Abstract
Inter- and intragenic noncoding transcription is widespread in eukaryotic genomes; however, the purpose of these types of transcription is still poorly understood. Here, we show that intragenic sense-oriented transcription within the budding yeast ASP3 coding region regulates a constitutively and immediately accessible promoter for the transcription of full-length ASP3. Expression of this short intragenic transcript is independent of GATA transcription factors, which are essential for the activation of full-length ASP3, and independent of RNA polymerase II (RNAPII). Furthermore, we found that an intragenic control element is required for the expression of this noncoding RNA (ncRNA). Continuous expression of the short ncRNA maintains a high level of trimethylation of histone H3 at lysine 4 (H3K4me3) at the ASP3 promoter and makes this region more accessible for RNAPII to transcribe the full-length ASP3. Our results show for the first time that intragenic noncoding transcription promotes gene expression.
Keywords: noncoding RNA, intragenic transcript, chromatin remodeling, nitrogen starvation, ASP3
INTRODUCTION
Noncoding RNAs (ncRNAs) are widespread transcripts occurring from yeast to human, but their functions remain unclear (Kapranov et al. 2002; Rinn et al. 2003; Yelin et al. 2003; Cheng et al. 2005; Davis and Ares 2006; Neil et al. 2009). Recent findings in human, mouse, and Saccharomyces cerevisiae demonstrate that these ncRNAs play roles in the regulation of gene expression (Azzalin et al. 2007; Houseley et al. 2007; Rinn et al. 2007; Luke et al. 2008; Nagano et al. 2008; Pandey et al. 2008; Schoeftner and Blasco 2008). ncRNAs exhibit their functions through either the transcribed RNAs (Camblong et al. 2007; Martianov et al. 2007; Berretta et al. 2008; Nishizawa et al. 2008; Wang et al. 2008) or the action of transcription (Martens et al. 2004; Bird et al. 2006; Hongay et al. 2006; Uhler et al. 2007; Houseley et al. 2008; Hartzog and Martens 2009). Although ncRNAs are predominantly transcribed in the antisense orientation to repress sense transcripts through RNA-mediated transcriptional gene silencing (Hongay et al. 2006; Camblong et al. 2007, 2009; Berretta et al. 2008; Houseley et al. 2008), other types of ncRNA-mediated gene regulation have been identified. ncRNAs can be transcribed upstream of a promoter in the sense orientation to repress the downstream gene expression by a transcriptional interference mechanism (Martens et al. 2004; Bird et al. 2006). Additionally, antisense ncRNAs can promote gene expression through a chromatin-remodeling mechanism (Uhler et al. 2007). However, different regulations and functions have mainly been revealed for ncRNAs that are transcribed from intergenic regions. Little is known about the functions of the numerous ncRNAs transcribed from intragenic regions.
Nitrogen regulation (nitrogen catabolite repression [NCR]) in S. cerevisiae has been shown to be controlled by GATA family transcription factors. Upon depletion of rich nitrogen sources (e.g., glutamine and ammonia), the NCR genes are activated by the GATA factors Gat1p and Gln3p (Mitchell and Magasanik 1984; Courchesne and Magasanik 1988; Stanbrough et al. 1995). In the presence of rich nitrogen sources, both GATA factors are phosphorylated by TOR kinase and are restricted to the cytoplasm through the interaction with Ure2p (Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999).
Asparaginase II is a periplasmic enzyme in yeast, hydrolyzing both D- and L-asparagine to aspartate and ammonium cation (Dunlop et al. 1978). ASP3, an NCR-regulated gene that encodes asparaginase II, comprises four identical copies located near a ribosomal DNA cluster region on chromosome XII and is activated in a nitrogen-limited environment (Dunlop et al. 1980; Kim et al. 1988). The GATA activators Gat1p and Gln3p are required for up-regulation of ASP3 (Oliveira et al. 1999, 2003; Scherens et al. 2006). A large-scale transcriptome study showed that yeast cells alter the structure of the 5′ untranslated region of mRNA to enhance gene expression in response to environmental changes (Law et al. 2005). This study also detected a short nc transcript within the open reading frame (ORF) of ASP3. So far, however, no investigation regarding the regulation and function of this ncRNA has been undertaken. Here we study the phenotypes and functions of this ncRNA. We show that the intragenic transcript of ASP3 is expressed in the sense orientation both under nitrogen-rich conditions and under nitrogen starvation. Expression of this transcript is not mediated by GATA transcription factors and independent of RNA polymerase II (RNAPII). This intragenic transcription facilitates transcriptional initiation of RNAPII at the ASP3 promoter, thus enhancing the expression of full-length ASP3.
RESULTS
The intragenic transcript of ASP3 is expressed under nitrogen starvation and under nitrogen-rich conditions
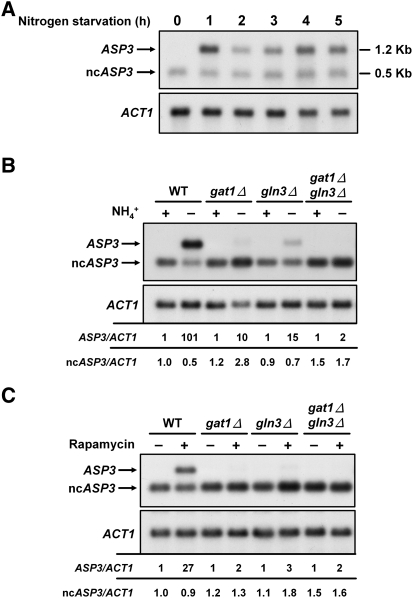
In a previous study, ASP3 has been shown to produce two transcripts with different 5′ termini: The short form of ASP3 was only detected under regular growth conditions, and the full-length transcript was only expressed when nitrogen was limited (Law et al. 2005). We analyzed the expression of both the short transcript and full-length transcript of ASP3 under different nitrogen conditions by Northern blotting. The short form of ASP3, termed ncASP3, was detected in the presence of ammonium-containing medium and continuously expressed under nitrogen starvation for 5 h (Fig. 1A). We should note that the faster migrating band assigned to ncASP3 was not detected in the asp3Δ strain (see below), suggesting that the signal is indeed a variant transcript of ASP3 and not due to a cross-hybridizing species. In contrast to the short transcript, full-length ASP3 was only induced in the nitrogen-limiting medium, as previously reported (Kim et al. 1988). Its activation was strong within 1 h after nitrogen starvation and fluctuated somewhat afterward. Compared to the expression status of full-length ASP3, the level of the short transcript remained essentially unchanged after nitrogen starvation for the indicated time points (Fig. 1A). These data indicate that the regulation of the short transcript is different from that of full-length ASP3.
FIGURE 1.
Intragenic transcription from the ASP3 ORF is detected in both ammonium-containing medium and under nitrogen starvation. (A) The wild-type strain was pre-grown in ammonium-containing medium. Cells were then washed with phosphate buffer and transferred to a 3% glucose solution for nitrogen starvation for 2 h. Northern analysis of the two different transcripts of ASP3 was conducted using an ASP3 probe. The blot was subsequently probed against ACT1 as a loading control. The sizes of transcripts are labeled at right. (B,C) Intragenic transcription is independent of GAT1 and GLN3. (B) The wild-type, gat1Δ, gln3Δ, and gat1Δ gln3Δ mutant strains were treated as described in A, and then analyzed by Northern blotting. (C) Strains described in B were pre-grown in ammonium-containing medium, treated with rapamycin, and then analyzed by Northern blotting. In B and C, ASP3 and ncASP3 signals were normalized to ACT1 signals. Values are displayed below the blots.
Transcription of full-length but not short-form ASP3 is dependent on GAT1 and GLN3
Nitrogen-dependent ASP3 expression was suggested to be mediated by two GATA-type transcription factors, Gat1p and Gln3p, which are restricted to the cytoplasm by Ure2p-mediated recruitment in the presence of rich nitrogen sources (Beck and Hall 1999; Oliveira et al. 2003). With the addition of rapamycin, which inhibits TOR kinase activity and mimics nitrogen starvation conditions, the NCR-related genes are activated by both Gat1p and Gln3p (Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Bertram et al. 2000). To test whether the short form of ASP3 is also regulated by the nitrogen regulatory mechanism, the expression of ASP3 in wild-type, gat1Δ, gln3Δ, and gat1Δ gln3Δ strains was determined by Northern analysis in ammonium-containing medium and under nitrogen starvation conditions. As shown in Figure 1B, expression of full-length ASP3 was significantly decreased in gat1Δ and gln3Δ strains and almost abolished in the gat1Δ gln3Δ double deletion strain. These data agree with previous studies reporting the requirement of Gat1p and Gln3p for full-length ASP3 gene activation after nitrogen starvation (Oliveira et al. 2003; Scherens et al. 2006). Analogous regulatory mechanisms for full-length ASP3 expression were observed in cells treated with rapamycin, which induces a condition mimicking nitrogen starvation (Fig. 1C). In contrast to transcription of full-length ASP3, expression of the short transcript was not significantly altered in GATA-deleted strains (Fig. 1B,C). These data reveal that the regulation of the short-form ASP3 is different from that of the full-length transcript and is independent of nitrogen availability.
Intragenic transcription of ncASP3 is RNAPII-independent
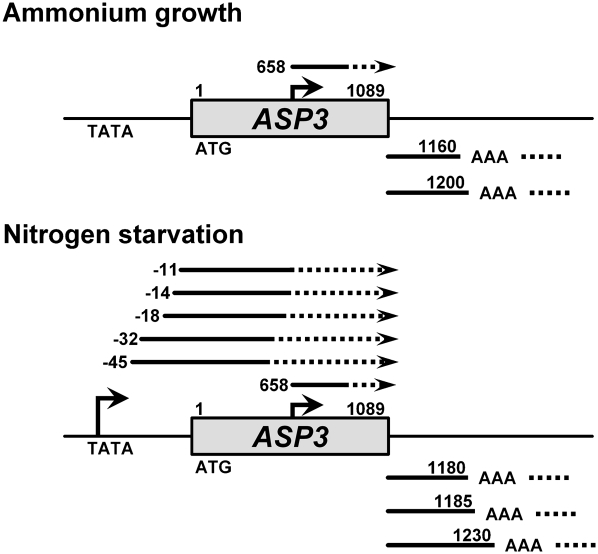
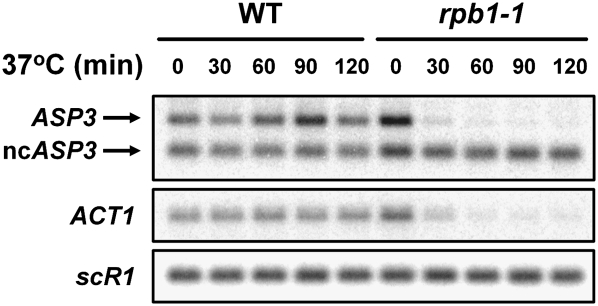
The 5′ terminus of the short transcript was identified to start 657 nt downstream relative to the initiation codon of ASP3 under nitrogen-rich conditions (Law et al. 2005). To test whether the continuously expressed short transcript that we observed under nitrogen starvation (Fig. 1) is transcribed from the same position, we performed 5′ and 3′ RACE followed by sequencing analysis (Fig. 2). Under both nitrogen-rich and nitrogen-depleted conditions, the short transcripts were internally initiated at position +658, while the full-length ASP3 had several transcription start sites in the ASP3 promoter region, with a dominant position at −18 nt (data not shown). DNA sequencing data from 3′ RACE showed that the transcripts obtained under both conditions are polyadenylated to similar extents (Fig. 2). The RACE data furthermore indicated that the transcription start site but not the 3′-termination sequence contributes to the length of the short transcript. To examine whether the short transcript of ASP3 is synthesized by RNAPII, RNA from a strain with a temperature-sensitive mutant (rpb1-1) of RPO21 (Nonet et al. 1987), which encodes the largest subunit of RNAPII, was subjected to Northern analysis (Fig. 3). When cells were shifted to non-permissive temperature, the full-length ASP3 mRNA was dramatically decreased, resembling the ACT1 mRNA control. However, unlike the decline of the ASP3 mRNA, the level of ncASP3 transcript was not affected when RNAPII was inactivated. The pattern of ncASP3 was similar to that of an RNAPIII-dependent transcript, scR1, which was also not affected in the rpb1-1 mutant. These data demonstrate that transcription of the short form of ASP3 is RNAPII-independent. Given that there is no Kozak consensus sequence (Kozak 1987) in the 5′ region and many stop codons are located in the vicinity downstream from the first ATG, the short transcript should be an RNAPII-independent ncRNA from the ASP3 coding region.
FIGURE 2.
5′ and 3′ RACE analyses of transcripts obtained in ammonium-containing medium or under nitrogen starvation. A schematic diagram shows the results of 5′ (upper arrows) and 3′ (lower bars) RACE from sequencing analyses. The intragenic transcript is initiated at position 658 nt of the ASP3 coding sequence and polyadenylated under both conditions.
FIGURE 3.
RNAPII-independent transcription of the short form of ASP3. Northern analysis of total RNA was performed under RNAPII inactivation. The rpb1Δ single copy–ASP3 cells containing either wild-type or rpb1-1 mutant plasmid were pre-grown at 23°C, and then shifted to 37°C for the indicated times. ACT1 is an RNAPII-dependent transcript, and scR1 is an RNAPIII-dependent transcript.
An intragenic control element regulates ncASP3 expression
Our results showed that the short transcript of ASP3 is internally initiated in both ammonium-containing medium and under nitrogen starvation. To determine the promoter of ncASP3, we measured the β-galactosidase activity in the strains containing either empty plasmid or containing the plasmid with the ASP3(306-657) coding region upstream of the lacZ reporter gene. For the latter strain, a fivefold increase in β-galactosidase activity was detected; this increase was not dependent on nitrogen availability (Fig. 4A). These data are consistent with the finding that expression of the short transcript is unaffected under conditions containing or lacking nitrogen nutrients (Fig. 1A). Next, to gain further insight into the regulatory mechanism underlying the formation of the short transcript, the ASP3 locus containing the ASP3 coding sequence together with 755-nt upstream and 500-nt downstream flanking sequences was cloned into a CEN plasmid and expressed under the control of its own promoter. This plasmid was transformed into the asp3 deletion strain in which all four copies of ASP3 have been removed. Northern analysis of the transformants showed that both ASP3 and the ncRNA signals disappeared in the asp3Δ strain, supporting the conclusion that the faster-migrating band was not a cross-hybridizing signal (Fig. 4B). The expression levels of full-length and short-form ASP3 were slightly lower than those in the wild-type strain; however, the ratio of the two transcripts was preserved (Fig. 4B), indicating that transcription from this plasmid was regulated in the same way as that in the wild-type strain. To further define the control element for ncASP3, we tested two mutant plasmids: one containing a deletion of the putative TATA-box (tataΔ, deletion from −112 to −91 nt relative to the ASP3 coding region), and the other containing a deletion of the potential intragenic control element (ICE) within the ASP3 gene (iceΔ, deletion from 402 to 609 nt). Northern analysis showed that expression of full-length ASP3 was considerably decreased in the tataΔ construct, while the expression of the short transcript remained unchanged (Fig. 4B), indicating that the regulation of the short transcript is different from that of full-length ASP3. However, transcription of ncASP3 was completely abolished in cells containing the iceΔ construct. Together, these data strongly suggest that there is a control element for ncASP3 located within the ASP3 coding region and that the regulation of ICE is independent of nitrogen availability.
FIGURE 4.
Expression of the short transcript is regulated by an intragenic control element but not the TATA element of ASP3. (A) Reporter assays of the wild-type strain with an empty vector (pLG669Z-UASΔ) or a plasmid containing the ASP3(306-657) coding region upstream of the LacZ reporter gene (pLG669Z-PncASP3) were conducted under ammonium-containing or nitrogen starvation conditions. The β-galactosidase activities shown are means of three independent measurements. (B) Total RNAs were extracted from wild-type and asp3Δ mutant strains containing an empty vector (Vec), wild-type (ASP3), TATA box-deleted (tataΔ), or intragenic control element-deleted (iceΔ) expressing plasmid (pRS316 backbone) in ammonium-containing medium and under nitrogen starvation. The ASP3 and ncASP3 signals determined by Northern blotting were normalized to ACT1 and are displayed below the blots.
Intragenic transcription mediates ASP3 expression in cis
Our data show that transcription of the short form of ASP3 requires the ICE within the ASP3 coding sequence. To our surprise, without the expression of the short transcript, the transcription of full-length ASP3 was decreased to about two-thirds of the wild-type level in cells containing the iceΔ construct (Fig. 4B). These data imply that either intragenic transcription or the control element within the coding region can promote full-length gene expression. Gene activation in cis mediated through a histone-remodeling mechanism by intergenic nc transcription was discovered previously (Uhler et al. 2007). To test whether such activation also exists intragenically, we engineered a strain containing a single chromosomal copy of ASP3 and deleted the intragenic control element within ASP3 coding region (iceΔ). In this strain, the short transcript was no longer detected, and the level of the full-length ASP3 was decreased by one-third under nitrogen starvation (Fig. 5A), similar to the result observed in the asp3Δ strain containing the plasmid expressing asp3-iceΔ (Fig. 4B). These data further confirmed the essential role of the ICE in intragenic transcription.
FIGURE 5.
Intragenic transcription promotes full-length ASP3 gene expression in cis but not in trans. (A) Northern analysis of the single copy–ASP3 wild-type and iceΔ strains in ammonium-containing medium and under nitrogen starvation. The ASP3 and ncASP3 signals were normalized to that of ACT1 and are shown below as the average of three independent experiments. (B) Total RNAs were extracted from the wild-type strain containing the empty vector or ncASP3-expressing plasmid under the control of the ADH1 promoter in ammonium-containing medium and under nitrogen starvation. Following Northern blotting, the ASP3 and ncASP3 signals were normalized to the signal of ACT1. Values are displayed below the blots.
To address the question whether the short transcript can also act in trans to promote gene expression, we exogenously expressed the short form of ASP3 in a plasmid under the control of the ADH3 promoter. Northern analysis revealed no significant alteration of full-length ASP3 expression in cells overexpressing the short transcript compared to that in the strain with the empty vector (Fig. 5B). These results suggest that expression of full-length ASP3 is not promoted by the ncRNA itself, but by the action of transcription in cis.
Disruption of the intragenic transcription hampers chromatin remodeling and transcriptional initiation of ASP3
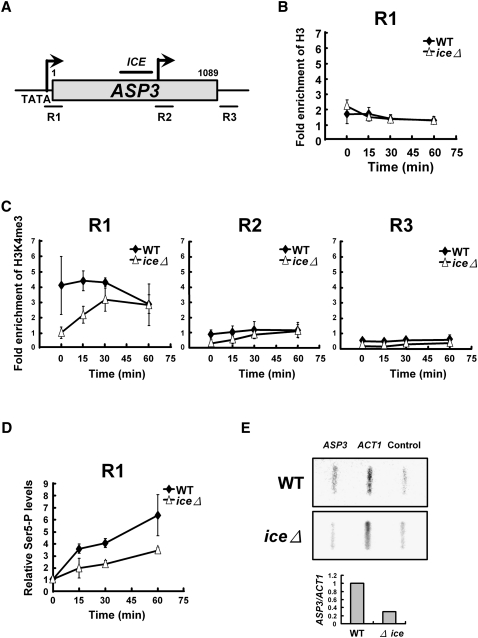
Nucleosomes are composed of histone proteins and DNA, and the transcriptional activity of RNA polymerase on nucleosomes is correlated with the patterns of post-translational modification on histones (Strahl and Allis 2000; Iizuka and Smith 2003). Trimethylation of histone H3 at lysine 4 (K4) is found in the 5′ region of transcribed genes and represents a hallmark of highly active genes (Santos-Rosa et al. 2002; Ng et al. 2003; Sims et al. 2003); this modification has been proposed to facilitate transcriptional elongation (Gerber and Shilatifard 2003). To determine the mechanism underlying the transcriptional modulation of full-length ASP3 expression, we performed chromatin immunoprecipitation (ChIP) and analyzed coprecipitated DNA by quantitative real-time PCR using primer pairs that amplify different regions of the ASP3 locus (Fig. 6A). Unlike the chromatin remodeling mechanism in which the nc transcript accelerated histone loss from the promoter region (Uhler et al. 2007), histone H3 eviction from the ASP3 promoter displayed no difference between wild-type and iceΔ strains after nitrogen starvation (Fig. 6B). To determine whether histone modification is altered in response to intragenic transcription, we used ChIP to determine the level of H3K4me3 at the ASP3 locus using an antibody specific for H3K4me3. Surprisingly, H3K4me3 was present in the wild-type strain at the 5′ end of ASP3 under nitrogen-rich conditions and maintained at about the same level after nitrogen starvation for 60 min (Fig. 6C, left graph). In the iceΔ strain, the level of H3K4me3 was not enriched in this region under nitrogen-rich conditions, yet it gradually increased to that of the wild type after nitrogen starvation for 60 min. The enrichment of H3K4me3 was only detected in the promoter region of ASP3 but not in the coding or the 3′ downstream region (Fig. 6C). These data show that this histone modification, which is specifically enriched at active sites of transcription, was already present at the ASP3 promoter in the wild-type strain before nitrogen starvation. Similar observations were reported in mammalian genes activated by the transcription factor NF-κB in response to inflammation. A subset of NF-κB target genes that contained high levels of H3K4me3 at promoter regions before stimulation was immediately transcribed upon activation (Saccani et al. 2001; Natoli 2009). Next, we used ChIP to investigate whether initiation of transcription by RNAPII at ASP3 is affected in the absence of the short transcript under nitrogen starvation. The carboxy-terminal domain (CTD) of RNAPII is phosphorylated at serine 5 upon initiation and phosphorylated at serine 2 during elongation (Komarnitsky et al. 2000). By using ChIP with specific antibodies against CTD serine 5P, we determined the efficiency of initiation of ASP3 transcription. Consistent with the predominant enrichment of H3K4me3 in the promoter region of the wild-type strain, the phosphorylation of serine 5 within the RNAPII CTD occurred much faster in the wild-type strain at the ASP3 promoter than in the iceΔ strain after nitrogen starvation (Fig. 6D).
FIGURE 6.
Disruption of the intragenic transcription hampers chromatin remodeling and transcriptional initiation of ASP3. (A) Schematic diagram of the ASP3 locus. The three regions used for PCR amplification in the ChIP assay are marked below. (B–D) ChIP analyses performed at the ASP3 locus. Samples were collected at the indicated time points after nitrogen starvation. Regions 1–3 used for the PCR amplification are indicated. (B) ChIP analyses of histone H3 occupancy. (C) ChIP analyses of H3K4me3 at the ASP3 locus. (D) Rate of RNAPII-mediated initiation of transcription at the TATA-box of ASP3 was assayed by ChIP, using antibody against Ser5-phosphorylated CTD. Data are shown as relative levels compared to signals measured at time 0. (E) Nuclear run-on analysis was conducted to detect the transcription rate of ASP3 in the wild-type and iceΔ strains after nitrogen starvation. Fragments of ACT1 and the multiple cloning sites of the pBSKS plasmid were used as positive and negative controls, respectively. The ASP3 signals were normalized to that of ACT1 and are shown below.
Elevated mRNA levels may be caused by an increased transcription rate and/or a reduced mRNA degradation rate. To examine whether the elevated level of full-length ASP3 mRNA in the wild-type strain resulted from an increase in the transcription rate, we performed nuclear run-on experiments. The nuclear run-on assay is often employed to specifically determine the activity of RNA polymerase complex on the DNA template. As shown in Figure 6E, while the signal of ASP3 was readily detected in the wild-type strain, the signal in the iceΔ strain was reduced and barely visible. These data suggest that continuous transcription of the ncRNA from the intragenic region of ASP3 facilitates transcriptional initiation of RNAPII at the promoter region of ASP3. As a result, the rate of transcription is increased, leading to an accumulation of the full-length ASP3 mRNA. In summary, our results demonstrate that transcription from the intragenic coding region makes the upstream promoter region more accessible to RNAPII to transcribe the full-length ASP3. Furthermore, our data suggest that the intragenic transcription-regulated ASP3 expression occurs only in cis but not in trans.
DISCUSSION
Environmental changes have been shown to instigate alterations in the structure of the 5′ untranslated region of mRNA in cells, leading to changes in gene expression (Law et al. 2005). Here we demonstrate that the internal initiation of ncASP3 transcription occurs in both nitrogen-rich medium and under nitrogen starvation. Expression of this short transcript is independent of GATA-type transcription factors, which are necessary for full expression of NCR-regulated genes, as we observed for the full-length ASP3. We also provide evidence showing that the short form of ASP3 is not synthesized by RNAPII. To gain insight into the roles of the short transcript, we identified an intragenic control element within the ASP3 coding region, which is necessary for the expression of this ncRNA. In the absence of intragenic transcription, expression of the full-length ASP3 and RNAPII recruitment to the promoter region were decreased; concomitantly, a lower H3K4me3 histone occupancy at the ASP3 promoter was detected. Taken together, these findings suggest that the transcription of short RNA in cis maintains a more accessible promoter for the RNAPII recruitment and facilitates the full-length ASP3 expression.
Production of short RNA due to internally initiated transcription induced by changes in the cells' environment was previously reported. A short transcript inside the ORF of the PRY3 gene was detected after treating yeast cells with mating pheromone (Bickel and Morris 2006). Multiple elements within the PRY3 locus were determined that affect expression of both full-length and short transcripts. The pheromone-induced transcription factor Ste12p was found to bind to upstream elements at the PRY3 promoter, impeding full-length transcription and simultaneously inducing initiation of the short transcript. The daughter-specific transcription factor Ace2p is required for expression of both full-length and short transcripts, and its binding sites are found in the PRY3 promoter region. In addition, a putative TATA-box was discovered within the coding region for the short transcript. However, for transcription of ASP3, apparently a different mechanism applies. Several potential binding sites for GATA-type transcription factors are present at the ASP3 promoter (Oliveira et al. 2003; Scherens et al. 2006). However, our results show that Gat1p and Gln3p are only required for the expression of full-length ASP3 but not of the short nc transcript. Despite analyzing a series of deletion mutants by Northern blotting, we did not find a control element within the ASP3 promoter regulating the expression of the short transcript (data not shown). Instead, we discovered that deletion of a 208-nt sequence within the ASP3 coding region (+402 to +609) abolishes ncASP3 transcription. The deleted DNA region, which we define as the intragenic control element (ICE), contains two separate TATA-like sequences. However, mutations of these TATA-like sequences did not change ncASP3 transcription compared with that of the wild-type sequence (data not shown).
Genome-wide studies established the presence of numerous unannotated transcripts within the yeast genome (Davis and Ares 2006; Neil et al. 2009). Several recent studies revealed that transcription of ncRNAs plays certain functional roles. The effects of these transcripts could be repressive (Martens et al. 2004; Bird et al. 2006; Hongay et al. 2006; Camblong et al. 2007; Houseley et al. 2008) or stimulatory (Uhler et al. 2007) for transcription. The gene PHO5 represents the first example of transcriptional activation by a ncRNA: In this case, the transcription of an noncoding antisense RNA initiated at the 3′ end of the gene activates full-length PHO5 expression (Uhler et al. 2007). This antisense transcription promotes the recruitment of RNAPII through accelerating histone loss from the promoter region. Different from the regulation of PHO5, we found that the ncRNA in ASP3 is transcribed within the coding region in the sense orientation. Furthermore, we provide evidence for a histone-remodeling mechanism in which full-length gene activation is mediated by H3K4 trimethylation. Such post-translational modification was not observed in the activation of PHO5 (Uhler et al. 2007).
The efficient expression of necessary genes in response to environmental changes is an essential requisite of life. In mammalian cells, the transcription factor NF-κB controls the induction of many inflammatory genes to cope with microbial infection. Two classes of NF-κB-regulated genes that possess different chromatin configurations and induction kinetics were identified. One class of genes contains high levels of H3K4me3 before stimulation and provides NF-κB and RNAPII with constitutive and immediate access (Saccani et al. 2001; Natoli 2009). Here we discovered that a similar mechanism is present in yeast. Continuous transcription of the short form of ASP3 maintains H3K4me3 in the promoter region even under nitrogen-rich conditions (Fig. 7). This covalent modification may facilitate the binding of chromatin-remodeling ATPase to disrupt the nucleosomes (Santos-Rosa et al. 2003). Therefore, the promoter region is slightly “open” and more accessible for transcription. Upon nitrogen starvation, RNAPII is immediately recruited to the ASP3 promoter without requiring progressive chromatin remodeling to enhance accessibility. The rapid induction of full-length ASP3 may therefore be of benefit to adapt to nutrient deficiency. In conclusion, our findings illustrate a different example of transcriptional regulation: Activation of gene expression is achieved by intragenic transcription, which leads to accelerated RNAPII recruitment through a chromatin-remodeling mechanism.
FIGURE 7.
Model for the chromatin status at the ASP3 promoter in the presence and absence of intragenic transcription. The chromatin status in wild-type (left) and iceΔ (right) strains is shown under nitrogen-rich conditions (upper) and under nitrogen starvation (bottom). With the help of intragenic transcription (light-blue lines), the promoter region of ASP3 is immediately accessible for RNAPII upon nitrogen starvation. In the absence of intragenic transcription, the promoter region is more condensed, thus requiring chromatin-remodeling ATPase to enhance the accessibility of this region to RNAPII for ASP3 transcription (red lines) after nitrogen depletion.
MATERIALS AND METHODS
Plasmids, yeast strains, and growth conditions
To exogenously express ASP3, the ASP3 ORF along with 1000-bp upstream and 500-bp downstream flanking sequence was amplified by PCR from S. cerevisiae genomic DNA and subsequently cloned into pGEMTeasy (Promega) to obtain pGEMTeasy-ASP3. The HindIII–SacII DNA fragment, which contains the 755-bp ASP3 promoter and 500-bp downstream sequence from pGEMTeasy-ASP3, was then subcloned into pRS316 to obtain pRS316-ASP3. The XhoI–SacII DNA fragment from pRS316-ASP3 was further cloned into pRS306 to generate pRS306-ASP3. The pYM1 plasmid (Knop et al. 1999) was treated with BglII–Klenow–SacI to obtain KanMX6, and this fragment was then ligated with SacII-T4 polymerase-SacI-treated pRS306-ASP3 to obtain pRS306-ASP3-KanMX6. pRS316-asp3(iceΔ) and pRS306-asp3(iceΔ)-KanMX6 plasmids were generated by using mutagenic PCR to delete the ASP3(385-625) DNA sequences in pRS316-ASP3 and pRS306-ASP3-KanMX6, respectively. pRS316-asp3(tataΔ) was generated by using mutagenic PCR to delete nucleotide −129 to −76 of ASP3 in pRS316-ASP3. pRS315-rpb1-1 (expressing a temperature-sensitive Rpb1p) was created by cloning the HindIII fragment from pRP1-1 (kindly provided by Richard Young [Massachusetts Institute of Technology]) into the same site of pRS315, and pRS315-RPB1 (expressing the wild-type Rpb1p) was generated by cloning the HindIII–Klenow–XbaI fragment from RY2049 (kindly provided by Richard Young) into the SalI–Klenow–XbaI-treated pRS315-rpb1-1. pLG669Z-UASΔ was obtained by deletion of the XhoI fragment containing the upstream activation sites (UAS) of pLG669Z (Guarente and Ptashne 1981). The pLG669Z-PncASP3 plasmid used for the reporter assay was created by insertion of the ASP3(306-657) DNA fragment into pLG669Z-UASΔ. All primer sequences for PCR and mutagenesis are available upon request.
All yeast strains used in this study were isogenic to BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0). The gat1, gln3, and gat1 gln3 mutants in the BY4741 background were obtained from the KanMX6-deletion library (Invitrogen). There are four copies of ASP3 genes in the BY4741 strain. For simplification of the procedure, all four copies of ASP3 were deleted by double crossover of the ASP3 locus with an asp3∷KanMX6 fragment, which was PCR-amplified from the pYM1 plasmid, and selected for G418-resistant clones for the asp3 deletion mutant. Single-copy ASP3 wild-type and iceΔ strains were created by single crossover of the MluI-linearized pRS306-ASP3-KanMX6 or pRS306-asp3(iceΔ)-KanMX6 into the asp3∷KanMX6 strain and selected for Ura+ colonies. The wild-type and RPB1 temperature-sensitive mutant strains were created in the single-copy ASP3 BY4743 strain by deletion of one allele of RPB1 with HIS3MX6, transformation with pRS315-RPB1 or pRS315-rpb1-1, and tetradissection to obtain Ura+, Leu+, and His+ colonies.
Nitrogen starvation was performed for 2 h as previously described (Oliveira et al. 2003) unless indicated otherwise. Rapamycin treatment was conducted as described (Scherens et al. 2006) in synthetic complete medium with 2% glucose for 20 min.
RNA analysis
For total RNA extraction, cells were grown to mid-log phase, harvested, lysed in 500 μL of TRIzol solution (Invitrogen) with glass beads, and vigorously vortexed for 5 min at room temperature. The following manipulations were conducted as described by the manufacturer. For Northern analysis, 5 μg of total RNA was separated on a 1.2% agarose gel containing 2.1% formaldehyde in MOPS/formaldehyde buffer. RNA was transferred to a nylon membrane (Perkin-Elmer) and hybridized with [α-32P]dCTP-labeled probes for the full-length and short forms of ASP3 and ACT1. scR1 was detected with oligonucleotide 5′-ATCCCGGCCGCCTCCATCAC-3′ labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB).
RACE analysis was performed by using the RACE cDNA Amplification Kit (Clontech). RNAs were prepared from ammonium-containing and nitrogen-starved cells. For sequencing of the RACE products, products were cloned into pGEMTeasy plasmid and then sequenced by SP6 or ASP3 primers.
β-Galactosidase assays
Cell pellets were collected from cells grown in ammonium-containing medium or grown under nitrogen starvation. Assays were performed as described (Guarente and Ptashne 1981) with chloroform and 0.1% SDS to permeabilize cells. One unit of β-galactosidase activity is defined as the optical density of product at A420 per milliliter of culture at A600 in a 30-min assay. The values shown represent the means of three independent measurements.
Chromatin immunoprecipitation (ChIP) analysis
Yeast cells were grown overnight in 10 mL of synthetic medium with 2% glucose and refreshed in 200 mL of synthetic medium with 2% glucose until A600 reached 0.5–0.8. An aliquot of 50 mL culture was collected and designated as time 0, and the remaining culture was centrifuged for 1 min at 14,000g. Nitrogen starvation was performed for 15, 30, and 60 min. All collected 50-mL cultures were fixed with 1% formaldehyde, and ChIP assay was performed as described (Ezhkova and Tansey 2006) using anti-H3 (ab1791; Abcam), anti-H3K4me3 (9751; Cell Signaling), or anti-CTD-Ser5-P (05-623; Millipore) antibodies. DNA was analyzed by quantitative real-time PCR using PCR primers specific to the TATA-box, the intragenic region or to the downstream sequence of ASP3. The control primers amplified an intergenic region of chromosome V that lacks any open reading frame (Komarnitsky et al. 2000). Results represent means from at least three independent experiments.
Nuclear run-on analysis
The nuclear run-on assay was performed as described (Elion and Warner 1986) with the following modifications. Cultures containing 5 × 107 cells were collected after nitrogen starvation for 1 h. Cells were permeabilized in 0.5% of sodium N-lauroylsarcosine. Transcription was carried out for 30 min at 30°C in the presence of 100 μCi of [α-32P]rUTP. Total RNAs were isolated, denatured for 5 min at 95°C, and mixed with hybridization buffer. Membranes with immobilized DNAs were hybridized with labeled RNAs for 16 h at 60°C. The DNA fragments corresponding to ASP3(1-347), ACT1(391-1108), and the multiple cloning region of pBSKS (Stratagene) were used as the probes.
ACKNOWLEDGMENTS
We thank Richard Young and Leonard Guarente for providing plasmids and Heiko Kuhn for manuscript editing. This work is supported by the National Science Council (NRPGM-98-3112-B-002-039) and the National Health Research Institute of Taiwan (NHRI-EX98-9727BI) to S.-C.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2177410.
REFERENCES
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692 [DOI] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem 275: 35727–35733 [DOI] [PubMed] [Google Scholar]
- Bickel KS, Morris DR 2006. Role of the transcription activator Ste12p as a repressor of PRY3 expression. Mol Cell Biol 26: 7901–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AJ, Gordon M, Eide DJ, Winge DR 2006. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J 25: 5726–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F 2009. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev 13: 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Courchesne WE, Magasanik B 1988. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: Roles of the URE2 and GLN3 genes. J Bacteriol 170: 708–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop PC, Meyer GM, Ban D, Roon RJ 1978. Characterization of two forms of asparaginase in Saccharomyces cerevisiae. J Biol Chem 253: 1297–1304 [PubMed] [Google Scholar]
- Dunlop PC, Meyer GM, Roon RJ 1980. Nitrogen catabolite repression of asparaginase II in Saccharomyces cerevisiae. J Bacteriol 143: 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA, Warner JR 1986. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol 6: 2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP 2006. Chromatin immunoprecipitation to study protein–DNA interactions in budding yeast. Methods Mol Biol 313: 225–244 [DOI] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem 278: 26303–26306 [DOI] [PubMed] [Google Scholar]
- Guarente L, Ptashne M 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci 78: 2199–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci 96: 14866–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Martens JA 2009. ncRNA transcription makes its mark. EMBO J 28: 1679–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745 [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Smith MM 2003. Functional consequences of histone modifications. Curr Opin Genet Dev 13: 154–160 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919 [DOI] [PubMed] [Google Scholar]
- Kim KW, Kamerud JQ, Livingston DM, Roon RJ 1988. Asparaginase II of Saccharomyces cerevisiae. Characterization of the ASP3 gene. J Biol Chem 263: 11948–11953 [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GL, Bickel KS, MacKay VL, Morris DR 2005. The undertranslated transcriptome reveals widespread translational silencing by alternative 5′ transcript leaders. Genome Biol 6: R111 doi: 10.1186/gb-2005-6-13-r111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Mitchell AP, Magasanik B 1984. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol 4: 2758–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Natoli G 2009. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol 1: a000224 doi: 10.1101/cshperspect.a000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh EA 2008. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol 6: 2817–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira EM, Carvajal E, Bon EP 1999. L-Asparaginase II of Saccharomyces cerevisiae. Activity profile during growth using an ure2 mutant P40-3C and a P40-3C + Ure2p strain. Appl Biochem Biotechnol 77–79: 311–316 [DOI] [PubMed] [Google Scholar]
- Oliveira EM, Martins AS, Carvajal E, Bon EP 2003. The role of the GATA factors Gln3p, Nil1p, Dal80p and the Ure2p on ASP3 regulation in Saccharomyces cerevisiae. Yeast 20: 31–37 [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, Harrison PM, Nelson FK, Miller P, Gerstein M, et al. 2003. The transcriptional activity of human Chromosome 22. Genes Dev 17: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2001. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med 193: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell 12: 1325–1332 [DOI] [PubMed] [Google Scholar]
- Scherens B, Feller A, Vierendeels F, Messenguy F, Dubois E 2006. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term. FEM Yeast Res 6: 777–791 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Nishioka K, Reinberg D 2003. Histone lysine methylation: A signature for chromatin function. Trends Genet 19: 629–639 [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Rowen DW, Magasanik B 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci 92: 9450–9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ 2007. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci 104: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R 2008. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379–386 [DOI] [PubMed] [Google Scholar]