucleosides; 3-methylpseudouridine (m3Ψ), 3-methyluridine (m3U), and 2-methylguanosine (m2G) are indicated. Nonspecific peak resulting from experimental conditions is marked with an X.
TABLE 1.
Quantification of nucleosides in 23S rRNA fragment 1777–1922
For quantitative assessments of the substrate preferred by RlmH in vitro, 70S ribosomes isolated from the ΔrluD, ΔrlmH/ΔrluD, and ΔrlmH strains were treated with purified RlmH and analyzed by RP-HPLC as described above. Even though ribosomes were incubated for 1 h with ∼200-fold molar excess of RlmH protein, the level of U1915 methylation in ΔrluD and ΔrlmH/ΔrluD ribosomes was relatively low (∼20–30%), whereas under the same conditions, the level of Ψ1915 methylation in ΔrlmH ribosomes was ∼90% (see Fig. 1; Table 1).
Taken together, these results show that RlmH methylates pseudouridine more efficiently than uridine at position 1915 of 23S rRNA both in vitro and in vivo.
Ribosome specificity of RlmH
Our previous studies showed that purified RlmH protein preferred 70S ribosomes over free 50S subunits as the substrate in vitro (Ero et al. 2008). However, the RlmH protein with an N-terminal His-tag was used in the aforementioned studies. In the current study, we used the native RlmH protein in order to exclude the effect of the affinity tag.
70S ribosomes and free 50S subunits from an rlmH knockout strain (ΔrlmH) were incubated for 1 h with ∼200-fold molar excess of purified native RlmH protein, subjected to rRNA fragment isolation and HPLC analysis of nucleosides as described above. As already mentioned, ∼90% of ΔrlmH 70S ribosomes were methylated by native RlmH as compared with m3Ψ levels of wt 70S ribosomes (Fig. 1; Table 1). Unlike the His-tagged RlmH protein, the native RlmH protein was shown to methylate ∼80% of the free 50S subunits when excess amounts of protein and long incubation times are used (Table 1). We suspected that the activity of RlmH on 50S results from the presence of trace amounts of 30S subunits in the 50S subunit preparation.
It is known that 70S ribosomes dissociate at Mg2+ concentrations below 2 mM, whereas free 30S and 50S subunits associate to form 70S ribosomes at Mg2+ concentrations above 6 mM (Blaha et al. 2002). Testing RlmH activity within this range of magnesium ion concentrations allows determination of whether the free 50S subunits or the 70S ribosomes are used as the substrate.
The methyltransferase activity of RlmH at various Mg2+ concentrations was studied using either ΔrlmH 70S ribosomes or free 50S and 30S subunits in experiments. RlmH-dependent incorporation of [3H]-methyl groups into ribosomes was monitored by TCA precipitation and scintillation counting. Methyl group incorporation activity of RlmH followed the same Mg2+ dependency pattern as ribosome subunit association/dissociation (Fig. 2). Hence, the true substrate of RlmH is indeed the 70S ribosome.
FIGURE 2.
Association of 30S and 50S subunits is necessary for RlmH activity. ΔrlmH 70S ribosomes (filled circles and solid line) or 50S and 30S subunits (open circles and dotted line) were methylated at various Mg2+ conditions using [3H]-SAM and purified RlmH as described in Materials and Methods. Incorporation of [3H]-methyl groups into TCA insoluble matter was determined. One experiment representative of three repeats is shown.
Enzymatic properties of RlmH
We determined the apparent KM and kcat values for 70S ribosomes and SAM using purified RlmH protein and radioactively labeled SAM as described in Materials and Methods. Preliminary experiments determined the assay conditions in which the rate of [3H]-methyl group incorporation from [3H]-SAM into 70S ribosomes (ΔrlmH strain) was proportional to the concentration of RlmH and linear over more than 5 min (data not shown). Thus, the initial rate measurements represented the true initial velocity of the reaction, and the dependence of the rate on the substrate concentration could be measured. For 70S ribosomes, initial rates were determined at high SAM concentration (100 μM) and varying concentrations of 70S ribosomes (Fig. 3). The apparent KM value determined under these conditions was 0.51 ± 0.06 μM and the kcat value was 4.95 ± 1.10 min−1 for native RlmH (Table 2). To determine the apparent KM and kcat for SAM, ΔrlmH 70S ribosomes were present at 2 μM and the SAM concentration was varied (Fig. 3). For native RlmH, the KM value determined for SAM was 27 ± 3 μM and the kcat value was 6.41 ± 1.33 min−1 (Table 2). kcat values determined for both RlmH substrate 70S ribosomes and the cofactor SAM correlated relatively well (varying <20%), indicating that the kinetic constants were obtained under appropriate conditions. It should be noted that the N-terminally His-tagged RlmH protein is significantly less efficient compared with the native RlmH protein (Table 2).
FIGURE 3.
Characterization of the RlmH-directed methylation. Determination of catalytic parameters of native RlmH for substrate 70S ribosomes (top panel) and cofactor S-adenosyl-L-methionine (SAM) (bottom panel). The initial rate of methylation was measured in the presence of 84 nM purified RlmH, 100 μM [3H]-SAM and increasing concentrations of ΔrlmH 70S ribosomes, or increasing concentrations of [3H]-SAM and 2 μM ΔrlmH 70S ribosomes, respectively. The amount of [3H]-methyl groups incorporated was monitored by acid precipitation and scintillation counting. One experiment representative of three repeats is shown.
TABLE 2.
Kinetic parameters of RlmH and RluD
Enzymatic properties of RluD
RluD isomerizes all three uridines (U1911, U1915, and U1917) in the stem–loop 69 of 23S rRNA to pseudouridines. The substrate of RluD protein is the 50S subunit (Leppik et al. 2007; Vaidyanathan et al. 2007). The observation that the RlmH protein preferentially methylates pseudouridines gives us the opportunity to use a coupled enzyme assay to determine the kinetic parameters of U1915 pseudouridylation by RluD. Rate equations for consecutive irreversible reactions were used (McClure 1969; Storer and Cornish-Bowden 1974).
In the current work, RlmH is the reporter enzyme in a two-step coupled enzyme assay with RluD. In the first step, 50S subunits of the ΔrlmH/ΔrluD strain were incubated with purified RluD protein for 20–60 sec. In the second step, the preincubated mix consisting of 30S subunits, purified RlmH protein (at saturating concentration), and [H3]-SAM was added to the first mix and incubated for 15 sec. Incorporation of [H3]-methyl groups into ribosomes was determined by TCA precipitation. The two-step reaction scheme used:
RlmH-directed methylation of ribosomes without the prior incubation with RluD protein was marginal (<3% of the RluD reaction, data not shown). Isomerization of uridine 1915 is the rate-limiting step and methylation of Ψ1915 is the coupling reaction. It must be noted that association of ribosomal subunits is a fast reaction and occurs in less than a second (Antoun et al. 2004; Hennelly et al. 2005). Control experiments demonstrated that in the presence of 30S subunits, there is no detectable RluD-directed pseudouridylation during the short time of the second incubation step (data not shown). Preliminary experiments determined the appropriate concentrations of both RluD and RlmH proteins, as well as the suitable incubation times for the coupled enzyme assay (data not shown). The initial rates of RluD were obtained with a fixed concentration of purified RluD protein and varying concentrations of ΔrlmH/ΔrluD 50S subunits (Fig. 4). Pseudouridine methylation reaction by RlmH is assumed to follow first-order kinetics. The initial rate experiments of RluD revealed an apparent KM value of 0.98 ± 0.18 μM for 50S ribosomal subunits and a kcat value of 1.97 ± 0.46 min−1 (Table 2). This method allowed us to follow the RluD-directed formation of pseudouridine at position 1915.
FIGURE 4.
Characterization of the RluD-directed pseudouridine synthesis. Determination of catalytic parameters of RluD for substrate 50S subunits. The initial rate of pseudouridylation was measured in the presence of 190 nM purified RluD and increasing concentrations of ΔrlmH/ΔrluD 50S subunits. RlmH-dependent pseudouridine methylation was used as the reporter reaction for monitoring pseudouridine synthase activity of RluD in a coupled enzyme assay. The amount of [3H]-methyl groups incorporated was monitored by acid precipitation and scintillation counting. One experiment representative of three repeats is shown.
The questions whether the three pseudouridines in stem–loop 69 are synthesized in an ordered way and whether U1911 and U1917 are isomerized at similar rates as U1915 remain open. We tested the order of uridine isomerization by RluD using the chemical modification and primer extension method (Ofengand et al. 2001). 50S subunits of the ΔrlmH/ΔrluD strain were incubated with low concentrations of RluD protein for various time periods. RNA was isolated and treated with CMCT and alkali, followed by a reverse transcriptase directed primer extension. Reverse transcriptase stop signals corresponding to pseudouridines at positions 1911, 1915, and 1917, appear simultaneously over time (Fig. 5). This suggests that all three pseudouridines in the stem–loop 69 of 23S rRNA are made concurrently. Therefore, the kinetic parameters determined for the synthesis of pseudouridine at position 1915 can probably be extrapolated for synthesis of pseudouridines at positions 1911 and 1917 as well.
FIGURE 5.
Time course of isomerization of U1911, U1915, and 1917 by RluD. Primer extension analysis was used to detect the formation of pseudouridines in the stem–loop 69 of 23S rRNA. 23S rRNA was extracted from ΔrlmH/ΔrluD 50S subunits treated with purified RluD protein for various times at 25°C. 23S rRNA was analyzed for pseudouridines by CMCT/alkali treatment and reverse transcriptase directed primer extension. Plus lane indicates +CMCT/alkali-treated RNA and minus lane indicates –CMCT/alkali-treated RNA. Sequence of 23S rRNA around stem–loop 69 is shown at right. Bands corresponding to the 23S rRNA positions 1911, 1915, and 1917 are indicated. Note that CMCT induces a reverse transcriptase stop one nucleotide before the actual site of pseudouridylation.
DISCUSSION
The results obtained in this study demonstrate that the substrate nucleotide for RlmH-directed methylation is the 23S rRNA pseudouridine 1915 both in vitro and in vivo (Table 1). This finding is also supported by the observation that every genome that contains an rlmH ortholog also contains an ortholog of rluD (Purta et al. 2008). However, when RlmH protein is in excess and the substrate pseudouridine is not available, it is also able to methylate uridine at the same position both in vitro and in vivo (Fig. 1; Table 1). A small amount of m3U was detected in 23S rRNA from the rluD-deficient strain even without overexpression of RlmH (Table 1). RlmH protein is prone to mistake uridine for pseudouridine as their chemical structures are very similar and the methyl group acceptor N3 is in the same position in both bases. It remains to be determined what the exact mechanism is that allows RlmH to distinguish between pseudouridine and uridine. It likely involves recognizing N1 nitrogen that is part of the glycosidic bond in uridine but swaps places with C5 carbon upon pseudouridine formation. Nonetheless, there must be a reason why m3U1915 is usually not formed in cells, evidenced by the fact that none was detected in 23S rRNA isolated from 70S ribosomes of wild-type cells (Fig. 1; Table 1).
The substrate of RlmH is the 70S ribosome, illustrated by the fact that the methyl group incorporation activity of RlmH followed the same Mg2+ dependency pattern as ribosome subunit association/dissociation (Fig. 2). Other rRNA modification enzymes, shown to be able to modify 70S ribosomes, are RluD (Vaidyanathan et al. 2007), RsmG (GidB) (Okamoto et al. 2007), and RlmE (RrmJ and FtsJ) (Caldas et al. 2000a; Bügl et al. 2000). However, RlmH is the only known rRNA modification enzyme to modify exclusively 70S ribosomes, as RsmG also modifies 30S subunits, and both RlmE and RluD modify 50S subunits (Bügl et al. 2000; Caldas et al. 2000a; Okamoto et al. 2007). This is in very good agreement with the docking data of the RlmH crystal structure into the 70S ribosome showing extensive contacts of RlmH with both ribosome subunits (Purta et al. 2008). No other rRNA modification enzyme is known to make simultaneous contacts with both ribosomal subunits. Interestingly, according to the model of Purta et al. (2008), RlmH is positioned in such a way that it does not interfere with tRNA binding to the ribosomal P site, but it would hinder tRNA binding to the A site (Purta et al. 2008). This might indicate that the physiological substrate of RlmH is the ribosomal initiation complex with fMet-tRNA in the P site of ribosome. RlmH-dependent modification could provide proof that the last steps of ribosome biogenesis overlap with the first steps of translation (Ero et al. 2008; Purta et al. 2008).
Analysis of the pseudouridylation pattern of 23S rRNA from various ribosome assembly precursor particles indicated that the three highly conserved pseudouridines (1911, 1915, and 1917) in the stem–loop 69 are formed during the late assembly steps (Leppik et al. 2007). This finding is supported by the in vitro studies showing that the purified RluD protein is far more efficient in modifying 50S subunits than free 23S rRNA (Vaidyanathan et al. 2007). While the exact mechanism of substrate nucleotide (U1911, U1915, and U1917) recognition and pseudouridine synthesis by RluD is not known, it was shown that the formation of Ψs at positions 1911 and 1917 is autonomous of each other and independent of Ψ1915 formation (Leppik et al. 2007). Here we show that all three Ψs in stem–loop 69 appear concurrently over time upon RluD treatment of the 50S subunit, meaning that they are synthesized at a similar rate and stochastically rather than in any specific order (Fig. 5). RluD is, according to current knowledge, the only pseudouridine synthase acting during late stages of ribosome assembly (Siibak and Remme 2010).
The observation that RluD acts on 50S subunits, whereas RlmH methylates 70S ribosomes explains why the RluD-dependent formation of pseudouridine at position 1915 of 23S rRNA occurs before RlmH-dependent methylation at the same position. This can also explain why no m3U1915 is formed in wild-type cells, whereas a low level of m3U1915 is detected in 70S ribosomes isolated from the rluD deletion strain. Taken together, RluD- and RlmH-directed modifications of rRNA are both late events in ribosome large subunit maturation and occur in a specific order.
The rate-limiting step of the ribosome large subunit biogenesis is the final maturation of 50S particles which takes 1–2 min at 37°C (Lindahl 1975) and 5 min at 25°C (Peil et al. 2008). This time is probably needed to make the late assembly-specific modifications of rRNA and r-proteins.
The kinetic constants of tRNA pseudouridine synthases can be determined by measuring the [3H] release from a [5-3H]-uridine–labeled transcript of a synthetic gene upon pseudouridine formation (Arluison et al. 1999; Ramamurthy et al. 1999; Hamilton et al. 2005). However, the complexity of the ribosome precludes the use of this method for determination of kinetic parameters of rRNA pseudouridine synthases. To date, no kinetic parameters for rRNA pseudouridine synthases are available. Luckily, another method can be employed to assess the efficiency of Ψ1915 formation. Since RlmH protein is efficient and specific to Ψ1915, it can be used as a reporter enzyme in a coupled enzyme assay with RluD. Here we show that the rate of RluD-directed pseudouridylation in vitro is sufficient for this modification to take place during the time frame of final 50S maturation (Table 2).
Methylation of pseudouridine 1915 in stem–loop 69 by RlmH, synthesized only on 70S ribosomes, is very likely the last modification incorporated into ribosomes. Synthesis of this modification probably coincides with the translation initiation. We show that the RlmH-directed methylation is a relatively fast process with a kcat value of ∼5–6 min−1 and apparent KM values of ∼0.5 and ∼27 μM for 70S ribosomes and cofactor SAM, respectively (Table 2). N-terminally His-tagged RlmH protein was significantly less efficient than the native protein (Table 2). We speculate that either one or both of the His-tags of RlmH dimer can hinder substrate recognition and/or RlmH binding into the 70S ribosome (which is a snug fit based on the docking data of Purta et al. [2008]) by sterically clashing with the 30S subunit.
Only a few rRNA methyltransferases have previously been kinetically characterized. These enzymes include RsmE (YggJ), RlmD (YgcA and RumA), and RlmE (FtsJ and RrmJ). The kcat value determined for RlmD, which synthesizes m5U1939 on 23S rRNA, is 3.6 min−1 (Agarwalla et al. 2002). This is in good agreement with the early assembly-specific modification enzymes being relatively fast, as the time frame when their substrate is available, is limited in cells. The kcat values for RsmE, which synthesizes m3U1498 on 30S subunits, and RlmE, responsible for the synthesis of Um2552 on 50S subunits, are 0.078 and 0.064 min−1, respectively (Hager et al. 2004; Basturea and Deutscher 2007). It is unlikely that RsmE and RlmE enzymes act this slow in vivo. More likely, their in vivo substrate is different from the substrate tested in vitro, or, alternatively, the purified enzymes have lost a cofactor that enhances their activity in cells.
RlmH is the first pseudouridine-specific methyltransferase to be enzymatically characterized. Furthermore, RlmH served as a useful tool for the first-time enzymatic characterization of a ribosomal RNA pseudouridine synthase, RluD. Understanding the kinetics of rRNA modification, an integral part of ribosome maturation, enables a glimpse into the complex process of how ribosomes are synthesized in cells.
MATERIALS AND METHODS
Strains and plasmids
rlmH gene knockout mutant (strain no. JW0631) of E. coli strain BW25113 was obtained from Nara Institute of Science and Technology (Keio collection) (Baba et al. 2006). The ΔrlmH (rlmH gene replaced with the kanamycin resistance cassette) phenotype was transferred to E. coli MG1655 strain (Blattner et al. 1997) as described in Ero et al. (2008). The ΔrluD (rluD gene replaced with the chloramphenicol resistance cassette) knockout strain was generated as described in Leppik et al. (2007). The ΔrlmH/ΔrluD double knockout strain (rlmH gene replaced with the kanamycin resistance cassette and rluD gene replaced with the chloramphenicol resistance cassette) was generated as described in Ero et al. (2008). E. coli strain MG1655 was used as wild-type control.
For generation of RlmH protein expression vector, the rlmH gene was amplified by PCR from genomic DNA of E. coli MG1655 strain and cloned into pBAD/Myc-His A expression vector (Invitrogen Life Technologies) between NcoI and BglII restriction sites. Constructed plasmid (pBAD-rlmH) was verified by sequencing. Plasmid pQE60-rluD–expressing RluD protein with C-terminal His-tag was constructed as described in Ero et al. (2008). Standard techniques were employed for DNA manipulations, plasmid DNA isolation, and E. coli transformation (Sambrook et al. 1989).
Preparation of ribosomes
70S ribosomes were prepared as described in Ero et al. (2008). For preparation of 50S and 30S subunits, 70S ribosomes were suspended in buffer TKNM (1 mM MgCl2, 60 mM NH4Cl, 60 mM KCl, 20 mM Tris/HCl at pH 8.0, and 6 mM β-mercaptoethanol) and layered onto a 10%–25% (w/w) sucrose gradient in TKNM buffer followed by centrifugation at 19,500 rpm for 18 h in a Beckman SW-28 rotor (ω2t = 2.8 × 1011). Sucrose gradients were analyzed with continuous monitoring of absorbance at 254 nm. Ribosomal subunits from sucrose gradient fractions were concentrated with Amicon Ultra 100k filters (Millipore); buffer was exchanged to TKNM-10 (10 mM MgCl2, 60 mM NH4Cl, 60 mM KCl, 20 mM Tris/HCl at pH 8.0, and 6 mM β-mercaptoethanol) and stored at −80°C.
rRNA was purified from ribosome subunits by extraction with phenol and chloroform followed by ethanol precipitation. rRNA was dissolved in water and stored at −80°C.
HPLC analysis
Preparation of rRNA fragment
A fragment of 23S rRNA corresponding to nucleotides (nt) 1777–1922 was excised by RNase H using oligonucleotides complementary to nt 1760–1777 (primer C4: 5′-CAGTTGCAGCCAGCTGG-3′) and 1922–1942 (primer U1 mini: 5′-TTTCGCTACCTTAGGACCG-3′) of E. coli 23S rRNA essentially as described by Douthwaite and Kirpekar (2007). In the denaturation step, 300 pmol of rRNA was mixed with a 10-fold molar excess of both oligodeoxynucleotides and heated for 3 min at 100°C in 270 μL of 1 mM EDTA. Denatured RNA probes were placed on ice and 30 μL of 10× buffer (600 mM HEPES at pH 7.0 and 1.25 M KCl) were added. In the hybridization step, the reactions were heated for 1 min at 90°C and cooled in a water bath over the period of 2 h to 45°C. The resulting RNA-DNA hybrids were digested with 10 units of RNase H (Fermentas Life Sciences) in the presence of 8 mM MgCl2 and 1 mM DTT for 30 min at 37°C to cleave RNA in the RNA/DNA heteroduplexes. Nuclease-treated RNA was phenol-extracted and recovered by ethanol precipitation, and the 145-nt-long rRNA fragment was gel-purified using a 5% LE TOP agarose gel. The RNA fragment was excised and extracted from the gel by overnight incubation in 450 μL of 2M NH4OAc (pH 6.0) at 4°C. The RNA fragment was precipitated with two volumes of a 1:1 mixture of ethanol and isopropanol, collected by centrifugation, and dissolved in water.
High-performance liquid chromatography
For HPLC analysis, 100–200 pmol of gel-purified RNA fragment were digested with nuclease P1 (MP Biochemicals) and bacterial alkaline phosphatase (Fermentas Life Sciences) according to the method of Gehrke and Kuo (1989). Nucleoside composition was determined by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 μm) equipped with a precolumn (20 × 4.6 mm) at 30°C on a SHIMADZU Prominence HPLC system. RP-HPLC analysis was performed using the gradient conditions of Gehrke and Kuo (1989). Nucleoside absorbance profiles were recorded at 260 nm, and peak areas were integrated. The relative amounts of nucleosides were calculated in relation to the 100% value of wild-type ribosomes. In the case of m3U, which was not detected in the wild-type ribosomes, the theoretical amount (one uridine out of the 28 uridines in the RNA fragment is completely methylated, assuming that absorbance of m3U is equal to U) corresponds to 100%.
Purification of proteins
Recombinant N-terminal His6-tagged RlmH and C-terminal His6-tagged RluD proteins were prepared as described in Ero et al. (2008).
Nontagged RlmH protein was purified from E. coli TOP10 cells (Invitrogen Life Science), harboring the pBAD-rlmH plasmid and grown in 2×YT liquid media in the presence of 10 mM arabinose. Cells were harvested, suspended in buffer (20 mM Tris/HCl at pH 7.5, 200 mM NaCl, 10% glycerol, 0.5 mM PMFS, and 1 mM DTT), and passed through a French pressure cell at 18,000 psi. The supernatant was applied to a Q-Sepharose anion-exchange column (GE Healthcare) in order to bind the contaminants while the RlmH protein appeared in the flow-through. The fractions enriched with RlmH were pooled and concentrated with Amicon Ultra 3k filters (Millipore), and the buffer was exchanged to 50 mM sodium acetate (pH 5.8), 125 mM KCl, 5% glycerol, 1 mM DTT, and 0.5 mM PMFS. Protein mixture was applied to an SP-Sepharose cation-exchange column (GE Healthcare). Proteins were eluted from the column with a linear gradient of 0–0.5 M NaCl over 20 column volumes. Fractions containing purified RlmH protein were pooled and concentrated with Amicon Ultra 3k filters (Millipore); buffer was exchanged to 50 mM Tris/HCl (pH 7.5), 200 mM KCl, 10% glycerol, and 1 mM DTT. Purity of RlmH protein was assessed by SDS-PAGE and mass-spectrometry. Protein concentration was determined by the Bradford method (Bradford 1976). Protein was flash-frozen and stored at −80°C.
In vitro methylation assays
In vitro modification of ribosomes for HPLC analysis was performed as follows. Reaction mixture (75 μL) containing 240 pmol of 70S ribosomes or 50S subunits (purified from ΔrlmH, ΔrluD, or ΔrlmH/ΔrluD strains), 5 μg of purified nontagged RlmH protein, 100 μM S-adenosyl-L-methionine in methylation buffer (50 mM Tris/HCl at pH 8.0, 100 mM NH4Cl, 20 mM MgCl2, and 1 mM DTT) was incubated for 1 h at 37°C. rRNA was purified by extraction with phenol and chloroform followed by ethanol precipitation. rRNA was dissolved in water and used for 23S rRNA fragment generation and HPLC analysis as previously described.
RlmH activity at various magnesium concentrations was tested as follows. Reaction mixture (50 μL) contained 24 pmol of ΔrlmH 70S ribosomes or 24 pmol ΔrlmH 50S subunits and 40 pmol wild-type 30S subunits, 2 μg of purified nontagged RlmH protein, 100 μM [3H]-S-adenosyl-L-methionine (Amersham Pharmacia Biotech), in methylation buffer with MgCl2 concentrations 1, 2.5, 6, 7.5, and 10 mM. Reaction was started with addition of ribosomes and carried out for 10 min at 37°C. Reaction products were precipitated ice-cold 5% TCA and collected on glass fiber filters (Whatman). Radioactivity was determined by scintillation counting using Optiphase HiSafe III scintillator (PerkinElmer).
For determination of kinetic parameters for RlmH, initial reaction rates of RlmH were measured by varying one substrate concentration at a constant saturating concentration of the other substrate. 0.96 μM His-tagged RlmH, or 0.084 μM native RlmH proteins in methylation buffer (final volume 25 μL) was used for kinetic assays. To determine the apparent KM for substrate 70S ribosomes, 100 μM [3H]-SAM and RlmH were preincubated for 10 min at 37°C. [3H]-methyl group incorporation was measured after addition of 0.5–4.3 μM ΔrlmH 70S ribosomes and incubation for 20/40/60 sec. at 37°C. For determination of the apparent KM for cofactor SAM, 10–75 μM [3H]-SAM and RlmH were preincubated for 10 min at 37°C. [3H]-methyl group incorporation was measured after addition of 2 μM ΔrlmH 70S ribosomes and incubation for 15/30/45 sec. at 37°C. Reaction products were precipitated with TCA and radioactivity was determined as described before. The slopes of methyl group incorporation versus time plots were calculated, and the initial rates obtained were plotted against the respective substrate concentrations. From fitting the Michaelis-Menten equation to the data, apparent KM and kcat values were determined.
In vitro pseudouridylation assays
In vitro pseudouridylation was tested as follows. 50S subunits (1.44 μM) dissociated from ΔrlmH/ΔrluD 70S ribosomes were incubated with N-terminally His-tagged RluD protein (0.29 μM) in TKNM buffer (60 mM KCl, 60 mM NH4Cl, 50 mM Tris/HCl [pH 8], 6 mM MgSO4, 6 mM β-mercaptoethanol) for 60/80/100/120/140 sec at 25°C. Reaction was stopped by addition of 1 mL of PN solution (Qiagen). rRNA extraction, CMCT-alkali treatment, and primer extension were done as described in Leppik et al. (2007).
Coupled enzyme assay was employed to determine the kinetic parameters for RluD. RlmH-dependent pseudouridine-specific methylation was used as the reporter reaction for measuring the pseudouridine synthase activity of RluD. To determine the apparent KM for 50S subunits, 0.6–10 μM ΔrlmH/ΔrluD 50S subunit mixtures and methylation mixtures (2–30 μM wild-type 30S subunits, 200 μM [H3]-SAM [1000 DPM/pmol], and 2 μM nontagged RlmH) were preincubated in buffer (50 mM Tris/HCl at pH 8.0, 100 mM NH4Cl, 10 mM MgCl2, and 1 mM DTT) for 10 min at 37°C. Pseudouridylation reaction was started by addition of purified RluD protein (final concentration 0.19 μM,) to ΔrlmH/ΔrluD 50S subunit mixture (final reaction volume 25 μL) and after 20/40/60 sec of incubation at 37°C, 25 μL of methylation mixture was added. Reaction mixture was incubated for 15 sec at 37°C. Reaction products were precipitated with TCA and radioactivity was determined by scintillation counting. Apparent KM and kcat value for RluD were calculated in similar fashion as described before and based on the assumption that RlmH-directed methylation reaction follows the first-order kinetics.
ACKNOWLEDGMENTS
We are grateful to Kalle Kipper for valuable advice and criticism, Triinu Siibak and Lauri Peil for help, and Joachim Gerhold (all University of Tartu) for linguistic proofreading. This work was supported by Estonian Science Foundation Grants ETF 7884 (A.L.) and 7509 (J.R.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2234310.
REFERENCES
- Agarwalla S, Kealey JT, Santi DV, Stroud RM 2002. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J Biol Chem 277: 8835–8840 [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J 2004. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci 101: 8900–8905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Refaii A, Alix JH 2009. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol Microbiol 71: 748–762 [DOI] [PubMed] [Google Scholar]
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF 2006. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23: 865–874 [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Tenson T, Ehrenberg MM 2004. Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol Proced Online 6: 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arluison V, Buckle M, Grosjean H 1999. Pseudouridine synthetase Pus1 of Saccharomyces cerevisiae: Kinetic characterisation, tRNA structural requirement and real-time analysis of its complex with tRNA. J Mol Biol 289: 491–502 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2: doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturea GN, Deutscher MP 2007. Substrate specificity and properties of the Escherichia coli 16S rRNA methyltransferase, RsmE. RNA 13: 1969–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Burkhardt N, Nierhaus KH 2002. Formation of 70S ribosomes: Large activation energy is required for the adaptation of exclusively the small ribosomal subunit. Biophys Chem 96: 153–161 [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 5: 1453–1474 [DOI] [PubMed] [Google Scholar]
- Bradford MM 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP 1996. Modulation of chemical composition and other parameters of the cell growth rate. In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed (ed. Neidhardt FC et al. ), pp. 1553–1568 American Society for Microbiology, Washington, DC [Google Scholar]
- Bügl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U 2000. RNA methylation under heat shock control. Mol Cell 6: 349–360 [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G 2000a. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem 275: 16414–16419 [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Richarme G 2000b. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem Biophys Res Commun 271: 714–718 [DOI] [PubMed] [Google Scholar]
- Douthwaite S, Kirpekar F 2007. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol 425: 1–20 [DOI] [PubMed] [Google Scholar]
- Ejby M, Sorensen MA, Pedersen S 2007. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc Natl Acad Sci 104: 19410–19415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ero R, Peil L, Liiv A, Remme J 2008. Identification of pseudouridine methyltransferase in Escherichia coli. RNA 14:2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P 2000. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell 100: 537–549 [DOI] [PubMed] [Google Scholar]
- Gegenheimer P, Watson N, Apirion D 1977. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem 252: 3064–3073 [PubMed] [Google Scholar]
- Gehrke CW, Kuo KC 1989. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr 471: 3–36 [DOI] [PubMed] [Google Scholar]
- Gutgsell NS, Deutscher MP, Ofengand J 2005. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 11: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager J, Staker BL, Jakob U 2004. Substrate binding analysis of the 23S rRNA methyltransferase RrmJ. J Bacteriol 186: 6634–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CS, Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG 2005. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. Arch Biochem Biophys 433: 322–334 [DOI] [PubMed] [Google Scholar]
- Hennelly SP, Antoun A, Ehrenberg M, Gualerzi CO, Knight W, Lodmell JS, Hill WE 2005. A time-resolved investigation of ribosomal subunit association. J Mol Biol 346: 1243–1258 [DOI] [PubMed] [Google Scholar]
- Hirabayashi N, Sato NS, Suzuki T 2006. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J Biol Chem 281: 17203–17211 [DOI] [PubMed] [Google Scholar]
- Holmes KL, Culver GM 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol 11: 179–186 [DOI] [PubMed] [Google Scholar]
- Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene PJ, Santi DV 1998. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry 37: 15951–15957 [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipper K, Hetenyi C, Sild S, Remme J, Liiv A 2009. Ribosomal intersubunit bridge B2a is involved in factor-dependent translation initiation and translational processivity. J Mol Biol 385: 405–422 [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Bruenger E, Hashizume T, Peltier JM, Ofengand J, McCloskey JA 1996. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res 24: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppik M, Peil L, Kipper K, Liiv A, Remme J 2007. Substrate specificity of the pseudouridine synthase RluD in Escherichia coli. FEBS J 274: 5759–5766 [DOI] [PubMed] [Google Scholar]
- Lewicki BT, Margus T, Remme J, Nierhaus KH 1993. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol 231: 581–593 [DOI] [PubMed] [Google Scholar]
- Lindahl L 1975. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol 92: 15–37 [DOI] [PubMed] [Google Scholar]
- McClure WR 1969. A kinetic analysis of coupled enzyme assays. Biochemistry 8: 2782–2786 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Osswald M, Brimacombe R 1992. Identification of intermolecular RNA cross-links at the subunit interface of the Escherichia coli ribosome. Biochemistry 31: 3004–3011 [DOI] [PubMed] [Google Scholar]
- Nierhaus KH 1991. The assembly of prokaryotic ribosomes. Biochimie 73: 739–755 [DOI] [PubMed] [Google Scholar]
- O'Connor M, Dahlberg AE 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol 254: 838–847 [DOI] [PubMed] [Google Scholar]
- Ofengand J 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett 514: 17–25 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M 2004. Modified nucleosides in Escherichia coli ribosomal RNA. In EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology (ed. Curtiss R), Chap. 4.6.1 ASM Press, Washington, DC: [DOI] [PubMed] [Google Scholar]
- Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, Peil L, Kaya Y 2001. Pseudouridines and pseudouridine synthases of the ribosome. Cold Spring Harb Symp Quant Biol 66: 147–159 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol 63: 1096–1106 [DOI] [PubMed] [Google Scholar]
- Peil L, Virumae K, Remme J 2008. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J 275: 3772–3782 [DOI] [PubMed] [Google Scholar]
- Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Douthwaite S 2008. YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA 14: 2234–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V, Swann SL, Spedaliere CJ, Mueller EG 1999. Role of cysteine residues in pseudouridine synthases of different families. Biochemistry 38: 13106–13111 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Conrad J, Hall BG, Ofengand J 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4: 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Siibak T, Remme J 2010. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA doi: 10.1261/rna.2160010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D 1989. Processing pathway of Escherichia coli 16S precursor rRNA. Nucleic Acids Res 17: 1649–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer AC, Cornish-Bowden A 1974. The kinetics of coupled enzyme reactions. Applications to the assay of glucokinase, with glucose 6-phosphate dehydrogenase as coupling enzyme. Biochem J 141: 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan PP, Deutscher MP, Malhotra A 2007. RluD, a highly conserved pseudouridine synthase, modifies 50S subunits more specifically and efficiently than free 23S rRNA. RNA 13: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]