Abstract
Triggering and propagating an intracellular innate immune response is essential for control of viral infections. RNase L is a host endoribonuclease and a pivotal component of innate immunity that cleaves viral and cellular RNA within single-stranded loops releasing small structured RNAs with 5′-hydroxyl (5′-OH) and 3′-monophosphoryl (3′-p) groups. In 2007, we reported that RNase L cleaves self RNA to produce small RNAs that function as pathogen-associated molecular patterns (PAMPs). However, the precise sequence and structure of PAMP RNAs produced by RNase L is unknown. Here we used hepatitis C virus RNA as substrate to characterize RNase L mediated cleavage products [named suppressor of virus RNA (svRNA)] for their ability to activate RIG-I like receptors (RLR). The NS5B region of HCV RNA was cleaved by RNase L to release an svRNA that bound to RIG-I, displacing its repressor domain and stimulating its ATPase activity while signaling to the IFN-β gene in intact cells. All three of these RIG-I functions were dependent on the presence in svRNA of the 3′-p. Furthermore, svRNA suppressed HCV replication in vitro through a mechanism involving IFN production and triggered a RIG-I-dependent hepatic innate immune response in mice. RNase L and OAS (required for its activation) were both expressed in hepatocytes from HCV-infected patients, raising the possibility that the OAS/RNase L pathway might suppress HCV replication in vivo. It is proposed that RNase L mediated cleavage of HCV RNA generates svRNA that activates RIG-I, thus propagating innate immune signaling to the IFN-β gene.
Keywords: 3′-phosphate, Hepatitis C virus, RIG-I, RNase L, innate immunity
INTRODUCTION
Viral RNAs, often in the form of cytoplasmic 5′-triphosphorylated, double-stranded, or uridine and adenosine-rich viral RNAs, are pathogen-associated molecular patterns (PAMPs) that trigger innate immunity through RIG-I-like receptors (RLR), a familty of cytoplasmic pathogen recognition receptors (PRRs) (Horner and Gale 2009; Rehwinkel and Reis e Sousa 2010; Ting et al. 2010; Yoneyama and Fujita 2010). These RNA PAMPs interact with either of two RLRs, RIG-I and MDA5, containing N-terminal caspase activation and recruitment domains (CARD) and C-terminal DExD/H Box RNA helicase motifs (Yoneyama et al. 2004; Kato et al. 2005, 2006; Gitlin et al. 2006). Subsequently, RIG-I and MDA5 interact with another CARD protein, IPS-1 (MAVS, VISA, Cardif), in the mitochondrial membrane (Kawai et al. 2005; Meylan et al. 2005; Seth et al. 2005; Xu et al. 2005; Loo et al. 2006). IPS-1 then relays the signal to the kinases, IKKε and TBK1 that phosphorylate transcription factor IRF-3. Transcription factor NF-κB is simultaneously activated through IPS-1. The homodimerized and phosphorylated IRF-3 relocalizes to the nucleus along with activated NF-κB and independently or together activate different target genes, including the IFN-β gene.
The IFN response against RNA viruses is frequently mediated by RNase L, part of a ribonucleolytic pathway containing the PRR, 2′-5′-oligoadenylate synthetase (OAS) (Silverman 2007). Type I IFNs induce at the transcriptional level a group of OAS proteins that are activated by viral dsRNA PAMPs to produce 2-5A [px5′A(2′p5′A)n; x = 1–3; n ≥ 2] from ATP (Hovanessian et al. 1977; Kerr and Brown 1978; Hovanessian and Justesen 2007). OAS activators include viral replicative intermediates, ds RNA genomes, annealed ss RNAs of opposite polarity and highly structured ss RNA. 2-5A is the ligand and activator of RNase L, a ubiquitous enzyme in mammalian cells, including primary human hepatocytes, that lies dormant until viral infections occur (Zhou et al. 2005). Human RNase L is a 741 amino acid polypeptide containing, from N- to C-termini, nine ankyrin repeats, several protein kinase-like motifs, and the ribonuclease domain (Hassel et al. 1993; Zhou et al. 1993). 2-5A binds to ankyrin repeats 2 and 4 (Tanaka et al. 2004), causing catalytically inactive RNase L monomers to form activated dimers with potent ribonuclease activity (Dong and Silverman 1995; Cole et al. 1996). Once activated, RNase L cleaves single-stranded regions of viral and host RNAs, principally at UpAp and UpUp dinucleotides, leaving 3′-phosphoryl and 5′-hydroxyl groups at the termini of the RNA cleavage products (Floyd-Smith et al. 1981; Wreschner et al. 1981). Interestingly, cleavage of cellular (self) RNA by RNase L results in the production of short RNAs that activate RNA helicases, RIG-I and MDA5, and the adapter IPS-1 resulting in activation of the IFN-β gene (Malathi et al. 2005, 2007). As a result, circulating levels of IFN-β production were reduced several fold in Sendai virus- or encephalomyocarditis virus-infected RNase L−/− mice compared with infected wild-type mice. In addition, 2-5A treatment of wild-type mice, but not of RNase L−/− mice, resulted in IFN-β production. However, the sequence and structure of the small RNAs produced by RNase L, their interactions with RIG-I and/or MDA5, and their role in mediating antiviral innate immunity have remained largely unexplored.
Hepatitis C virus (HCV), Hepacivirus genus of the Flaviviridae family, is a virus that has infected about four million adults in the United States and is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (Armstrong et al. 2006). During HCV infections, the viral PAMP that triggers type I IFN production is the polyuridine tract (poly-U/UC) in the 3′ nontranslated region of the viral genomic RNA (Saito et al. 2008). Poly-U/UC requires a 5′-triphosphate to activate RIG-I in the cytoplasm of infected cells. Here we use HCV genomic RNA as a model substrate to characterize the requirements for RLR signaling by RNase L cleavage products. Our findings demonstrate a requirement for the 3′-monophosphate and complex features in the RNA cleavage product responsible for potent PAMP activity.
RESULTS
Identification of HCV RNA cleavage products that bind RIG-I
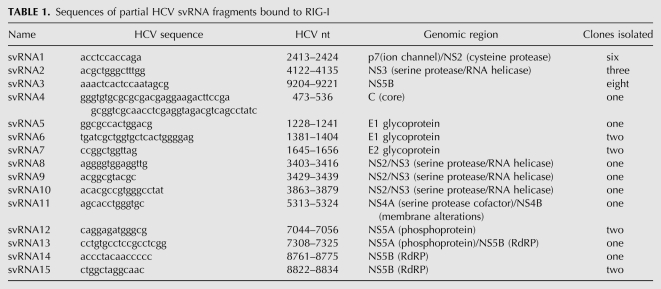
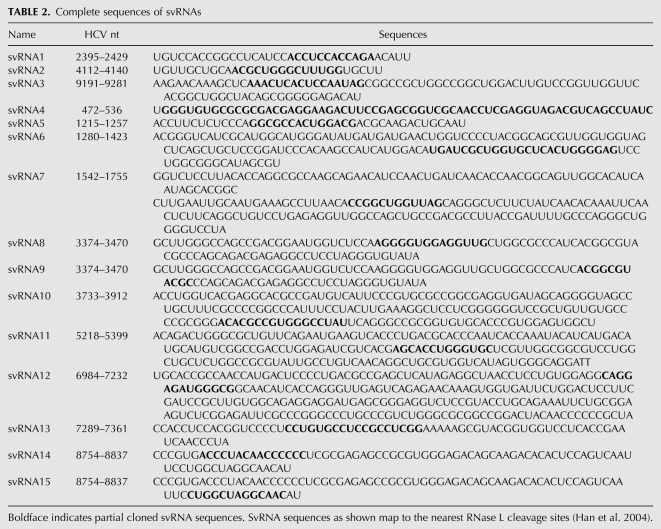
Here we investigated whether RNase L processes HCV genomic RNA into small RNAs with PAMP activity (designated “suppressor of virus RNA” or svRNA). HCV RNA was selected as a substrate because the RNase L cleavage sites had been previously determined (Han et al. 2004), thus allowing the termini of cleavage products to be precisely mapped. In addition, an M-fold secondary structure prediction of the entire HCV H77 genomic RNA (performed as in Palmenberg and Sgro 1997) was used to identify structural domains in the HCV genomic RNA. Based on both known and predicted structural features in the HCV RNA, we selected eight regions spanning the entire genome as substrates for digestion by RNase L (Fig. 1A, lower). The eight HCV RNA segments were generated by in vitro transcription, purified, and cleaved by RNase L. RNA cleavage products <200 nucleotides (nt) in length were isolated, bound to flag tagged-RIG-I or -MDA5, and cloned (Supplemental Fig. S1). Fifteen small RNAs with affinity for RIG-I were cloned, while no clones were obtained using MDA5 (Table 1). It was apparent from the comparison of the cloned sequences with previously determined RNase L cleavage sites (Han et al. 2004) that all were partial clones of the small RNAs (Table 2).
FIGURE 1.
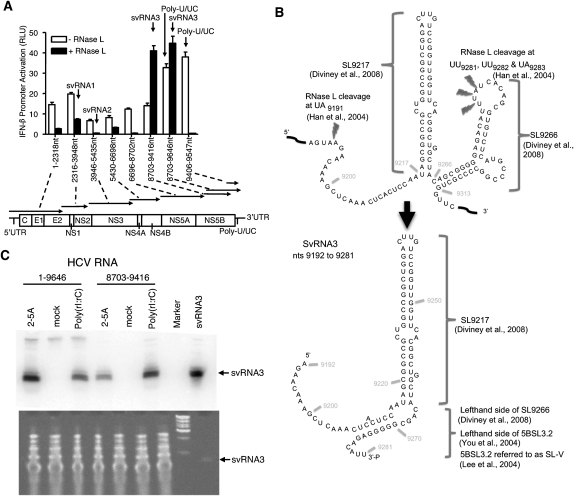
Cleavage of HCV RNA by RNase L produces small RNAs with PAMP activity. (A) Activation of the IFN-β promoter after 18 h in Huh7 cells in response to transfection with undigested or RNase L-digested HCV RNA segments. (B) RNase L releases svRNA3 from two adjacent stem–loops in the NS5B region of the HCV open reading frame. (C) Detection of RNase L-mediated cleavage product of HCV RNA (svRNA3) in Huh7.5 cells at 96 h after electroporated with full-length HCV genomic RNA or a region of HCV RNA encoding svRNA3 (nt 8703–9416) as determined in a Northern blot (upper) (Materials and Methods). Small RNAs, <200 nt (10 μg), and 100 ng of svRNA3 were stained with gelstar (Cambrex Bio Science) (lower).
TABLE 1.
Sequences of partial HCV svRNA fragments bound to RIG-I
TABLE 2.
Complete sequences of svRNAs
Prior to investigating the small RNAs, we determined whether the uncleaved HCV RNA segments had PAMP activity. The RNAs were individually transfected into human hepatoma Huh7 cells containing the human IFN-β promoter fused to firefly luciferase cDNA. As reported previously (Saito et al. 2008), some of these RNA fragments induced the IFN-β promoter, especially fragments 8703–9416 and 8703–9646 nt, which contains the poly-U/UC region (9406–9547 nt) (Fig. 1A). In contrast, the small RNAs generated using RNase L had little or no ability to induce the IFN-β promoter, except for the small RNAs generated from fragments 8703–9416 and 8703–9646 nt (Fig. 1A). Remarkably, cleavage of the 8703–9416 nt fragment with RNase L caused a large increase in the ability to induce the IFN-β promoter, whereas RNase L cleavage of the 3′-extended fragment (8703–9646 nt) containing the poly-U/UC region resulted in a modest increase in PAMP activity (p < 0.0006 and p < 0.038, respectively). In contrast, PAMP activity associated with poly-U/UC was destroyed by digestion with RNase L because of the sequence preferences of RNase L for UU and UA dinucleotides in single-stranded RNA (Fig. 1A, rightmost pair of columns). The RNA fragment (8703–9416 nt) that produced the highest level of PAMP activity upon RNase L cleavage yielded three clones of RIG-I-bound RNAs: svRNA3, svRNA14, and svRNA15 (Tables 1, 2). SvRNA14 and svRNA15 originated from an identical 82-nt fragment of the NS5B gene, but lacked significant structure with a maximum of four predicted consecutive base pairs (as determined using MFOLD) (Zuker 2003), and were not pursued further. In contrast, svRNA3 (9192–9281 nt also from the NS5B gene) is a highly structured RNA, including a stretch of 11 consecutive base pairs. Cleavage of HCV RNA by RNase L at nt 9191 (UA9191) and nt 9281 (UU9281) releases a 90-base fragment of HCV RNA from the NS5B coding region that refolds into svRNA3 (Fig. 1B).
SvRNA3 is formed from HCV genomic RNA through the action of RNase L in intact human hepatoma cells
To establish if svRNA3 could be demonstrated to form in intact cells, full-length genomic RNA or HCV RNA fragment 8703–9416 nt, both from HCV 1a, strain H77, were electroporated into Huh7.5 cells. After 24 h, transfection of flag-RNase L cDNA was performed to elevate levels of RNase L followed by treatment with IFN-β to elevate OAS levels (Supplemental Fig. S2A). Production of HCV core antigen was observed in cells containing full-length HCV RNA demonstrating translation and processing of the HCV polyprotein, (Supplemental Fig. S2B). Furthermore, RNase L activation was observed in flag-RNase L expressing cells as measured by the appearance in RNA chips of specific and characteristic rRNA cleavage products when cells were treated with either 2-5A (activator of RNase L) or poly(rI:rC) (activator of OAS) (Supplemental Fig. S2C; Silverman et al. 1983). We observed that svRNA3 was generated from HCV RNA in the intact cells upon activation of RNase L by 2-5A or poly(rI):(rC) as determined in Northern blots (Fig. 1C). Results show that HCV RNA (either the full-length genomic RNA or fragment 8703–9416) is processed into svRNA3 when OAS and RNase L are present and active.
Activation of the RNA helicase, RIG-I, by svRNA3 is dependent on its 3′-p group
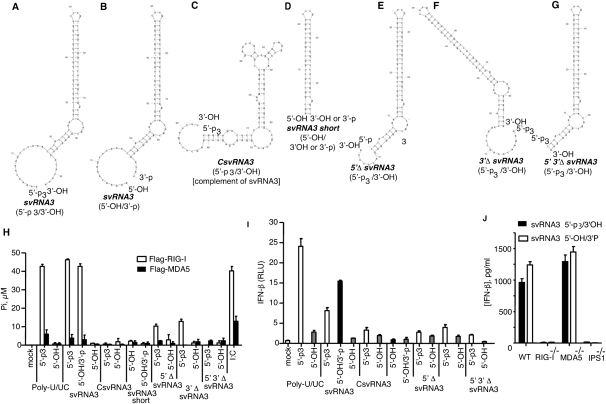
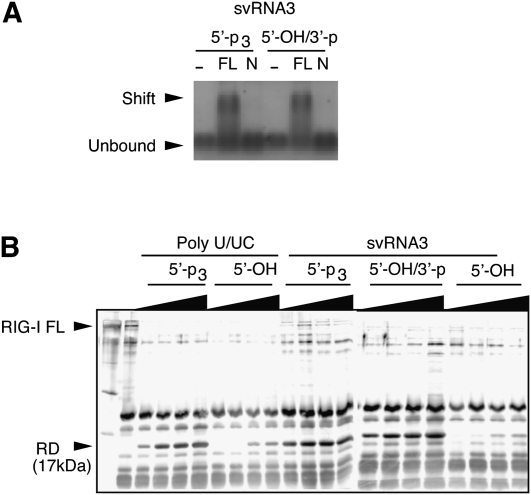
A method was devised for producing svRNA3 (5′-OH/3′-p) in which a precursor (with 5′-p3 and a 3′-extension of UUA) was synthesized with T7 RNA polymerase (Fig. 2A). The 5′-p3 was removed with calf intestinal phosphatase (CIP), while the UUA sequence, and any possible aberrant extension of the 3′-terminus (Cazenave and Uhlenbeck 1994; Schlee et al. 2009), was removed by RNase L, which also generated a 3′-p (Fig. 2B). In every instance, 5′-p3/3′-OH svRNA3 refers to the precursor, whereas 5′-OH/3′-p is the mature RNase L product. In addition, to determine the minimal structural requirements in svRNA3 that contribute to signaling, we also generated derivatives of svRNA3 (5′-p3 forms) [a complementary form (CsvRNA3), an isolated stem–loop (svRNA3 short), and deletion of the 5′, 3′, or both overhangs (5′-Δ svRNA3, 3′-Δ svRNA3, and 5′-, 3′-Δ svRNA3)] (Fig. 2C,–G). Activation of the ATPase function of RIG-I was obtained with poly-U/UC (5′-p3/3′-OH) and with svRNA3 (5′-p3/3′-OH or 5′-OH/3′-p), but not with the 5′-OH/3′-OH forms of these RNAs (Fig. 2H). Interestingly, the 5′-OH/3′-p and 5′-p3/3′-OH forms of svRNA3 were equally active in this assay. Weak activation of RIG-I ATPase was also obtained with the 5′- and 3′-deleted forms of svRNA3 (5′-p3/3,-OH), but not with any of the other RNAs. The only RNA that activated the MDA5 ATPase, albeit modestly, was poly(rI):(rC) (Fig. 2H).
FIGURE 2.
Activation of RIG-I ATPase activity and IFN-β induction by svRNA3 and its derivatives. Predicted RNA secondary structure of (A) svRNA3 (5′-p3/3′-OH); (B) svRNA3 (5′-OH/3′-p); (C) CsvRNA3 (5′-p3/3′-OH), complement of svRNA3; (D) svRNA3 short (5′-OH/3′-OH or 5′-OH/3′-p), the main stem–loop structure of svRNA3; (E) 5′ΔsvRNA3 (5′-p3/3′-OH), svRNA3 deleted for the 5′-ss overhang; (F) 3′ΔsvRNA3 (5′-p3/3′-OH), svRNA3 deleted for the 3′-ss overhang; (G) 5′ 3′ΔsvRNA3 (5′-p3/3′-OH), svRNA3 deleted for both the 3′- and 5′-ss overhangs. (H) Activation of RIG-I or MDA5 ATPase (20 min at 37°C) by the indicated RNAs. (I) Activation of the IFN-β promoter in Huh7 cells after 18 h by transfection of the indicated RNAs. (J) Induction of IFN-β by svRNA3 in mouse embryo fibroblasts (mef). IFN-β levels in supernatants of WT, RIG-I-deficient, MDA5-deficient, or IPS1-deficient mouse embryo fibroblasts 18 h after transfection with 30 pmol of svRNA3. IFN-β protein levels were measured by ELISA. RNA structures were as predicted by mfold software (Zuker 2003).
The 5′-OH/3′-p form of svRNA3 stimulated the IFN-β promoter activity in Huh7 cells to 190% of the level obtained with the 5′-p3/3′-OH form, whereas the 5′-OH/3′-OH form was inactive (Fig. 2I). The 5′-p3/3′-OH form of poly-U/UC stimulated the IFN-β promoter to 155% of the level obtained with 5′-OH/3′-p svRNA3. These findings show that a 3′-p group on svRNA3 could effectively substitute for a 5′-p3 group to signal to the IFN-β gene. None of the other RNAs induced IFN-β promoter activity. These results suggest that 5′- and 3′-ss regions of svRNA3 and both the upper and lower stems were required for optimal RIG-I activation.
IFN-β induction was compared in wild-type (WT) and gene deficient mouse embryo fibroblasts (mef) treated with svRNAs1, -2 (Supplemental Fig. S3), and svRNA3 (Fig. 2J) (all with 5′-OH/3′-p termini). While svRNA1, svRNA2, and svRNA3 all required RIG-I and IPS-1 expression (but not MDA5) for IFN-β induction, svRNA3 was about 30-fold more active. Furthermore, the 5′-p3/3′-OH and 5′-OH/3′-p versions of svRNA3 were equally potent PAMPs in WT mef (Fig. 2J).
Viral RNA PAMPs that signal through RIG-I induce conformational changes that displace the C-terminal repressor domain (RD) allowing interaction with the adapter protein, IPS-1 (Saito et al. 2007). SvRNA3 with either 5′-p3/3′-OH or with 5′-OH/3′-p formed a stable complex with full-length RIG-I, but not with an N-terminal polypeptide of RIG-I (Fig. 3A). Efficient displacement of the RIG-I RD was obtained with 5′-p3-polyU/UC and either 5′-p3 or 3′-p forms of svRNA3 (Fig. 3B). The dephosphorylated forms of these RNAs lacked activity.
FIGURE 3.
SvRNA3 binds RIG-I causing the release of its repressor domain (RD). (A) RIG-I binding of svRNA3 with different termini (as shown) as determined by gel-shift analysis of full-length (FL) RIG-I or its N-terminal (N) polypeptide (1–228 amino acids). (B) Release of RIG-I RD by partial trypsin digestion. The silver-stained gel image shows trypsin digestion products of Flag-RIG-I incubated with 3, 6, 15, or 30 pmol of the indicated RNA.
SvRNA3 induces a hepatic innate immune response in mice
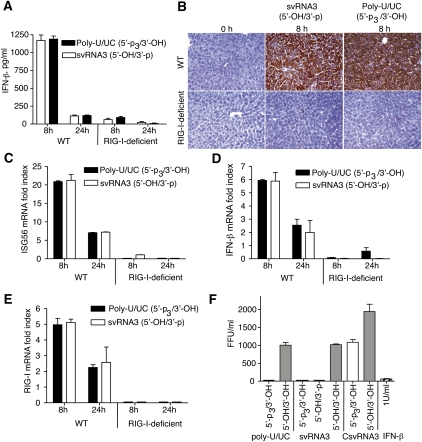
To study the effects of svRNA3 on hepatic innate immunity in vivo, WT or RIG-I-deficient mice were treated by hydrodynamic tail vein injection with either svRNA3 (5′-OH/3′-p) or, as a positive control, poly-U/UC (5′-p3/3′-OH). This procedure efficiently introduces the HCV RNA and its subgenomic counterparts into the mouse hepatocytes, thus modeling the viral RNA–host interactions of an acute HCV infection (Saito et al. 2008). Both svRNA3 (5′-OH/3′-p) and the poly-U/UC (5′-p3/3′-OH) RNA were equally potent in the RIG-I-dependent induction of IFN-β by 8 h in WT, but not in RIG-I-deficient mice, as determined by specific ELISAs performed on sera (Fig. 4A). By 24 h post-treatment circulating IFN-β levels returned to basal levels. A dramatic induction of ISG54 protein was observed in the liver after 8 h of treatment with svRNA3 or poly-U/UC in WT mice, but not in RIG-I-deficient mice (Fig. 4B) indicative of paracrine signaling of IFN-β. In addition, levels of mRNAs for ISG56, IFN-β, and RIG-I were highly induced at 8 h in WT, but not in RIG-I-deficient mice, and were declining by 24 h post-treatment with either svRNA3 or poly-U/UC (Fig. 4C,–E).
FIGURE 4.
HCV svRNA3 induces a hepatic innate immune response and suppresses HCV replication by a paracrine mechanism. (A–E) Hydrodynamic tail vein injection of poly-U/UC (5′-p3/3′-OH) or svRNA3 (5′-OH/3′-p) in WT and Rig-I−/− mice (n = 3). (A) Induction of circulating IFN-β by poly-U/UC or svRNA3. (B) Hepatic induction of ISG54 protein as determined by immunohistochemistry (IHC) by poly-U/UC or svRNA3. (C–E) Hepatic induction of Isg56, Ifnb, and Rig-i mRNAs by poly-U/UC or svRNA3 as determined by qRT-PCR. (F) Paracrine inhibition of HCV infections by poly-U/UC (5′-p3/3′-OH) or svRNA3 (5′-OH/3′-p). Huh7.5 cells (in triplicate) were treated for 12 h before HCV infection with medium containing IFN-β or conditioned medium from Huh7 cells transfected with the indicated RNAs. The graph shows the number of infected cells as determined by focus-forming unit (FFU) assay at 48 h after infection.
SvRNA3 or 2-5A inhibits HCV replication in Huh7 cells
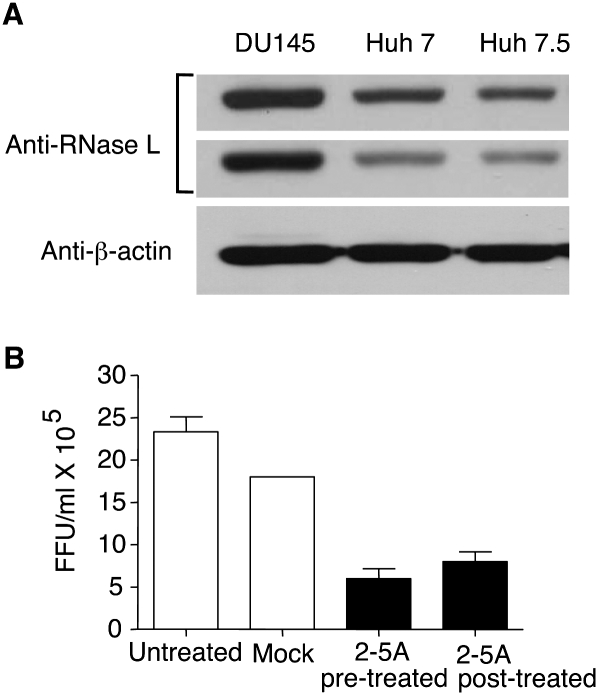
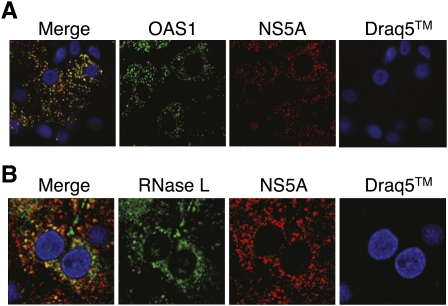
To determine if svRNA3 could inhibit HCV replication in vitro through a paracrine mechanism, conditioned media from Huh7 cells transfected with svRNA3, CsvRNA3, or poly-U/UC were added to Huh7.5 cells (harboring a defective form of RIG-I) (Sumpter et al. 2005) for 12 h prior to infection with HCV strain JFH-1 for 48 h (Wakita et al. 2005). Focus-forming unit (FFU) assays with anti-HCV antibody showed antiviral activity with poly-U/UC (5′-p3/3′-OH) and svRNA3 (5′-p3/3′-OH or 5′-OH/3′-p), but not with the same RNAs containing 5′-OH and 3′-OH termini (Fig. 4F). The complement of svRNA3, CsvRNA3, was inactive in these assays, while IFN-β, included as a positive control, inhibited HCV infection. In addition, direct activation of RNase L in Huh7 cells reduced HCV yields to 33% ± 11 (p = 0.0091) of that obtained for the control infected cells as measured by FFU, even though these cells have a relatively low level of RNase L (Fig. 5). Presumably, the anti-HCV effect would be even greater in human hepatocytes in vivo, which are known to express RNase L at higher levels (Zhou et al. 2005). We therefore evaluated OAS1 and RNase L expression in human HCV patients via confocal microscopy analysis of immunostained liver sections. Both OAS1 and RNase L were abundant in HCV-infected hepatocytes from patients (Fig. 6). The merged images suggest, but do not prove, colocalization with the viral NS5A protein. However, NS5A does mark intracellular sites of HCV replication and was previously shown to be a binding partner of OAS1 (Taguchi et al. 2004; Wakita et al. 2005).
FIGURE 5.
Inhibition of HCV replication in response to 2-5A activation of RNase L. (A) RNase L levels in lysates (30 μg) of DU145 prostate cancer cells and hepatoma cell lines Huh7 and Huh7.5 as determined in immunoblots (different exposures of the same blot are shown). Blots were probed with monoclonal antibody reactive to human RNase L (Dong and Silverman 1995) and compared to anti-β-actin as control. (B) Huh7 cells were either mock transfected or transfected with 2-5A trimer (p35′A2′p5′A2′p5′A) before or after HCV infection (JFH1 strain) for 8 h. Supernatants were harvested at 48 h post-infection and titered for HCV by focus-forming assays.
FIGURE 6.
Hepatic expression of HCV NS5A, OAS1, and RNase L in HCV-infected liver. Images show a 0.15 μm optical section of liver biopsy specimen stained with Draq-5 to show nuclei (blue), and immunostained with antibodies specific to NS5A (red) and (A) OAS1 (green) or (B) RNase-L (green). The left panel of each set features the merged image showing sites of protein codistribution (yellow). Images represent a single patient and are representative of analyses from six different patients with chronic HCV infection (data not shown). 60X magnification.
DISCUSSION
Our results define features in an RNase L-mediated RNA cleavage product necessary for RIG-I activation. The finding of a previously unrecognized role for a 3′-p in RIG-I activation, effectively substituting for a 5′-p3 group, is a novel paradigm for an RNA PAMP. The predicted structure of svRNA3 includes a broken-stem–loop with 5′ and 3′overhangs, both of which contribute to the activation of RIG-I. Surprisingly, the main double-stranded stem of svRNA3 (svRNA3 short) including a 3′-p lacked PAMP activity. Results indicate that in addition to 3′-p a higher order structure is necessary for recognition by and activation of RIG-I. For instance, we have observed that svRNA4 (65 nt) bearing 3′-p or 5′-p3 did not stimulate RIG-I activity.
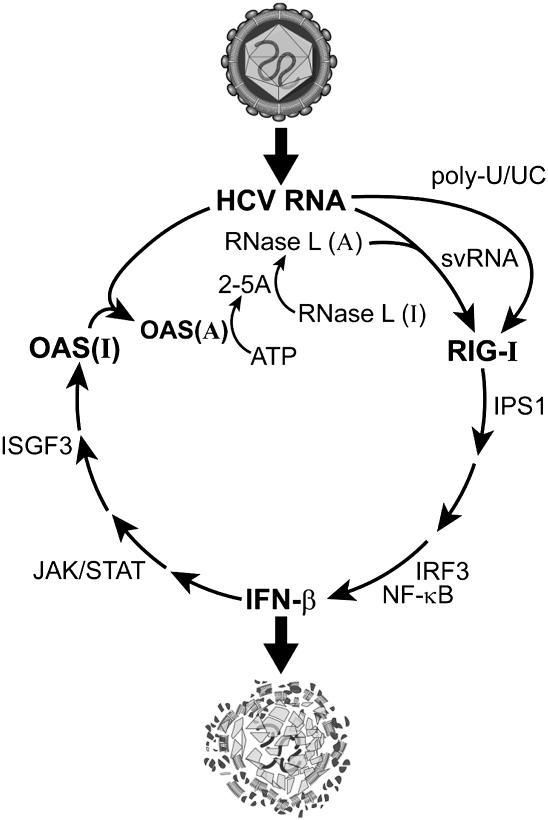
These findings suggest the following scenario for innate immunity against HCV (Fig. 7). At early times of infection, RIG-I detects and responds to the HCV PAMP, poly-U/UC in the HCV 3′-UTR in concert with a 5′-p3 group, leading to activation of IRF3 and synthesis of type I IFN (Saito et al. 2008). IFN-β induces tissue-wide transcriptional activation of OAS genes and viral dsRNA activates OAS to produce 2-5A (Han and Barton 2002), which activates RNase L. Specifically, RNase L cleaves ss regions of HCV RNA on the 3′ side of UUp and UAp dinucleotides, including destroying the poly-U/UC PAMP (Fig. 1A). While it is premature to conclude that svRNA will amplify innate immune signaling against HCV, our data suggest this possibility. However, in addition, the intracellular host innate immune response is blunted by the action of different HCV proteins, including the NS3/4A protease complex that cleaves IPS-1, the HCV core protein that binds STAT1, and NS5A protein that suppresses activation of PKR (for review, see Horner and Gale 2009).
FIGURE 7.
Hypothetical temporal appearance and roles of the PAMPs, poly-U/UC, and svRNA3, in the innate immune response against HCV infections. Active (A) and inactive (I) forms of OAS and RNase L are indicated.
RNase L cleaves at sites throughout the HCV genomic RNA producing many small RNAs, but svRNA3 is by far the most active PAMP that is released during this process. Remarkably, IFN sensitivity of HCV strains in vivo nevertheless correlates with the number of potential RNase L cleavage sites (Han et al. 2004). Accordingly, there are fewer potential RNase L cleavage sites in IFN-resistant genotype 1a and 1b compared to IFN-sensitive genotypes 2a, 2b, 3a, and 3b (Han and Barton 2002). Our new findings indicate that an RNase L cleavage product of HCV RNA is able to stimulate RIG-I signaling. SvRNA3, composed of RNA sequences from two adjacent stem–loop structures within the NS5B portion of the HCV open reading frame, refolds into a RIG-I activator after its release by RNase L (Fig. 1B). HCV1a RNA is cleaved efficiently by RNase L at UA9191, UU9281 UU9282, and UA9283 (Han et al. 2004), liberating svRNA3 with a 3′ phosphoryl group (Fig. 1B). Importantly, RNA sequences and structures in this region of HCV RNA are phylogenetically conserved and they are required for HCV RNA replication (Tuplin et al. 2002, 2004; Lee et al. 2004; You et al. 2004; Diviney et al. 2008). Single-stranded UA and UU dinucleotides are phylogenetically conserved at nt 9281–9283 in HCV genotypes 1, 2, 3, 4, 5, and 6 (Diviney et al. 2008, Fig. 6A). The 9284UCACAGC9290 sequences immediately adjacent to these single-stranded UA and UU dinucleotides are invariant in HCV genotypes 1–6 and they form a kissing interaction with complementary sequences in the HCV 3′ NTR (Diviney et al. 2008, Fig. 8). The 9300GCCCG9304 sequences in the subsequent loop are also invariant in HCV genotypes 1–6 and are predicted to form a pseudoknot via base-pairing to 9108CGGGC9112 sequences upstream in the NS5B ORF (Diviney et al. 2008). The RNA sequences and structures in this region of HCV NS5B are well conserved across all six HCV genotypes. Finally, because RNase L has a relatively broad antiviral activity for many RNA viruses (Silverman 2007), chemical features of svRNA such as, but not limited to, the 3′-p group, or methods that will activate RNase L in vivo may lead to broad-spectrum antiviral agents active against HCV and other important viral pathogens.
MATERIALS AND METHODS
Mice, cells, and virus
Rig-i−/− mice and mef and Ips1−/− mef were provided by S. Akira (Osaka, Japan) (Kato et al. 2005 ; Kumar et al. 2006) and Mda5−/− mef were provided by M. Colonna and M. Diamond (St. Louis, Missouri) (Gitlin et al. 2006). Huh7 and Huh7.5 (T55I mutant RIG-I) (Sumpter et al. 2005) human hepatoma cells, and plasmid p90/HCV FL-long pU (AF009606) encoding a full-length HCV genome, genotype 1a (strain H77), were kindly provided by C. Rice (Rockefeller University) (Blight et al. 2003). A clone of HCV JFH-1 was provided by T. Wakita (Tokyo, Japan) (Wakita et al. 2005). 293T and DU145 cells were obtained from American Type Culture Collection.
Synthesis and purification of HCV RNA
Subgenomic HCV RNA fragments were produced from T7 promoter-linked PCR products generated from HCV H77 plasmid. The HCV DNA was transcribed using the T7-Megascript kit according to the manufacturer's protocol (Applied Biosystems). RNA was purified according to the protocol provided with the Megaclear kit (Applied Biosystems). As an alternative to in vitro synthesis, some of the smaller RNAs (<50 nt) with either a 3′-OH or a 3′-p group were chemically synthesized at IDT, Inc. The presence of 3′-p group was confirmed by mass spectrophotometric analysis.
Cleavage of HCV RNA by RNase L
HCV RNA (100 μg per reaction) was digested in vitro with purified recombinant human RNase L (2 μg) (Dong et al. 1994) and unfractionated 2-5A (10 μM) prepared as described (Thakur et al. 2007). Control reactions were with RNase L in the absence of 2-5A. Cleavage of the RNA was monitored in reactions that included trace amounts (0.1 nM) of an RNA FRET probe containing multiple cleavage sites for RNase L (Thakur et al. 2007). Samples were taken from 0 to 60 min at 22°C to measure fluorescence as an indicator of RNase L activity. To generate small RNAs with 3′-p ends, the RNA were digested in vitro with purified RNase L and crude 2-5A as above. Small RNAs lacking the 3′-p or 5′-p3 were generated by incubating with 10 units CIP (NEB) for 1 h at 37°C and 10 min for 75°C. Removal of 5′-p3 was monitored in a parallel reaction using 5′-32P-end labeled svRNA3 and CIP. Reactions were monitored up to 1 h until removal of radiolabeled 32P from svRNA3 was complete as monitored on a 12% sequencing gel (Malathi et al. 2007). Small RNA cleavage products (<200 nt) were purified using a solid-phase fractionation method (mirVana miRNA Isolation Kit, Ambion). To confirm complete cleavage of svRNA3, trace amounts of dephosphorylated svRNA3 and svRNA3′-p were labeled at its 5′-OH with [γ32-P]-dATP and T4 polynucleotide kinase (NEB, MA) and separated on 12% sequencing gels and subjected to autoradiography. RNA digestions were also monitored by analysis on RNA chips (Agilent Bioanalyzer). To map the RNase L cleavage site in svRNA3, the RNA product was converted to cDNA using ExactSTART Small RNA Cloning Kit (EPICENTRE Biotechnologies) and sequenced. Briefly, the RNAs were tailed with polyA polymerase and converted to cDNA using oligo-dT(12-18) and MMLV reverse transcriptase (Epicentre Biotechnologies) and sequenced.
Additional plasmids, reagents, and transfection
Plasmids pEF-TAK Flag-RIG-I, pEF-TAK Flag-MDA5, recombinant full-length (RIG-I) and N-RIG (encoding amino acids 1-228) were described previously (Saito et al. 2007, 2008). In vitro transcribed HCV RNA (1 μg) or svRNAs (30 pmol or as indicated) were transfected using Lipofectamine 2000 as per maufacturer's protocol. Cells were transfected with 5 μM 2-5A complexed with Lipofectamine 2000 (Invitrogen) or poly(rI:rC) (1 μg/mL) complexed with Fugene 6 (Roche) as described previously (Malathi et al. 2007). For IFN-β promoter assays, Huh7 cells (2 × 105 per well in six-well plates) were transiently transfected with hIFN-β-luc (1 μg) encoding firefly luciferase cDNA under the control of the human IFN-β promoter (Sumpter et al. 2005) and a Renilla luciferase control plasmid (pRL-TK) (0.1 μg). After 24 h, 1 μg HCV RNA segment, 30 pmol of HCV svRNA3 or its derivatives, were complexed with lipofectamine 2000 and transfected. Control cells were treated with lipofectamine 2000 only. After 18 h, samples were subjected to dual luciferase assays (Promega).
Expression and purification of Flag-RIG-I and Flag-MDA5
293T cells (4 × 106) were transfected using lipofectamine 2000 (Invitrogen) with 10 μg of FLAG-tagged human RIG-I (pEF-BOS Flag-RIG-I) or human MDA5 (pEF-BOS Flag-MDA5) or vector alone. At 48 h post-transfection, cells were lysed and Flag-RIG-I or Flag-MDA5 immunoprecipitated as described (Sumpter et al. 2005; Plumet et al. 2007). To purify Flag-RIG-I or Flag-MDA5 proteins, the immunoprecipitated beads were incubated with 100 μg/mL of Flag peptide in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl for 20 min at 25°C. The Flag-tagged proteins were concentrated using Microcon centrifugal devices with a 30-kDa MWCO (Millipore) and the purity of the protein determined by SDS/polyacrylamide gel electrophoresis.
Cloning and sequencing of svRNAs
The small RNAs generated by RNase L-digestion of HCV RNA (pooled) (16 μg) were added to the immunoprecipitated Flag-RIG-I or Flag-MDA5 (1–2 μg) in 100 μL final volume. The mixtures were stirred for 4 h at 4°C, the complex was collected by a brief (30 sec) centrifugation at 2000 g. The beads were washed twice in the same buffer (500 μL) containing 100 mM NaCl. The bound RNAs were recovered after acid phenol extraction using the mirVana miRNA Isolation Kit (Ambion) and cloned using the miRCat-33 microRNA Cloning Kit Integrated DNA Technologies (IDT). The 3′ cloning linkers were ligated to small RNA species in preparation for cDNA synthesis and amplification. Reverse transcription of the linkered RNA species was followed by PCR amplification and cloning of the PCR the amplicons using TOPO-TA Cloning kit (Invitrogen). Plasmid DNA was prepared and sequenced to identify the HCV RNA fragments. Subsequently, the precise ends of the fragments were determined by comparing to the RNase L-mediated cleavage sites in HCV H77 RNA (Han et al. 2004).
RIG-I binding and activation assays by gel-shift analysis and partial trypsin digestion
Complex formation between 10 pmol of purified N-RIG (RIG-I amino acids 1–228, control) or full-length RIG-I (FL) and 6 pmol of indicated RNAs was determined by incubating for 15 min at 37°C in binding buffer (20 mM Tris-HCl pH 8.0, 1.5 mM MgCl2, 1.5 mM dithiothreitol), followed by electrophoresis on a 2% agarose gel and staining with Sybr Green II RNA Gel stain kit (Lonza) (Saito et al. 2008). The gel-shift was visualized using a UV illuminator (302 nm) with a Sybr Green detection filter. Effect of RNA on RIG-I activation was determined by limited trypsin digestion of the RIG-I /RNA complex. The complex formed between 15 pmol of purified RIG-I protein and increasing amounts (3, 6, 15, or 30 pmol) of polyU/UC or svRNA3 containing the indicated ends was digested with trypsin for 15 min at 37°C. After inactivation of trypsin, one-tenth of the reaction mix was separated on 4%–15% gradient SDS polyacrylamide gel and silver-stained (Saito et al. 2008; Takahasi et al. 2008).
ATPase activation assays
ATPase assays were performed in helicase buffer (25 mM Tris-HCl, pH 7.4, 3 mM dithiothreitol) in the presence of 2 mM ATP, 3 mM MgCl2 as described (Gee et al. 2008). The standardized reactions contained 225-nM full-length Flag-RIG-I in a 20 μL reaction at 37°C typically ranging from 5 to 90 min. Reaction samples were stopped by rapid dilution (20-fold) in acidic malachite green solution (Cytoskeleton) supplemented with 10 mM EDTA and incubated for 15 min, and the absorbance was determined at 650 nM.
Western blots
Expression of Flag-hRNase L (48 h post-transfection) and HCV core protein (48 and 96 h post-electroporation) was determined on immunoblots using anti-FLAG monoclonal antibody (Sigma-Aldrich) or mouse monoclonal anti-HCV core antibody (Affinity Bioreagent, C7-50). Levels of expression of RNase L in Huh7, Huh7.5, and DU145 cells were determined on immunoblots using 30 μg of total cell lysates probed with anti-hRNase L monoclonal antibody (Dong and Silverman 1995). All secondary antibodies were purchased from Cell Signaling. Immunoreactive bands were detected using ECL reagents (GE Healthcare).
Hydrodynamic tail vein injections of mice
Two hundred μg of poly-U/UC (5′-p3/3′-OH) or svRNA3 (5′-OH/3′-p) RNA in PBS (50 μL) were mixed with 40 μL of transfection reagent (Altogen) and incubated 15–20 min at room temperature. A transfection enhancer reagent (10 μL) was added, vortexed gently, and incubated 10 min at room temperature. Two mL of 5% glucose was added and the solution was injected into the tail vein (Saito et al. 2008).
Immunohistochemistry
Mouse livers fixed in 4% paraformaldehyde were sectioned and immunostained using 1:1000 dilution of anti-mouse ISG54 antibody (provided by Dr. G. Sen, Cleveland Clinic) by the histology core at Cleveland Clinic. The sections were counterstained with hematoxylin. Liver biopsies recovered from patients with chronic hepatitis C virus infection (viral genotype 1b) were processed for immunostaining using monoclonal antibody specific to RNase L (Dong and Silverman 1995), OAS1 monoclonal antibody (a kind gift from Dr. Shawn Iadonato, Kineta, Inc. Seattle, WA), and polyclonal anti-NS5A (a gift from Dr. Jin Ye, University of Texas Southwestern Medical Center).
Quantitative RT-PCR analysis
Mouse liver RNA was extracted from tissue soaked in RNAlater reagent (Ambion) using RNeasy kit (Qiagen). One-step quantitative RT-PCR was performed using Applied Biosystems TaqMan Universal PCR master mix containing gene-specific primers for mouse Ifnb, rig-i, and isg56 and TaqMan probe (sequences shown in Supplemental Table S1). PCR was performed with an Applied Biosystems 7500 instrument and all data were presented as relative expression units after normalization to Gapdh mRNA.
ELISAs
Murine IFN-β from WT, Rig-i−/− mice, and culture supernatants derived from MEFs were measured by using ELISA kits purchased from PBL Biomedical Laboratories.
Detection of svRNA3 in intact cells
Huh7.5 cells (1 × 107) were electroporated with 30 μg of in vitro transcribed full-length HCV 1a RNA, RNA corresponding to nt 8703–9416 or mock treated using 0.4-cm gap cuvette (0.22 kV, 960 μF) and Gene Pulsar II from Bio-Rad. After 24 h, 8 μg of plasmid Flag-hRNase L was transfected using Fugene 6 reagent as per manufacturer's protocol. After 48 h (72 h post-electroporation), cells were treated with IFN-β (1000 IU/mL) and incubated for another 18 h. Poly(rI:rC) (1 μg/mL) or 5 μM of 2-5A was transfected using Fugene6 or lipofectamine 2000 reagent, respectively, for 6 h. Cell lysates were prepared from aliquots of samples for immunodetection of Flag-hRNase L and HCV core protein. Total RNA was isolated using the TRIZOL reagent. To monitor RNase L cleavage of RNA, RNA (4 μg) was separated and analyzed on RNA chips (Agilent BioAnalyzer) to monitor activation of RNase L. Small RNAs (<200 nt) were purified using a solid-phase fractionation method (mirVana miRNA Isolation Kit, Ambion). Small RNA (150 μg) from different treatments and 1 ng of svRNA3 (5′-OH/3′-p) was electrophoresed on 8% PAGE. The RNAs were transferred to BrightStar-Plus membrane (Ambion) and immobilized by UV cross-linking. Probe was synthesized using miRNA StarFire System corresponding to the sequence of svRNA3 (5′-gaaccaaccggacaagtccagccggccagcggccgctattggag-3′) with [α-32P]-dATP (6000 Ci/mmol, Perkin Elmer). Hybridization was done at 42°C in ULTRAhyb-Oligo Hybridization buffer (Ambion) for 18 h. RNA samples (10 μg) and 100 ng of svRNA3 (5′-OH/3′-p) were stained with Gel Star Nucleic acid stain (Lonza) to compare loading of the samples.
Ethics statement
Liver biopsies were obtained with informed, written consent from patients with chronic HCV infection with full approval from the University of Washington Institutional Research Board (IRB) on Human Subjects. Mouse experiments were conducted in accordance with guidelines set forth by the institutional Animal Care and Use Committee of the University of Washington.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants 1RC1A1086041 and CA44059 and from the Mal and Lea Bank Chair (to R.H.S.); Research Scholar Grant RSG-02-063-01-MBC from the American Cancer Society (to D.J.B.); and NIH grants DA024563 and AI060389 (to M.G., T.S., and N.C.). We thank Ann Palmenberg and Jean-Yves Sgro (Madison, WI) for providing a secondary structure prediction of HCV H77 RNA. We thank Babal Kant Jha (Cleveland) for preparing the RNase L and 2-5A and Craig P. Chappell (Seattle) for expert advice and assistance in hydrodynamic tail vein injections of mice.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2244210.
REFERENCES
- Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med 144: 705–714 [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Marcotrigiano J, Rice CM 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J Virol 77: 3181–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazenave C, Uhlenbeck OC 1994. RNA template-directed RNA synthesis by T7 RNA polymerase. Proc Natl Acad Sci 91: 6972–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JL, Carroll SS, Kuo LC 1996. Stoichiometry of 2′,5′-oligoadenylate-induced dimerization of ribonuclease L. A sedimentation equilibrium study. J Biol Chem 271: 3979–3981 [DOI] [PubMed] [Google Scholar]
- Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA–RNA interaction with upstream RNA sequences in NS5B. J Virol 82: 9008–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Silverman RH 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem 270: 4133–4137 [DOI] [PubMed] [Google Scholar]
- Dong B, Xu L, Zhou A, Hassel BA, Lee X, Torrence PF, Silverman RH 1994. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J Biol Chem 269: 14153–14158 [PubMed] [Google Scholar]
- Floyd-Smith G, Slattery E, Lengyel P 1981. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate–dependent endonuclease. Science 212: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Gee P, Chua PK, Gevorkyan J, Klumpp K, Najera I, Swinney DC, Deval J 2008. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J Biol Chem 283: 9488–9496 [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci 103: 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Barton DJ 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8: 512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Wroblewski G, Xu Z, Silverman RH, Barton DJ 2004. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J Interferon Cytokine Res 24: 664–676 [DOI] [PubMed] [Google Scholar]
- Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH 1993. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J 12: 3297–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Gale M Jr 2009. Intracellular innate immune cascades and interferon defenses that control hepatitis C virus. J Interferon Cytokine Res 29: 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian AG, Justesen J 2007. The human 2′-5′oligoadenylate synthetase family: Unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie 89: 779–788 [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Brown RE, Kerr IM 1977. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature 268: 537–540 [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23: 19–28 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Kerr IM, Brown RE 1978. pppA2′p5′A2′p5′A: An inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci 75: 256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, et al. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med 203: 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Shin H, Wimmer E, Paul AV 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol 78: 10865–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. 2006. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci 103: 6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Paranjape JM, Bulanova E, Shim M, Guenther-Johnson JM, Faber PW, Eling TE, Williams BR, Silverman RH 2005. A transcriptional signaling pathway in the IFN system mediated by 2′-5′-oligoadenylate activation of RNase L. Proc Natl Acad Sci 102: 14533–14538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M Jr, Silverman RH 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448: 816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Palmenberg AC, Sgro J 1997. Topological organization of picornaviral genomes: Statistical prediction of RNA structural signals. Seminars in Virology 8: 231–241 [Google Scholar]
- Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE 2: e279 doi: 10.1371/journal.pone.0000279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C 2010. RIGorous detection: Exposing virus through RNA sensing. Science 327: 284–286 [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci 104: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Silverman RH 2007. Viral encounters with OAS and RNase L during the IFN antiviral response. J Virol 81: 12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM 1983. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol 46: 1051–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M Jr 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79: 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi T, Nagano-Fujii M, Akutsu M, Kadoya H, Ohgimoto S, Ishido S, Hotta H 2004. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol 85: 959–969 [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M Jr, Inagaki F, Fujita T 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell 29: 428–440 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT 2004. Structural basis for recognition of 2′,5′-linked oligoadenylates by human ribonuclease L. EMBO J 23: 3929–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur CS, Jha BK, Dong B, Das Gupta J, Silverman KM, Mao H, Sawai H, Nakamura AO, Banerjee AK, Gudkov A, et al. 2007. Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc Natl Acad Sci 104: 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y 2010. How the noninflammasome NLRs function in the innate immune system. Science 327: 286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8: 824–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Evans DJ, Simmonds P 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol 85: 3037–3047 [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11: 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner DH, McCauley JW, Skehel JJ, Kerr IM 1981. Interferon action–sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289: 414–417 [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB 2005. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T 2010. Recognition of viral nucleic acids in innate immunity. Rev Med Virol 20: 4–22 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
- You S, Stump DD, Branch AD, Rice CM 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol 78: 1352–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Hassel BA, Silverman RH 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72: 753–765 [DOI] [PubMed] [Google Scholar]
- Zhou A, Molinaro RJ, Malathi K, Silverman RH 2005. Mapping of the human RNASEL promoter and expression in cancer and normal cells. J Interferon Cytokine Res 25: 595–603 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]