Abstract
S-adenosyl-(L)-homocysteine (SAH) riboswitches are regulatory elements found in bacterial mRNAs that up-regulate genes involved in the S-adenosyl-(L)-methionine (SAM) regeneration cycle. To understand the structural basis of SAH-dependent regulation by RNA, we have solved the structure of its metabolite-binding domain in complex with SAH. This structure reveals an unusual pseudoknot topology that creates a shallow groove on the surface of the RNA that binds SAH primarily through interactions with the adenine ring and methionine main chain atoms and discriminates against SAM through a steric mechanism. Chemical probing and calorimetric analysis indicate that the unliganded RNA can access bound-like conformations that are significantly stabilized by SAH to direct folding of the downstream regulatory switch. Strikingly, we find that metabolites bearing an adenine ring, including ATP, bind this aptamer with sufficiently high affinity such that normal intracellular concentrations of these compounds may influence regulation of the riboswitch.
Keywords: riboswitch, S-adenosylhomocysteine, X-ray crystallography, isothermal titration calorimetry, chemical probing
INTRODUCTION
S-adenosyl-(L)-methionine (SAM), one of the most utilized molecules in biology, serves as the primary methyl group donor in a diverse array of chemical reactions (Fontecave et al. 2004). Because of its central role in cellular homeostasis, the intracellular SAM concentration is strictly controlled by a variety of regulatory mechanisms. In many bacteria, the primary means of regulation are SAM-responsive riboswitches. These are cis-acting RNAs commonly found in the 5′-untranslated region (UTR) of bacterial mRNAs that directly bind SAM with high affinity and specificity to control transcription or translation (Grundy and Henkin 1998; Mironov et al. 2002; Winkler et al. 2003; Kazanov et al. 2007). The importance of this mode of SAM regulation in bacteria is underscored by the discovery that there are at least six classes of SAM sensitive regulatory elements distributed across a broad spectrum of bacterial clades (Wang and Breaker 2008; Weinberg et al. 2010).
The by-product of methylation by SAM, S-adenosyl-(L)-homocysteine (SAH) is a potent competitive inhibitor of many SAM-utilizing enzymes (Ueland 1982). At elevated concentrations, SAH is toxic and must be degraded by SAH hydrolase (ahcY) to adenosine and homocysteine as part of the SAM regeneration cycle (Loenen 2006). Recently, riboswitches that up-regulate expression of SAH hydrolase or 5-methyltetrahydrofolate-homocysteine methyltransferase (metH), another SAM recycling enzyme, were identified in proteobacteria and actinobacteria (Wang et al. 2008). These riboswitches promote expression of the mRNA via an SAH-stabilized conformation that either prevents formation of a rho-independent transcriptional terminator or exposes the Shine-Dalgarno sequence to permit ribosome binding (Wang et al. 2008). Intriguingly, the SAH riboswitch has been shown to be capable of regulating gene expression in human cells as an engineered ribosomal frame-shifting pseudoknot (Chou et al. 2010).
To properly regulate SAM metabolism, both SAM- and SAH-binding riboswitches must discriminate between these closely related compounds with high fidelity. The SAM-I riboswitch achieves a 550-fold level of discrimination for SAM over SAH through an electrostatic mechanism in which carbonyl oxygens in two universally conserved A–U pairs are placed adjacent to SAM's positively charged sulfonium ion (Montange and Batey 2006; Montange et al. 2010). Two other classes of SAM-sensing riboswitches whose structures have been solved in complex with SAM display similar mechanisms of selective binding (Gilbert et al. 2008; Lu et al. 2008). SAH riboswitches must be able to achieve equivalent levels of discrimination between these compounds. Comparison of the binding affinities of the Dechloromonas aromatica metH SAH riboswitch for SAM and SAH indicates that this RNA preferentially binds SAH with a 1000-fold higher affinity (Wang et al. 2008).
To determine the basis for ligand recognition and specificity by this regulatory RNA, we have solved the crystal structure of the SAH riboswitch aptamer domain from the plant pathogen Ralstonia solanacearum (Rso) in complex with SAH. This riboswitch is found upstream of ahcY and appears to control expression at the transcriptional level by promoting formation of a transcriptional antiterminator. The structure of its aptamer domain reveals an uncommon pseudoknot architecture with SAH bound to a cleft formed at the interface between the three helices that define the conserved secondary structure. Discrimination arises from packing of SAH's sulfoether moiety against the RNA such that SAM's additional methyl group creates a steric clash that precludes high affinity binding. Features of the RNA structure not predicted by a previous study (Wang et al. 2008) were validated by characterizing the SAH-binding activity of a set of mutants using isothermal titration calorimetry (ITC). Chemical probing and calorimetric analyses suggest that the free state of the riboswitch aptamer domain can access bound-like conformations, particularly at lower temperatures or higher magnesium concentrations. SAH binding stabilizes the overall RNA structure suggesting a regulatory mechanism in which SAH induces limited local conformational changes, but significantly stabilizes helical elements of the aptamer to preclude an alternative secondary structure that involves the 3′-helix of the aptamer. Regulation by SAH may be further complicated by the ability of the RNA to bind ATP with an equilibrium dissociation constant (KD,app) >100-fold lower than its typical cellular concentration (Bennett et al. 2009), suggesting that adenine-bearing metabolites may act as competitive inhibitors of this riboswitch in vivo.
RESULTS
RNA crystallization and structure determination
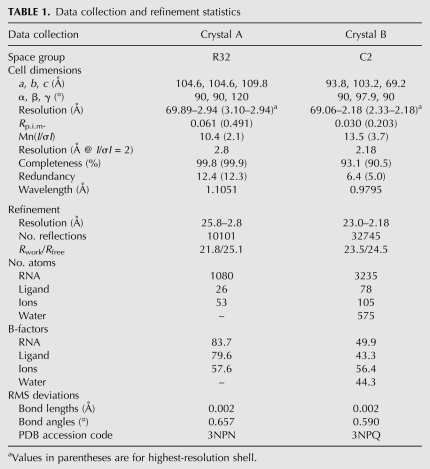
To obtain crystals of the riboswitch–SAH complex, we screened 10 of the smallest phylogenetic variants, all of which lacked a nonconserved helix (P3) that is absent from 34% of known sequences (Wang et al. 2008). A sequence from Rso that regulates the ahcY gene yielded initial hits in crystallization trials and this sequence was modified to produce crystals that improved diffraction quality. Several modifications of this RNA were made to promote crystallization and to solve the structure. The first contains a truncated P2 helix and a U13C point mutation that yielded diffraction to 2.8 Å (Table 1, Crystal A) and a suitable iridium (III) hexamine derivative for obtaining phase information. Higher resolution data (2.18 Å) (Table 1, Crystal B) were obtained by changing the tetraloop sequence engineered in L2 from GAAA to GAGA (sequence shown in Fig. 1A). Both sets of data yielded electron density maps of sufficient quality to build a model that contained the SAH ligand and all residues of the RNA except for four residues involved in a domain swap (nucleotides 39–42); crystallographic statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
FIGURE 1.
Structure of the SAH aptamer. (A) The secondary structure of the modified sequence of the Rso SAH riboswitch is shown with paired regions P1, P2, and P4 (blue), joining region J4/2 (orange), J2/1 (green), and J1/4 (cyan). The SAH ligand is depicted as a red icon and nucleotides that are disordered or involved in a domain swap are colored gray. Base interactions are represented using the notation of Leontis and Westhof (2001). (B) Cartoon representation of the aptamer tertiary architecture is shown with the color scheme used in A. SAH is shown in red and the dots represent the van der Waals surface of SAH. Gray dots depict nucleotides that could not be modeled into the electron density map. (C) The tertiary structure from B rotated 90° to emphasize that the adenine ring from SAH participates in the P2/P4 coaxial stack.
To verify that these alterations from the wild-type sequence did not significantly perturb the binding activity, we measured the affinity of these RNAs for SAH by isothermal titration calorimetry (ITC). The sequence containing all of the alterations used to obtain the highest resolution data set has a slightly higher affinity for SAH as compared to wild-type RNA (32 versus 18 nM for the wild-type and crystal sequences, respectively). These affinities are similar to those measured for SAH riboswitches from D. aromatica and Pseudomonas syringae, as measured by in-line probing (Wang et al. 2008), demonstrating that the binding characteristics of the Rso SAH riboswitch are representative of other phylogenetic variants.
Structure of the SAH riboswitch bound to SAH
The conserved secondary structure of the SAH riboswitch adopts a fold classified as an “LL-type” pseudoknot (Fig. 1A; Han and Byun 2003), a far more infrequent type than the classic “H-type” pseudoknot employed by the SAM-II (Gilbert et al. 2008) and pre-Q1 (Kang et al. 2009; Klein et al. 2009; Spitale et al. 2009) riboswitches. The three helices are arranged such that P2 and P4 coaxially stack and P1 lies perpendicular to them (Fig. 1B,C). The coaxial stack between P2 and P4 is facilitated by the formation of helix P2b, which is largely comprised of noncanonical base-pairing interactions between highly conserved nucleotides (U14•A29, C16–G31, A17•C28, and A18•C29) (Fig. 1A) in joining regions J2/1 and J4/2. Adenosine residues from J1/4 dock into the minor groove of P1, providing a structural anchor that stabilizes the overall topology, a common means of stabilizing RNA tertiary architecture (Nissen et al. 2001; Xin et al. 2008). The only other known structure of an LL-type pseudoknot is found in the hepatitis delta virus ribozyme, but a second internal pseudoknot (P1.1) further constrains the organization of the helices with respect to one another such that they are all oriented parallel to one another (Ferre-D'Amare et al. 1998). Thus, the SAH riboswitch aptamer likely represents the true architecture of an isolated LL-type pseudoknot.
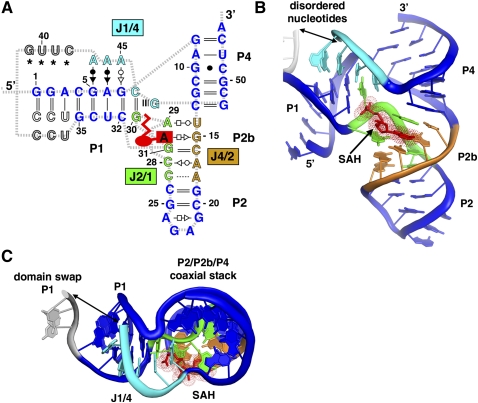
In the crystal lattice, each RNA is involved in a domain swap with a neighboring molecule such that the last three nucleotides on the 3′-side of the P1 helix are paired with the 5′-side of the P1 helix of the adjacent RNA (Fig. 2A). Because of the topological constraint of connecting P1 to P4, the fraying of P1 that permits the domain swap may reflect its structure in solution. To address this, we probed the RNA's structure using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Merino et al. 2005). This method interrogates the RNA structure using N-methylisatoic anhydride (NMIA), which reacts with 2′-hydroxyl groups within residues that can sample the C2′-endo sugar-pucker conformation (Gherghe et al. 2008). We probed both the wild-type and crystallized Rso aptamer sequences in the presence and absence of SAH. Within the P1 helix of both RNAs, the 2′-hydroxyls of the last two base pairs are more reactive than the rest of the helix, indicating the end of the 5′-end of the helix is frayed (Fig. 2B). These data reveal the P1 helix is partially melted, thereby increasing the effective length of the J1/4 linker. Notably, phylogenetic variants show highly divergent lengths of J1/4, with the majority containing an additional helix, P3, within J1/4, which may further facilitate pseudoknot formation (Wang et al. 2008).
FIGURE 2.
Domain-swapped dimer of the SAH riboswitch. (A) Nucleotides 36–38 form base-pairing interactions with nucleotides 1–3 of an adjacent molecule. Nucleotides 39–42 could not be clearly built into the electron density map, but the breaks in the chain are sufficiently close (double arrow) to allow for these nucleotides. Inspection of the lattice indicated no other possible way to connect two breaks with a spacing of <20 Å distance between them. Note that the architecture of the domain swap is a Holliday junction with no unpaired nucleotides in the four-way junction. (B) NMIA chemical probing of a crystallization construct (left) and the wild-type RNA (right) sequence shows that the last two nucleotides in P1 are frayed in solution regardless of SAH binding. Nucleotides 32–38 from the left panel correspond to positions 46–52 in the right panel.
SAH recognition and discrimination against SAM
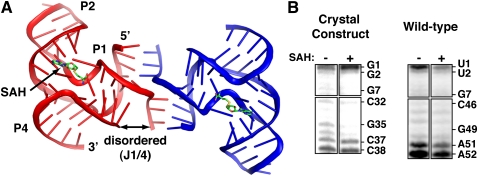
SAH is located within a cleft created by the minor grooves of P2b and P1. The binding site is lined by a cluster of highly conserved residues (>98% conservation) (Wang et al. 2008) that form hydrogen bonding interactions with the adenine ring and the methionine main chain atoms of SAH (Fig. 3A). The adenine ring intercalates between the nucleobases of A29 and C16 and forms a sheared G·A pair with G15 (Fig. 3B). The α-amine of SAH is close to N3 and a nonbridging phosphate oxygen of G30, indicating that this moiety forms a charge–charge interaction at the intermolecular interface. The carboxylate oxygens form hydrogen bonding interactions with the 2′-hydroxyl groups of G47 and G31 (Fig. 3A). It is important to note that the contacts between the homocysteine main chain atoms and RNA are slightly different in the three protomers in the asymmetric unit. Likely, the ligand does not form all of the possible hydrogen bonding interactions with the RNA as shown, but rather only a majority at any given time.
FIGURE 3.
SAH recognition. (A) The SAH binding pocket is shown with a color scheme consistent with Figure 1 with the exception that SAH is colored salmon. Hydrogen bonds are shown as black dotted lines and the black arrow shows the distance between the sulfur on SAH and O4′ of A29. (B) This orientation shows the top-down view of SAH in the binding pocket emphasizing the sheared G15·ASAH pair along with the hydrogen bonding interactions that G15 makes with the phosphate backbone. The orange mesh around SAH represents a “prime-and-switch” map, a means of reducing model bias (Terwilliger 2004), contoured at 1.0 σ to demonstrate that the presence and placement of the ligand is strongly supported by the crystallographic data. (C) The left panel shows SAH bound in the deep cleft created by the Rso riboswitch aptamer and the right panel shows SAH bound to a similar cleft created by the Mesembryanthemum crystallinum phenylpropanoid and flavonoid O-methyltransferase (Kopycki et al. 2008). The surface representation shown in blue depicts the van der Waals surface of the RNA or protein.
In the observed structure, SAH is 74% solvent inaccessible, with the 2′-OH, 3′-OH, and sugar edge of adenine projecting away from the RNA. Previously, it was found that alteration of these groups in SAH analogs severely impairs binding (Wang et al. 2008). We speculate that these effects reflect the sensitivity of the RNA to a change in sugar pucker of SAH as a result of 2′- or 3′-deoxy substitutions or weakening of base stacking by a 3-deaza substitution or addition of a 2-amino group. Another analog tested contains an arabinose sugar rather than ribose (Wang et al. 2008). While this sugar maintains the C3′-endo pucker, as observed in 9-β-D-arabinofuranosyladenine (Sundaralingam 1975), it also is severely impaired in binding. Modeling of this sugar into the structure indicated that the 2′-hydroxyl group would sterically clash with A29, explaining why this compound binds the riboswitch poorly. Interestingly, unlike the ligand binding pockets of other riboswitches that are extensively buried within the core of the RNA, this site is reminiscent of surface-accessible active sites often found in protein enzymes that use SAM as a cofactor (Fig. 3C; Kopycki et al. 2008).
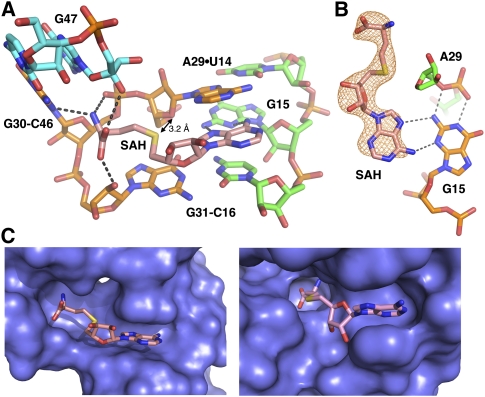
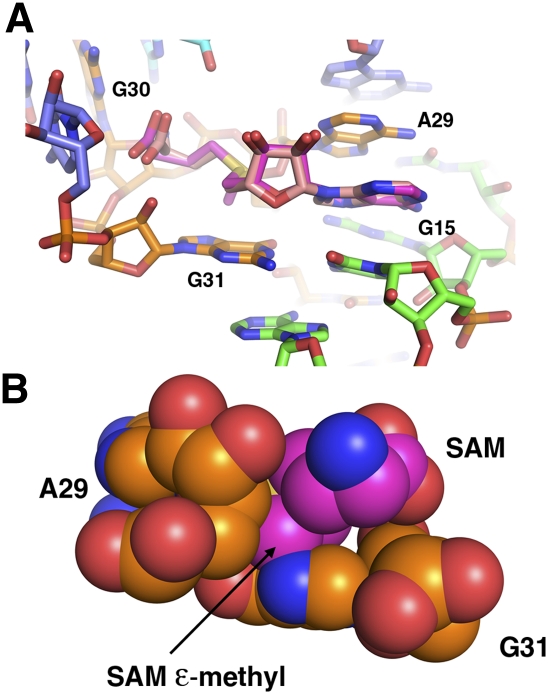
Discrimination between SAH and SAM is mediated by interactions between the ligand's sulfoether moiety and the RNA backbone. The sulfur atom of SAH is in van der Waals contact with O4′ of A29 (3.2 Å) (Fig. 3A). SAM chemically differs from SAH only by the presence of a methyl group on the sulfur atom and the associated positive charge (sulfonium cation). To illuminate how SAM might occupy this binding pocket, we superimposed it upon SAH (Fig. 4). If SAM binds this RNA in the same conformation as SAH, a steric clash would occur between the methyl group on SAM and the ribose C4′ of A29 and N7 of G31. This suggests that the primary mechanism for discrimination between SAH and SAM is by steric occlusion of the latter.
FIGURE 4.
Superposition of SAH and S-adenosylmethionine (SAM). (A) Close-up of the ligand binding pocket with SAH (mauve) and SAM (magenta) aligned. The chemical difference between the two compounds is the presence of a methyl group (epsilon) on SAM, as well as a positive charge on the sulfur atom. Note the projection of the methyl group toward the base of G31. (B) Back side of the binding pocket, with SAM, A29, and G31 shown as van der Waals spheres. The methyl group of SAM sterically clashes with C4′ of A29 and N7 of G31 if it is superimposed upon SAH, indicating SAH/SAM discrimination is through a steric mechanism.
Mutational analysis verifies base-pairing interactions in the binding pocket
The secondary structure of the SAH riboswitch was predicted from an alignment of 68 nonredundant sequences using the covariation model in CMfinder, an algorithm that predicts base pairs with 79% accuracy (Yao et al. 2006). Overall, the structure agrees well with this analysis. However, there are base pairs within the core that could not be accurately predicted due to a lack of sufficient covariation. For example, two predicted base pairs in the P4 helix (U14–G47 and G15–C46) are not present in the current structure. Instead, these four bases participate in interactions that line the binding pocket and form direct contacts with SAH. Moreover, we identified a base pair between C16 and G31 not predicted due to the 100% conservation of these nucleotides.
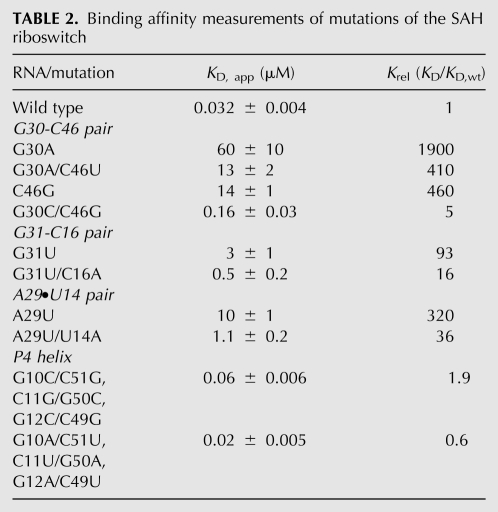
We verified each unpredicted base pairing interaction around the SAH binding pocket using a compensatory mutagenesis strategy with the wild-type Rso SAH aptamer. The apparent binding affinity (KD,app) of each RNA for SAH was measured using ITC (Table 2). Disrupting single point mutations of the G30–C46 and G31–C16 Watson-Crick pairs that flank SAH strongly disrupt binding activity, and in both cases the compensatory mutation yielded a moderate (approximately five- to 10-fold) improvement in SAH affinity. Interestingly, ∼7% of known sequences contain guanosine at both positions 30 and 46, suggesting that a G30·G46 pair is tolerated. The affinity of a C46G mutant was found to be the same as the G30A/C46U construct, consistent with the observed phylogenetic conservation pattern at this position. In one sequence from a Frankia species the C30–G46 transversion is observed. Introduction of this variation in the context of the Rso sequence shows a near wild-type binding affinity. The other base pair flanking the binding pocket, A29–U14, does not form a canonical Watson-Crick base pair, however, transversion of these positions increases the activity by 10-fold over the A29U mutation.
TABLE 2.
Binding affinity measurements of mutations of the SAH riboswitch
Unusually, the G10–C51 pair in the P4 helix is best modeled in a wobbled conformation in all three protomers in the asymmetric unit. In modeling the RNA structure, this cytosine was not assumed to be protonated, but nonetheless consistently adopted a pairing type that requires the cytosine N3 to be protonated in order to establish a hydrogen bond with guanosine O6. Moreover, this pair is part of a stretch of three contiguous pairs in the center of the P4 helix that has near 100% phylogenetic conservation, despite having no obvious role in SAH binding. To test their potential importance, we mutated this region of the P4 helix such that the G10–C51, C11–G50, and G12–C49 base pairs are either transversed or changed to A–U pairs (with purine-pyrimidine orientation preserved). In both cases, the SAH binding affinity is near wild type (Table 2). These data suggest that the identity and orientation of the pairs in the P4 helix is not important for SAH binding, but rather for the regulatory function. We speculate that the G10–C51 wobble may have a critical destabilizing effect on this helix, which facilitates the alternative secondary structure switch involving its 3′-side and downstream sequences.
Magnesium-dependent folding of the riboswitch aptamer
The central feature of riboswitches is the ability to fold into two mutually exclusive structures, dependent upon whether a ligand has bound to the aptamer. Therefore, to further understand SAH-dependent regulation by this RNA, the magnesium- and SAH-dependent folding of the riboswitch must be determined. We can observe the unfolding of this RNA with nucleotide resolution by probing its structure using NMIA chemistry as a function of temperature or magnesium concentration. For example, investigation of the unfolding of the guanine riboswitch aptamer domain in the presence and absence of its effector generated a clear map of the nucleotides involved in a conformational change involved in both ligand burial, as well as directing the downstream secondary structural switch (Stoddard et al. 2008).
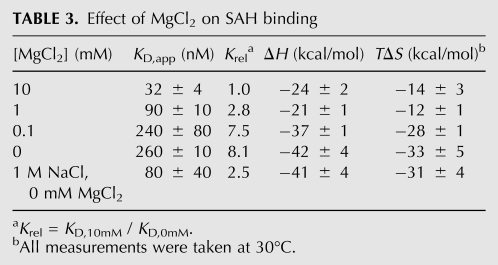
At 20°C, the reactivity pattern of the RNA is mostly unaffected by the presence of magnesium or SAH; the primary exception is J2/1 within the core of the SAH binding pocket. All three helical elements are protected from NMIA reactivity, while the joining regions and L2 show the most reactivity to NMIA. These data indicate that the Watson-Crick paired helical elements of the pseudoknot are established without MgCl2 or SAH, but that the P2b helix and the P2/P4 coaxial stack are not well structured and there is little interhelical organization. The addition of MgCl2 to a physiologically relevant concentration (1 mM MgCl2 in 100 mM NaCl) in the absence of SAH shows a highly localized change in the NMIA reactivity of A29, C28, and C27 (Fig. 5A). This region of the RNA contains two A–C pairs and the U14–A29 pair that comprise the P2b helix. The reactivity of U14, A17, and A18 also decreases, albeit to a lesser extent. Thus, magnesium appears to promote the formation of the P2b helix, one of the primary sites of interaction with SAH. Calorimetric titrations of SAH into the RNA at varying magnesium ion concentrations show that the favorable enthalpic contribution and entropic penalty both diminish with increasing magnesium. These data suggest that the degree of conformational change in the RNA upon SAH binding diminishes with increasing magnesium concentration (Table 3), substantiating a limited magnesium-induced stabilization of the aptamer.
FIGURE 5.
MgCl2 stabilizes the P2b helix. (A) NMIA chemical probing shows that nucleotides in the P2b helix become protected as a function of MgCl2 concentration. Addition of SAH in the absence of MgCl2 results in a similar and more intense protection pattern (right). (B) Cobalt (III) hexamine is found adjacent to the A18•C27 and A17•C28 base pairs in the crystal structure, suggesting the presence of a metal cation binding site. Hydrogen bonds are shown as gray dotted lines.
TABLE 3.
Effect of MgCl2 on SAH binding
Inspection of the crystal structure reveals a highly ordered cobalt (III) hexammine cation positioned within the major groove adjacent to the A17•C28 and A18•C27 pairs (Fig. 5B), consistent with the observation that metal binding facilitates P2b formation. Notably, high monovalent cation concentrations (1 M NaCl) allow the RNA to bind SAH with roughly the same affinity as that observed under 1 mM MgCl2 (Table 3). While trace contaminating divalent cations in the solution could account for this result, their concentration would have had to exceed 30 μM (the concentration of RNA in the experiment) to have influenced the results. These data suggest that P2b contains a nonspecific high affinity metal ion binding site occupied by a magnesium ion under physiological conditions. Since cobalt (III) hexammine does not completely mimic hexa-hydrated magnesium, with respect to how it specifically interacts with the RNA (Juneau et al. 2001; Batey and Doudna 2002), magnesium could preferentially occupy a slightly different position at this site. Despite the importance of metal ions for facilitating local folding of the P2b helix, the addition of SAH in the absence of magnesium still induces strong NMIA protections at these positions in P2b, demonstrating specific magnesium binding is not essential for SAH recognition.
SAH-dependent folding of the riboswitch
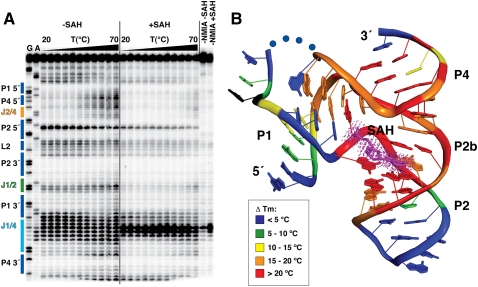
To examine the SAH-dependent folding of the riboswitch, NMIA probing was performed between 20°C and 70°C at 6 mM MgCl2 for the wild-type Rso ligand binding domain (Fig. 6A). For each nucleotide position, the band intensity versus temperature can be fit to a two-state binding model such that the transition melting temperature (TM) can be confidently calculated for nearly every position in the RNA using established methods (Wilkinson et al. 2005; Stoddard et al. 2008, 2010). In the unliganded RNA, nucleotides in J4/2, J1/4, and the P4 helix display clear melting transitions, with an average TM of 49 ± 2°C (Fig. 6A). In the presence of saturating SAH (500 μM), all of these nucleotides show a marked increase in their apparent melting transition (≥70°C). Thus, SAH has a significant stabilizing effect not only upon the RNA structure around the binding pocket, but also secondary structural elements including the P4 helix (Fig. 6B).
FIGURE 6.
Temperature-dependent NMIA reactivity of the SAH riboswitch aptamer domain. (A) Sequencing gel with secondary structure elements labeled and colored as in Figure 1. Sequencing lanes are labeled G and A for guanosine and adenosine positions, respectively. The temperature gradient is depicted as a black triangle above the gel with each lane representing a 5°C increase in temperature from 20 to 70°C. The left half of the gel shows reactions without SAH (−SAH), while the right side shows reactions with SAH (+SAH). Control reactions in which the NMIA reactant was omitted are labeled −NMIA −SAH (without SAH) and −NMIA +SAH (with SAH). (B) The difference in TM (TM,+SAH − TM,−SAH) are colored onto the tertiary structure. A negative value is shown in black, and a difference of <5°C, 5–10°C, 10–15°C, 15–20°C, or >20°C is colored blue, green, yellow, orange, or red, respectively. Blue dots correspond to nucleotides that could not be modeled into the electron density, and SAH is shown in magenta.
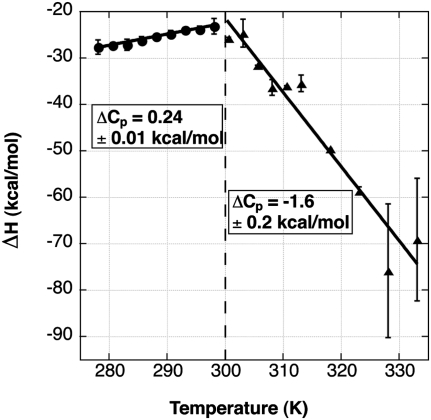
To provide another means of assessing ligand associated conformational changes in the RNA, we used ITC to measure the temperature dependence of the binding. In particular, a temperature-dependent heat capacity change, ΔCP = (ΔH/ΔT), can be diagnostic of coupled equilibria involving conformational flexibility of the unbound macromolecule (Ladbury 1995; Bruzzese and Connelly 1997). An observed temperature dependence of ΔCP is evidence for changes in how the ensemble of free-state conformers is populated as the temperature changes. Furthermore, several studies have generated a relationship between the sign of ΔCP and the polarity of surface area buried upon binding; positive and negative values are correlated with polar and nonpolar groups buried from water, respectively. Significant differences between measured and calculated values are generally associated with conformational changes due to an “induced fit” binding mechanism (Murphy and Freire 1992; Spolar and Record 1994).
Measurement of ΔH over a broad temperature range (5°C–60°C) by ITC reveals that the SAH riboswitch has two separate temperature regimes for heat capacity (Fig. 7). At low temperatures, a ΔCP,obs of 0.24 ± 0.01 kcal/mol·K is observed (Fig. 7) suggesting that the largest contribution to the heat capacity change is the burial of the polar groups on the amino acid main chain atoms from bulk solvent. Our experimental value is reasonably close to the ΔCP,calc obtained from the methods by Spolar and Record (1994) (0.085 kcal/mol·K) and Murphy and Freire (1992) (0.1 kcal/mol·K) that use the change in solvent accessible surface area assuming a rigid binding mechanism (no conformational changes during binding). The ∼2.5-fold discrepancy between ΔCP,obs and ΔCP,calc is likely due to a combination of two factors. First, the model compound data sets used to calculate ΔCP,calc behave differently than naturally occurring RNA–ligand interactions and may not be sufficient for accurate calculation of ΔCP,calc for this application (Mikulecky and Feig 2006). Second, the RNA likely undergoes subtle conformational changes during SAH binding and does not fit a strict lock-and-key binding mechanism. The close agreement between ΔCP,obs and ΔCP,calc, under a low temperature range, supports the SHAPE data, suggesting that at temperatures below 25°C and under a saturating magnesium concentration (>6 mM) the free RNA frequently samples bound-like states, which are represented by the crystal structure, and binding induces a limited conformational change.
FIGURE 7.
Isothermal titration calorimetry reveals two different heat capacity regimes. The graph shows the change in binding enthalpy (ΔH, kcal mol−1) plotted against temperature (K). A trend line is drawn through the data in each temperature regime, and the slope of this line defines the heat capacity change (ΔCP).
ITC data taken between 25°C and 60°C (Fig. 7) reveal an increasingly enthalpically driven binding event and the ΔCP,obs = −1.6 ± 0.2 kcal/(mol·K), suggesting that at higher temperatures burial of apolar groups is the most significant contribution to the heat capacity change. In light of NMIA chemical probing at temperatures >25°C, we support a model in which, at higher temperatures, the free RNA increasingly populates states in which the P2b helix and the P2/P4 coaxial stack are broken. SAH likely productively interacts with bound-like states of the RNA in which P2b and the P2/P4 stack are formed and stabilizes them, driving the ensemble population of RNA toward structured states. The large, negative value of the observed heat capacity change could thus be explained in part as due to formation of stacking interactions formed in the P2b helix and P2/P4 coaxial stack. This model is consistent with observations of the lysine and SAM-I riboswitches (Garst et al. 2008; Stoddard et al. 2010).
Adenosine analogs can bind the SAH riboswitch
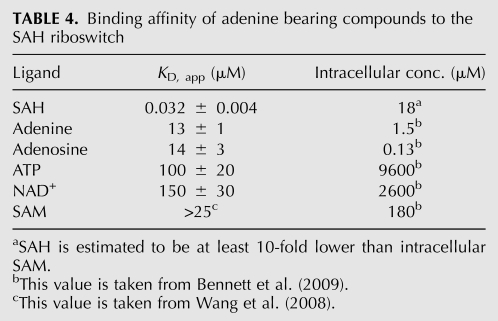
All of the hydrogen bonding interactions between SAH and the RNA are mediated through the adenine ring and the main chain atoms of methionine. Thus, this RNA could be thought of as having two distinct binding pockets, one for the adenine ring and one for the amino acid main chain. To determine whether adenine or methionine productively binds this RNA, we tested the binding of each by ITC. While adenine binds weakly to the RNA (∼10 μM affinity), we detected no binding by methionine or glycine. This is consistent with previous equilibrium dialysis measurements (Wang et al. 2008).
Interestingly, the binding of adenine was significantly tighter than the effective intracellular concentrations of adenine-bearing metabolites in the cell, such as ATP or NAD+ (9.6 or 2.6 mM, respectively) (Bennett et al. 2009). To further explore this issue, we tested the binding of other biologically relevant adenine containing compounds (Table 4). Adenine and adenosine bind with nearly identical affinity (13 and 14 μM, respectively). However, the measured intracellular concentration for Escherichia coli has been shown to be around 1.5 μM and 130 nM, respectively (Bennett et al. 2009). Therefore, these compounds are unlikely to influence gene regulation by SAH.
TABLE 4.
Binding affinity of adenine bearing compounds to the SAH riboswitch
Conversely, the measured KD for ATP binding is observed to be 100 μM, significantly lower than the intracellular concentration of ATP. This suggests that in vivo ATP could act upon the SAH riboswitch. NAD+ also has a sufficiently high affinity for the riboswitch such that it too might be expected to bind the riboswitch in the cell (Table 4). In addition, the measured affinity for SAM is observed to be >25 μM (Wang et al. 2008), and we estimate that it is close to our observed ATP binding affinity of 100 μM. These data, however, do not indicate whether binding of these compounds activate gene expression like SAH or act as competitive inhibitors to SAH binding. To provide some insight into this issue, we looked at the ability of adenine to stabilize the RNA, including the P4 helix using NMIA probing. Not surprisingly, we found that adenine binding protects nucleotides in J2/1 from NMIA modification, indicative of productive binding. However, adenine had a marginal effect on the stability of the RNA (the average TM of nucleotides in P4 and J1/2 is 49°C ± 2°C and 52°C ± 3°C in the absence and presence of adenine, respectively (data not shown). Therefore, adenine bearing compounds such as ATP and NAD+ may act as competitive inhibitors of SAH binding. The degree of competitive inhibition by ATP can be calculated using standard definitions for IC50 and competitive binding (Goodrich and Kugel 2007). Assuming that the riboswitch is under thermodynamic control, and given the intracellular concentrations of ATP and SAH (estimated to be 10–100-fold lower than SAM) and their affinities for the SAH riboswitch, we estimate that activation by SAH could be diminished by 10%–40%.
Discussion
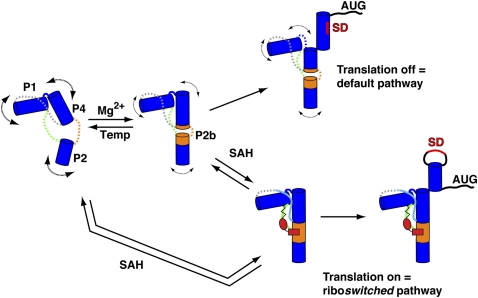
The SAH riboswitch family is unique among riboswitches because it appears to exclusively up-regulate gene expression in response to the buildup of its effector, SAH (Wang et al. 2008). Nonetheless, the mechanism by which the SAH riboswitch transduces ligand binding into a regulatory response is similar to other riboswitches (Wickiser et al. 2005; Fuchs et al. 2006; Ontiveros-Palacios et al. 2008). Following transcription of the aptamer domain, the riboswitch reaches a folding branch point, each leading to the formation of one of two mutually exclusive hairpin structures in the downstream expression platform (Fig. 8). The above structural and biophysical studies represent this time-point in the life of the riboswitch, which often has a programmed pause site to allow the aptamer to interrogate the cellular environment (Wickiser et al. 2005; Garst and Batey 2009). While cotranscriptional folding and SAH binding is certain to differ from our model derived from biochemical and structural data, it is highly likely that many of the features of this model are consistent with the biological process.
FIGURE 8.
Model of SAH riboswitch aptamer folding in Mg2+ or SAH. Blue cylinders represent the P1, P2, and P4 helices, and J4/2, J2/1, and J1/4 are shown in orange, green, and gray, respectively. In the absence of Mg2+ and SAH, the pseudoknot secondary structure is formed, the junction regions are dynamic (represented by dashed lines), and the P2b helix is not stably formed (left). In the presence of physiological magnesium concentrations (0.5–1 mM Mg2+), P2b helix formation becomes more favorable (shown as orange cylinders extending the P2 and P4 helices), but the J2/1 and J1/4 regions remain dynamic. SAH binding is supported in the presence or absence of magnesium; the structures of each are equivalent. SAH binding promotes a stable P2b helix and P2/P4 coaxial stack and the J2/1 and J1/4 regions become structured. Stabilization of the P4 helix determines the fate of the regulatory switch that either occludes or exposes the Shine-Dalgarno sequence (SD, red) for translational regulators in the absence or presence of SAH, respectively.
Our data support a model in which physiological magnesium and temperatures promote the formation of the P2b helix and the P2/P4 coaxial stack, but other elements such as the adenosine-minor triple interactions between J1/4 and P1 do not appear to be established (Fig. 8). The unliganded SAH riboswitch appears to sample a broad range of conformationally unique states, with magnesium biasing the ensemble toward bound-like states that are likely to be the binding-competent conformations of the aptamer. The interaction of SAH with the RNA stabilizes the entire aptamer domain, both secondary and tertiary structural elements (Fig. 6B). In particular, the P4 helix appears to be significantly stabilized, as assessed by its thermal stability. This helix is proposed to participate in a secondary structural switch in which the 3′-strand can form an alternative hairpin that either occludes the Shine-Dalgarno sequence to prevent translation of the message or creates a rho-independent transcriptional terminator to abort transcription (Fig. 8; Wang et al. 2008). This model is consistent with a common theme amongst the majority of riboswitches in which ligand binding directly establishes or stabilizes a structural element in the aptamer that is involved in a secondary structural switch (Batey et al. 2004; Garst and Batey 2009).
Another central feature of riboswitches is their ability to discriminate between chemically related metabolites. Previous studies of SAM-binding riboswitches demonstrated that discrimination between SAM and SAH is due to electrostatic interactions with the positively charged sulfonium ion of SAM (Gilbert et al. 2008; Lu et al. 2008; Montange et al. 2010). These studies have revealed that the SAH riboswitch achieves a similar level of discrimination between these compounds by employing a steric mechanism that excludes the methyl group on SAM. However, a potential challenge to the specificity of the SAH riboswitch for its effector in vivo is the observation that adenine and adenine-bearing compounds are able to bind with micromolar affinities. In particular, both ATP and NAD+ bind the aptamer domain with sufficiently high affinities that they would be expected to bind the riboswitch and thereby affect gene regulation. This assumes that the SAH riboswitch or at least a subset thereof, is under thermodynamic control in which ligand binding is sufficiently fast to reach equilibrium before a regulatory decision is made.
How ATP and other compounds affect regulation by the SAH riboswitch depends upon whether they can influence the downstream regulatory switch. Our NMIA chemical probing data with adenine suggest that binding of this moiety alone does not significantly stabilize the P4 helix. Thus, other cellular metabolites that are present in high concentrations could act as competitive inhibitors of SAH, requiring higher levels of SAH to achieve activation of gene expression. If this is the case, then it is likely even more potent competitive inhibitors can be developed that target the SAH riboswitch. Given their presence in medically important pathogens, such as Pseudomonas aeruginosa, and that they regulate a process that is important for removing a potent toxin (Wang et al. 2008), the SAH riboswitch presents itself as a strong candidate for future efforts toward the development of novel antimicrobial agents (Blount and Breaker 2006).
MATERIALS AND METHODS
RNA synthesis and purification
The RNA constructs used in this study were synthesized by in vitro transcription with T7 RNA polymerase and purified using previously described methods (Kieft and Batey 2004). Briefly, RNAs were transcribed using standard conditions and the product RNA purified on a 12% denaturing polyacrylamide gel. The RNA was eluted from the gel and concentrated/exchanged into 0.5X T.E. buffer with a 3000 MWCO centrifugal filter device. The concentration was calculated from the absorbance at 260 nm and the calculated molar extinction coefficient.
Isothermal titration calorimetry (ITC)
The affinity of each RNA for SAH, adenine, ATP, and NAD+ was measured using ITC following previously described methods (Gilbert and Batey 2009). RNA samples were prepared by dialyzing into a buffer containing 100 mM NaCl, 100 mM K+-HEPES pH 8.0, and 10 mM MgCl2 for ∼16 h at 4°C; samples for magnesium-dependent studies were dialyzed against the same buffer but with the appropriate MgCl2 concentration. To test if an elevated NaCl concentration could rescue ligand binding characteristics in the absence of MgCl2, the RNA was dialyzed against 1 M NaCl and 100 mM K+-HEPES pH 8.0. Following dialysis, the small molecule (all purchased from Sigma-Aldrich as dry powders) was dissolved in an aliquot of dialysis buffer and degassed at the appropriate temperature for 10 min. Twenty to 50 μM RNA was loaded into the sample cell while 200 μM–1 mM ligand was loaded into the syringe of the calorimeter. Each experiment was performed at the appropriate temperature with a 10–15 μL injection volume at a rate of 0.5 μL sec−1 and a reference power of 5 μcal sec−1. Data were fit with Origin ITC software (Microcal Software Inc.) to a single-site binding model to determine the apparent association constant, Ka.
To obtain the heat capacity change (ΔCp), the average ITC-determined ΔH°obs was plotted against the experimental temperature. The data were fit to a linear equation for two separate temperature regimes: 5°C–20°C and 25°C–60°C, with the observed heat capacity change at a constant pressure calculated as the slope of the fitted line.
Chemical probing with NMIA
RNA samples were prepared as described previously with the addition of 5′ and 3′ structure cassettes flanking the RNA sequence, and the NMIA modification reaction was carried out following established protocols (Mortimer and Weeks 2009). Two picomoles of RNA in 12 μL 0.5X T.E. buffer were incubated for 2 min at 90°C, immediately placed on ice, and incubated for 5 min. Following RNA refolding, 6 μL of 3.3X folding buffer (333 mM K-HEPES, pH 8.0, 20 mM MgCl2, and 333 mM NaCl) or 6 μL 3.3X folding buffer containing 1.7 mM SAH or 6.7 mM adenine (for a final concentration of 500 μM and 2 mM, respectively) were added to each sample and the RNA was incubated on ice for ∼5 min. Each reaction was split into two 9-μL aliquots in thin-walled PCR tubes and incubated at the desired temperature for 1 min. The temperature dependence of the pKa of HEPES buffer between 20°C and 70°C is not sufficient to alter the reactivity of NMIA or significantly influence RNA structure (Stoddard et al. 2008). One microliter of 130 mM NMIA in DMSO or neat DMSO was then added to each aliquot and the samples were incubated for five NMIA half-lives. Reactions performed in the absence of magnesium were later supplemented with 3 mM magnesium to allow for efficient reverse transcription.
Immediately following modification, the samples were subjected to a reverse transcription reaction. Three microliters of 32P 5′-end labeled DNA oligomer was added to each modified RNA sample. Reaction mixtures were incubated at 65°C for 5 min, 35°C for 5 min, and finally incubated at 53°C for 1 min to prepare for reverse transcription. Each sample was mixed with 6 μL of enzyme reaction buffer containing 250 mM KCl, 167 mM Tris-HCl, pH 8.3, 16.7 mM DTT, 1.67 mM each dNTP, and 0.33 units of Superscript III reverse transcriptase (Invitrogen). The reverse transcription reaction was incubated at 53°C for 10 min, and was stopped by the addition of 1 μL of 4.0 M NaOH followed by 5 min incubation at 95°C. The samples were then quenched with 29 μL acid stop mix and incubated at 95°C for 5 min. The reactions were resolved on a 12% denaturing polyacrylamide gel, electrophoresed at 75 W for 3 h and imaged using a Typhoon PhosphorImager (Molecular Dynamics).
X-ray crystallography, structure solution, and refinement
Crystallography constructs of Rso were prepared for crystallization by exchanging the RNA into 10 mM K+-HEPES pH 7.5 buffer containing 1.0 mM SAH. Crystallization trials were set up using the hanging drop/vapor diffusion method in which 1 μL of the complex was mixed with 1 μL of mother liquor and incubated at 25°C. The Rso–SAH complex crystallized in conditions containing 50 mM Na-cacodylate, pH 6.5, 12 mM CoCl2, 150 mM Mg(OAc)2, 20 mM cobalt hexamine, and 10% PEG-4000 (Crystal A) or 50 mM Na-cacodylate, pH 7.0, 2.5 mM spermine, 0.9 mM spermidine, 9 mM MgCl2, 2.5 mM cobalt hexammine, and 5% PEG-400 (Crystal B). For crystals used for collecting phase information, the cobalt hexammine was substituted for iridium hexammine (Keel et al. 2007). Prior to data collection, the crystals were exchanged into mother liquor supplemented to 20% PEG-4000 or PEG-400 and frozen in liquid nitrogen.
Structure solution and refinement
A complete data set at the iridium peak wavelength was taken at the National Synchrotron Light Source at the Brookhaven National Laboratory (Crystal A). The data were processed in the R32 space group with mosflm and scala of the CCP4 suite (Collaborative Computational Project, Number 4 1994). Intensity statistics analysis by phenix.xtriage (Adams et al. 2002) revealed no additional pseudosymmetry (translational or rotational) or twinning and significant anomalous signal to 4.0 Å. Iridium heavy atom location, refinement, phasing, and density modification were accomplished via hkl2map (Davis et al. 2004). The resulting experimental density map showed continuous density for the GAGA tetraloop and several nucleotides in helical regions. MRSAD was used to improve the experimental phases for completion of the model (Schuermann and Tanner 2003). Refinement via phenix.refine converged to an Rwork of 21.8% and Rfree of 25.1% at a 2.8-Å resolution. Nucleotides 39–42 between P1 and P4 were disordered and the structure includes a domain swap of nucleotides 36–39, base pairing with P1 of an adjacent molecule.
High-resolution refinement with pseudosymmetry
A high-resolution data set extending to 2.18 Å (Crystal B) was collected at beamline X25 at BNL-NSLS. While Crystal B was isomorphous to Crystal A, refinement of the model via phenix.refine in the R32 space group lead to unacceptable Rwork and Rfree (>0.30), despite acceptable merging statistics in scala. The true space group was found to be monoclinic, supported by low R-factors after rigid body refinement, and improved agreement between symmetry related intensities after integration and merging (Table 1) (Mn(I/σI) and Rp.i.m. at 2.18 Å resolution). Refinement of the atom positions and anisotropic displacement parameters was accomplished in phenix.refine using strict noncrystallographic symmetry, a TLS model encompassing each chain, and a twin law for a single twin domain (−1/2h−1/2k−l,-1/2h−1/2k+l,−1/2h+1/2k, refined twin fraction = 0.530). In spite of accounting for pseudosymmetry in the high-resolution set, no additional nucleotides could be built to account for the disorder between P1 and P4. The final refinement statistics are presented (Table 1). Analysis of the refined model in molprobity (Davis et al. 2004) indicates all helical base pairs obey known stereochemical and geometry restraints with the exception of the C51:G10 pair. These nucleotides form a noncanonical G:C wobble as previously observed in domain E of the Thermus flavus 5S rRNA (Perbandt et al. 2001).
Accession codes
Protein Data Bank: Coordinates and structure factors for the SAH riboswitch have been deposited with accession numbers 3NPN and 3NPQ.
ACKNOWLEDGMENTS
We are grateful to members of the Batey laboratory, Sunny Gilbert, Colby Stoddard, Jeffrey Kieft, and Jennifer Pfingsten, for their critical reading of this manuscript. This work was supported by the National Institutes of Health (R01 GM083953).
Authors' contributions: A.L.E. screened RNAs for crystallization, engineered variants, and collected initial diffraction data, as well as performed all of the biochemical experiments. A.H. collected all data sets in this manuscript. A.H. and F.E.R. solved and improved the experimental phases. F.E.R. and R.T.B. built and refined the model of the RNA–SAH duplex at 2.8 Å. F.E.R. refined the model at 2.18 Å. A.L.E. and R.T.B. wrote the paper with input from all authors.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2341610.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Batey RT, Doudna JA 2002. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry 41: 11703–11710 [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 [DOI] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR 2006. Riboswitches as antibacterial drug targets. Nat Biotechnol 24: 1558–1564 [DOI] [PubMed] [Google Scholar]
- Bruzzese FJ, Connelly PR 1997. Allosteric properties of inosine monophosphate dehydrogenase revealed through the thermodynamics of binding of inosine 5′-monophosphate and mycophenolic acid. Temperature dependent heat capacity of binding as a signature of ligand-coupled conformational equilibria. Biochemistry 36: 10428–10438 [DOI] [PubMed] [Google Scholar]
- Chou MY, Lin SC, Chang KY 2010. Stimulation of −1 programmed ribosomal frameshifting by a metabolite-responsive RNA pseudoknot. RNA 16: 1236–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC 2004. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Zhou K, Doudna JA 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395: 567–574 [DOI] [PubMed] [Google Scholar]
- Fontecave M, Atta M, Mulliez E 2004. S-adenosylmethionine: Nothing goes to waste. Trends Biochem Sci 29: 243–249 [DOI] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM 2006. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol 13: 226–233 [DOI] [PubMed] [Google Scholar]
- Garst AD, Batey RT 2009. A switch in time: Detailing the life of a riboswitch. Biochim Biophys Acta 1789: 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM 2008. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc 130: 12244–12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Batey RT 2009. Monitoring RNA–ligand interactions using isothermal titration calorimetry. Methods Mol Biol 540: 97–114 [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182 [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF 2007. Binding and kinetics for molecular biologist Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Grundy FJ, Henkin TM 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol 30: 737–749 [DOI] [PubMed] [Google Scholar]
- Han K, Byun Y 2003. PSEUDOVIEWER2: Visualization of RNA pseudoknots of any type. Nucleic Acids Res 31: 3432–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Podell E, Harrington DJ, Cech TR 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure 9: 221–231 [DOI] [PubMed] [Google Scholar]
- Kang M, Peterson R, Feigon J 2009. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell 33: 784–790 [DOI] [PubMed] [Google Scholar]
- Kazanov MD, Vitreschak AG, Gelfand MS 2007. Abundance and functional diversity of riboswitches in microbial communities. BMC Genomics 8: 347 doi: 10.1186/1471-2164-8-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel AY, Rambo RP, Batey RT, Kieft JS 2007. A general strategy to solve the phase problem in RNA crystallography. Structure 15: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Batey RT 2004. A general method for rapid and nondenaturing purification of RNAs. RNA 10: 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Edwards TE, Ferre-D'Amare AR 2009. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol 16: 343–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopycki JG, Rauh D, Chumanevich AA, Neumann P, Vogt T, Stubbs MT 2008. Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferases. J Mol Biol 378: 154–164 [DOI] [PubMed] [Google Scholar]
- Ladbury JE 1995. Counting the calories to stay in the groove. Structure 3: 635–639 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7:499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA 2006. S-adenosylmethionine: Jack of all trades and master of everything? Biochem Soc Trans 34: 330–333 [DOI] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A 2008. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol 15: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231 [DOI] [PubMed] [Google Scholar]
- Mikulecky PJ, Feig AL 2006. Heat capacity changes associated with nucleic acid folding. Biopolymers 82: 38–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT 2010. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol 396: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM 2009. Time-resolved RNA SHAPE chemistry: Quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc 4: 1413–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Freire E 1992. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv Protein Chem 43: 313–361 [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci 98: 4899–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros-Palacios N, Smith AM, Grundy FJ, Soberon M, Henkin TM, Miranda-Rios J 2008. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol 67: 793–803 [DOI] [PubMed] [Google Scholar]
- Perbandt M, Vallazza M, Lippmann C, Betzel C, Erdmann VA 2001. Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 Å resolution. Acta Crystallogr D Biol Crystallogr 57: 219–224 [DOI] [PubMed] [Google Scholar]
- Schuermann JP, Tanner JJ 2003. MRSAD: Using anomalous dispersion from S atoms collected at Cu Kα wavelength in molecular-replacement structure determination. Acta Crystallogr D Biol Crystallogr 59: 1731–1736 [DOI] [PubMed] [Google Scholar]
- Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE 2009. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem 284: 11012–11016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolar RS, Record MT, Jr, 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263:777–784 [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT 2008. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennely SP, Rambo RP, Sanbonmatsu KY, Batey RT 2010. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaralingam M 1975. Structure and conformation of nucleosides and nucleotides and their analogs as determined by x-ray diffraction. Ann N Y Acad Sci 255: 3–42 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC 2004. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr D Biol Crystallogr 60: 2144–2149 [DOI] [PubMed] [Google Scholar]
- Ueland PM 1982. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacol Rev 34: 223–253 [PubMed] [Google Scholar]
- Wang JX, Breaker RR 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168 [DOI] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR 2008. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell 29: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31 doi: 10.1186/gb-2010-11-3-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM 2005. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J Am Chem Soc 127: 4659–4667 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707 [DOI] [PubMed] [Google Scholar]
- Xin Y, Laing C, Leontis NB, Schlick T 2008. Annotation of tertiary interactions in RNA structures reveals variations and correlations. RNA 14: 2465–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo WL 2006. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22: 445–452 [DOI] [PubMed] [Google Scholar]