Abstract
The CRISPR–Cas system provides many prokaryotes with acquired resistance to viruses and other mobile genetic elements. The core components of this defense system are small, host-encoded prokaryotic silencing (psi)RNAs and Cas (CRISPR-associated) proteins. Invader-derived sequences within the psiRNAs guide Cas effector proteins to recognize and silence invader nucleic acids. Critical for CRISPR–Cas defense is processing of the psiRNAs from the primary transcripts of the host CRISPR (clustered regularly interspaced short palindromic repeat) locus. Cas6, a previously identified endoribonuclease present in a wide range of prokaryotes with the CRISPR–Cas system, binds and cleaves within the repeat sequences that separate the individual invader targeting elements in the CRISPR locus transcript. In the present study, we investigated several key aspects of the mechanism of function of Cas6 in psiRNA biogenesis. RNA footprinting reveals that Pyrococcus furiosus Cas6 binds to a 7-nt (nucleotide) sequence near the 5′ end of the CRISPR RNA repeat sequence, 14 nt upstream of the Cas6 cleavage site. In addition, analysis of the cleavage activity of P. furiosus Cas6 proteins with mutations at conserved residues suggests that a triad comprised of Tyr31, His46, and Lys52 plays a critical role in catalysis, consistent with a possible general acid–base RNA cleavage mechanism for Cas6. Finally, we show that P. furiosus Cas6 remains stably associated with its cleavage products, suggesting additional roles for Cas6 in psiRNA biogenesis.
Keywords: CRISPR RNAs, Cas proteins, endoribonuclease, RNA processing
INTRODUCTION
Recently, it has become clear that many prokaryotes possess a defense mechanism known as the CRISPR–Cas system that provides resistance to viruses and other potential genome invaders (Lillestol et al. 2006; van der Oost et al. 2009; Horvath and Barrangou 2010; Karginov and Hannon 2010; Marraffini and Sontheimer 2010a). Found in nearly all archaea and half of bacteria, this system is comprised of small prokaryotic silencing (psi)RNAs derived from clustered regularly interspaced short palindromic repeat (CRISPR) loci and Cas (CRISPR-associated) proteins (Jansen et al. 2002; Haft et al. 2005; Makarova et al. 2006). CRISPR loci contain series of short (typically 25–40 nt [nucleotide]) direct repeats separated by variable sequences of similar size (called “spacers” prior to the finding that some matched to known invader sequences) (Bolotin et al. 2005; Mojica et al. 2005; Pourcel et al. 2005; Lillestol et al. 2006; Makarova et al. 2006; Shah et al. 2009). A single CRISPR locus may contain as many as 200 variable sequence elements and repeats (Haft et al. 2005; Makarova et al. 2006). It has recently been shown that organisms with CRISPR sequence elements derived from a known phage or conjugative plasmid are resistant to infection by that phage or plasmid (Barrangou et al. 2007; Brouns et al. 2008; Deveau et al. 2008; Marraffini and Sontheimer 2008, 2010b). Phage infection triggers the acquisition of invader-derived sequences by the host CRISPR locus via an unknown mechanism (Barrangou et al. 2007; Deveau et al. 2008; Mills et al. 2009). Thus, the CRISPR–Cas system provides a defense that rapidly adapts to attacks on the host genome and has been shown to promote survival in natural microbial communities (Andersson and Banfield 2008; Tyson and Banfield 2008).
Transcripts of the CRISPR locus are processed into individual psiRNAs that comprise a single invader-targeting sequence and a short segment (typically 8 nt) of the repeat sequence at the 5′ end (Fig. 1; Tang et al. 2002, 2005; Lillestol et al. 2006; Brouns et al. 2008; Hale et al. 2008, 2009; Marraffini and Sontheimer 2008). Previously, we found that Cas6 binds and cleaves CRISPR RNAs within the repeat sequence (Carte et al. 2008). Cas6 cleavage generates the 5′ end of the mature psiRNAs, including the 8-nt 5′ repeat sequence (or psi-tag) that we hypothesize functions to identify psiRNAs within the cell (Carte et al. 2008; Hale et al. 2008, 2009). In Pyrococcus furiosus, psiRNAs also undergo 3′-end processing that results in two predominant species of each psiRNA—typically 39 nt and 45 nt in length (Hale et al. 2008, 2009).
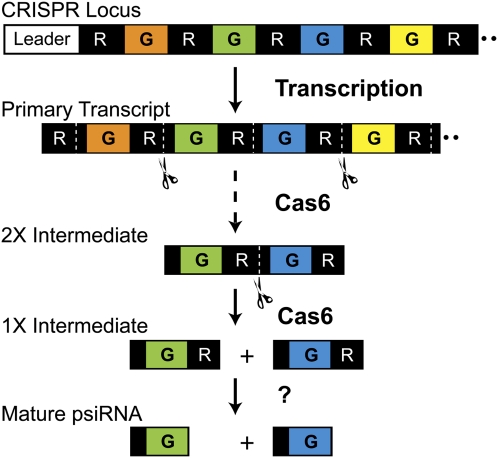
FIGURE 1.
psiRNA biogenesis. The primary CRISPR locus transcript contains multiple guide sequences (color segments, G) separated by a repeat sequence (black segments, R). Cas6 cleaves within the repeats of a CRISPR transcript (cleavage site labeled with dotted line), eventually producing individual RNAs comprised of an 8-nt repeat sequence (i.e., psi-tag), ∼37-nt guide sequence, and 22-nt repeat sequence (1× intermediate). These “1× intermediate RNAs” are the end-products of Cas6 cleavage. The 22-nt repeat sequence found at the 3′ end of the 1× Cas6 product is subsequently removed from the mature psiRNAs (that guide the Cmr complex to cleave target RNAs in P. furiosus) by an unknown activity. Cas6 cleavage proceeds through a series of intermediates including the 2× intermediate shown here.
Cas6 is one of the most widely distributed CRISPR-associated proteins and is found in bacteria and archaea (Haft et al. 2005; Makarova et al. 2006). The Cas6 protein contains a characteristic glycine-rich, GhGxxxxGhG motif (where h indicates a hydrophobic amino acid and xxxx includes at least one lysine or arginine) present at the C terminus (Haft et al. 2005). Cas6 cleavage activity is independent of divalent metal ions and generates products with 5′-hydroxyl and likely 2′–3′ cyclic phosphate ends (Carte et al. 2008), typical of metal-independent ribonucleases (Steyaert 1997; Raines 1998; Xue et al. 2006; Yajima et al. 2006; Keppetipola et al. 2009). Three conserved residues (Tyr, His, and Lys) that are separated from one another in the primary sequences of Cas6 proteins from diverse organisms were found to cluster in the crystal structure of Cas6 from P. furiosus (Carte et al. 2008). This same arrangement of residues makes up the catalytic active site of the metal-independent ribonuclease that removes introns from tRNAs in archaeal organisms (Xue et al. 2006). These observations suggested that these three conserved amino acids comprise a catalytic triad that would be required for cleavage of the CRISPR repeat RNA.
In the present study, Cas6 substrate recognition was examined at single-nucleotide resolution by RNA footprinting. The results indicate that Cas6 interacts directly with nucleotides 2–8 near the 5′ end of the P. furiosus CRISPR repeat. In addition, the role of the predicted catalytic triad amino acids in Cas6 function was tested through mutational analysis. Finally, native Cas6 (isolated from P. furiosus extract) was shown to cleave CRISPR repeat RNA and was found to co-purify with CRISPR RNA processing intermediates of the psiRNA biogenesis pathway.
RESULTS
Mapping the Cas6–CRISPR repeat RNA binding site
Previous RNA mutational analysis indicated that Cas6 interacts with sequence elements in the 5′ half of the CRISPR repeat RNA and that binding in this region is essential for cleavage (Carte et al. 2008). Substitution or deletion of nucleotides in this region (but not in the 3′ half of the RNA) disrupted recombinant P. furiosus Cas6 binding in gel shift assays (Carte et al. 2008). Moreover, an RNA comprised of the first 12 nt of the 30-nt P. furiosus CRISPR repeat was bound by Cas6 with similar affinity as full-length repeat RNA (Carte et al. 2008). Cas6 cleavage depends on the 5′ region required for binding, as well as sequences in the 3′ half of the CRISPR repeat (Carte et al. 2008).
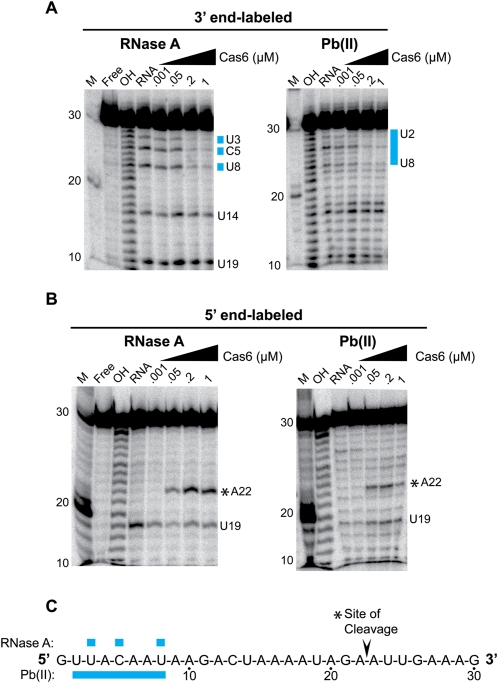
To investigate the molecular basis for Cas6 recognition of the CRISPR repeat RNA, we performed RNA footprinting with recombinant P. furiosus Cas6 protein and radiolabeled CRISPR repeat RNA (Fig. 2). Interactions were probed with lead (II) acetate, which cleaves within single-stranded and dynamic regions of RNA (Brunel and Romby 2000; Lindell et al. 2002), and RNase A, which cleaves after unpaired Cs and Us (Raines 1998). We used 5′- and 3′-end-labeled RNA to resolve interactions with the 3′ and 5′ regions of the repeat, respectively (Fig. 2A,B). We found that Cas6 provided strong concentration-dependent protection of nucleotides 2–8 of the CRISPR repeat from lead-induced cleavage (Fig. 2A,C). Similarly, RNase A cleavage at nucleotides 3, 5, and 8 was inhibited in the presence of Cas6 (Fig. 2A,C). No Cas6-dependent protection from either lead (II) acetate or RNase A cleavage was observed in other regions of the repeat RNA (Fig. 2A–C). Similar results were obtained using RNase T1 (data not shown). These findings indicate that the primary CRISPR RNA binding site for Cas6 is located within nucleotides 2–8 of the repeat and likely includes nucleotides 3, 5, and 8.
FIGURE 2.
Lead-induced and RNase A cleavage protection of CRISPR repeat RNA by P. furiosus Cas6. (A) 3′-end-labeled P. furiosus CRISPR repeat RNA was incubated in the absence (RNA) or presence of increasing concentrations of Cas6 (indicated as micromolar, μM) and subjected to RNase A cleavage (left panel) or lead-induced cleavage (right panel). RNAs were separated on 15% denaturing (7 M urea) polyacrylamide gels. Size markers include 5′-end-labeled RNA markers (M) and alkaline hydrolysis ladders (OH). (Blue bars) Sites of strong protection. (B) Cleavage protection assays performed as in A with 5′-end-labeled CRISPR repeat RNA. A summary of cleavage protections is displayed to the right of each gel. (C) A summary of observed cleavage protection is shown. The Cas6 cleavage site is indicated by an asterisk (*).
CRISPR repeat sequences are often palindromic, and the P. furiosus CRISPR repeat has the potential to form a weak stem–loop; however, it is a member of a group of repeat sequences that are predicted to be unstructured (Kunin et al. 2007). Analysis of the cleavage of the P. furiosus repeat RNA in the absence of proteins indicates that the RNA is predominantly unstructured in solution under the experimental conditions (Fig. 2A,B; data not shown).
Investigation of a putative catalytic amino acid triad
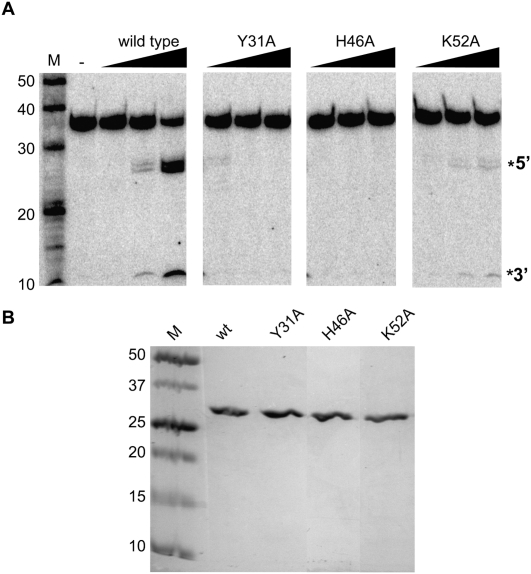
Based on the clustering of three conserved amino acids in the folded structure of the Cas6 protein (Carte et al. 2008) and the similarity of the configuration to archaeal tRNA splicing endonuclease (Xue et al. 2006; Calvin et al. 2008), we hypothesized that Cas6 cleavage involves a conserved Tyr, His, Lys triad (Carte et al. 2008). In order to investigate this possibility, we tested the ability of Cas6 proteins containing single amino acid substitutions in the putative catalytic triad residues (Y31, H46, K52) to cleave radiolabeled CRISPR repeat RNA (Fig. 3). The mutant proteins were expressed in Escherichia coli and purified (Fig. 3B).
FIGURE 3.
Cleavage activity of Cas6 mutants. (A) Uniformly 32P-labeled CRISPR repeat RNA was incubated in the absence (−) or presence of increasing concentrations of wild-type or mutant Cas6 (10, 50, and 500 nM) followed by separation on a 15% denaturing (7 M urea) polyacrylamide gel. The 5′ and 3′ cleavage products are indicated to the right. (B) Purified wild-type (wt) and mutant Cas6 proteins were analyzed by SDS-PAGE and Coomassie blue staining. Molecular weight markers are indicated in kilodaltons.
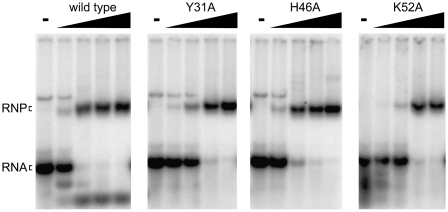
We found that substitution of any of the three putative triad amino acids with alanine led to a significant decrease in cleavage activity relative to wild-type Cas6 (Fig. 2A). No cleavage activity was observed with the Y31A and H46A Cas6 mutants. The cleavage activity was reduced ∼40-fold at the highest tested concentration (500 nM) of K52A Cas6 relative to wild-type Cas6. To determine whether the loss of cleavage activity was due to changes in the mutant proteins that disrupted RNA binding, we analyzed native gel mobility shift assays using each mutant protein and radiolabeled CRISPR repeat RNA (Fig. 4). We observed a slight decrease in the affinity of the K52A mutant for the repeat RNA relative to wild type, but all three mutant Cas6 proteins retained the ability to form a stable complex with the CRISPR repeat RNA (Fig. 4). The results indicate that Tyr31, His46, and Lys52 are not required for binding to CRISPR repeat RNA; however, these residues are required for efficient cleavage of the RNA. While both loss of cleavage activity and retention of protein binding activity associated with any of the mutants could reflect multiple possibilities, these findings provide additional support for the hypothesis that this triad of amino acids plays a direct role in catalysis of the cleavage reaction.
FIGURE 4.
Substrate binding activity of Cas6 mutants. Uniformly 32P-labeled CRISPR repeat RNA was incubated in the absence (−) or presence of increasing concentrations of wild-type or mutant Cas6 (1, 50, 200, and 500 nM). Formation of stable complexes was assessed by native gel mobility shift analysis. The positions of the free (RNA) and bound (RNP) substrate RNA are indicated.
Native Cas6 cleaves CRISPR repeat RNA and remains associated with its product
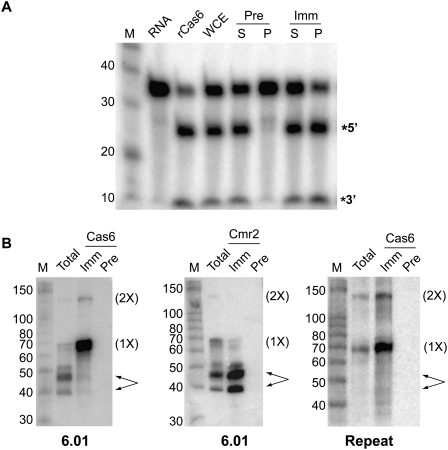
The previous Cas6 studies were performed with purified recombinant protein (Carte et al. 2008). To obtain native Cas6 protein, we raised polyclonal antibodies against recombinant Cas6 and immunoprecipitated the protein from extracts of P. furiosus. Samples immunoprecipitated with immune, but not preimmune, Cas6 antibodies, cleaved uniformly labeled CRISPR repeat RNA and generated the same size cleavage products as recombinant Cas6 (Fig. 5A). Remarkably, the Cas6 cleavage activity was also apparent in whole-cell extract under the standard assay conditions, which include 20 mM EDTA. Thus, the cleavage activity observed in P. furiosus extract is divalent metal ion–independent, as is the case for recombinant Cas6 (Carte et al. 2008).
FIGURE 5.
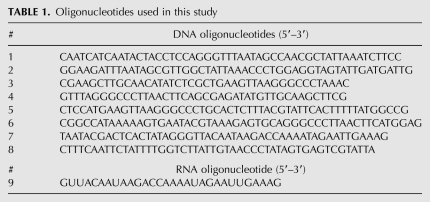
Immunopurified Cas6 cleaves CRISPR repeat RNA and associates with substrate and product RNAs. (A) Cleavage activity. Uniformly 32P-labeled CRISPR repeat RNA was incubated in the absence (RNA) or presence of recombinant Cas6 (rCas6), whole cell extract (WCE), or samples from immunoprecipitation reactions using Cas6 antibodies; (Pre) preimmune; (Imm) immune; (S) supernatant; (P) pellet. The RNAs were separated on a 15% denaturing, 7 M urea-containing polyacrylamide gel, along with 5′-end-labeled RNA markers (M). (B) Northern blot analysis of Cas6 immunoprecipitation. RNAs extracted from WCE or from IPs using preimmune (Pre) or immune (Imm) Cas6 or Cmr2 antibodies were separated on 15% denaturing, 7 M urea-containing polyacrylamide gels, along with 5′-end-labeled RNA markers (M). A 5′-end-labeled DNA oligonucleotide antisense to psiRNA 6.01-specific sequences, or to the repeat sequence of P. furiosus CRISPRs 1, 5, and 6 were used as probes, as indicated. The positions of the 2× intermediate (2×), 1× intermediate (1×), and mature psiRNAs (arrows) are indicated.
Cas6 cleaves within the repeat sequences found in precursor CRISPR RNAs, reducing the transcript step-by-step to “1× intermediate” RNAs comprised of the 8-nt psi-tag and an ∼37-nt invader-targeting sequence with the remaining 22 nt of repeat RNA at the 3′ end (Fig. 1; Hale et al. 2008). The repeat sequence at the 3′ end of the 1× intermediate RNA is trimmed by an unidentified nuclease to produce the mature psiRNA species present in the Cmr effector complex (Hale et al. 2008, 2009). To determine whether Cas6 is physically associated with any CRISPR RNA species, we probed the immunoprecipitated samples for RNAs containing common CRISPR repeat sequence and the specific invader-targeting sequence of the first psiRNA encoded by CRISPR locus 6 (repeat and 6.01) (Fig. 5B).
We found that Cas6 associates with its product as well as substrate RNA. Northern analysis indicates that Cas6 is specifically associated with the 2× intermediate, an intermediate product subject to further Cas6 cleavage (Fig. 5B). The 2× intermediate RNA is detected by both repeat and psiRNA 6.01 probes in total extract, and enriched in immunoprecipitations with immune Cas6 antibodies but not with preimmune antibodies or antibodies against the Cmr2 protein of the Cmr effector complex (Hale et al. 2009). The mature psiRNA species that function as part of the Cmr effector complex (Hale et al. 2009), coimmunoprecipitate with antibodies against Cmr2 but are not enriched in Cas6 immunoprecipitation samples. The most abundant CRISPR-derived RNA immunoprecipitated with Cas6 antibodies is the 1× intermediate RNA, which is the presumed end product of Cas6 processing of CRISPR transcripts (Carte et al. 2008; Hale et al. 2008).
DISCUSSION
The biogenesis of mature psiRNAs is critical to the function of the CRISPR–Cas system (Brouns et al. 2008). Separation of the individual targeting elements encoded in the host CRISPR locus is accomplished by endonucleolytic cleavage of the CRISPR RNA transcript within the intervening repeat elements by Cas6 in P. furiosus and other archaea and bacteria (Carte et al. 2008). This cleavage results in a 1× CRISPR RNA intermediate that retains 8 nt of the repeat at the 5′ end, and ∼22 nt of the next repeat at the 3′ end (Fig. 1). In P. furiosus, further 3′-end processing results in the generation of mature psiRNA species that retain the 5′ 8-nt, repeat-derived psi-tag that we hypothesize serves as a recognition sequence for other Cas proteins (Hale et al. 2008, 2009).
Not all organisms with the CRISPR–Cas system possess Cas6. In some of these organisms, Cse3 appears to be a functional homolog of Cas6. Cse3 was shown to be required for CRISPR RNA biogenesis in an E. coli K12 strain (Brouns et al. 2008). Cse3 is also a divalent metal-ion-independent endoribonuclease that cleaves within the 3′ region of CRISPR repeats. While the repeat sequence is distinct, Cse3 cleavage also leaves an 8-nt psi-tag on the 5′ end of the CRISPR RNAs (Brouns et al. 2008).
Interestingly, when coexpressed in E. coli, Cse3 was found to be a component of a large RNA–protein complex containing a number of other Cas proteins as well as mature psiRNA species (Brouns et al. 2008). In contrast, we have found that native P. furiosus Cas6 is not stably associated with the mature psiRNA species found in the Cmr effector complex in P. furiosus (Fig. 5B). Moreover, Cas6 was not co-purified with P. furiosus Cmr complexes (Hale et al. 2009). These results indicate that Cas6 is not likely a structural component of the Cmr effector complex in P. furiosus.
Cas6 was found associated with the 2× intermediate RNA that is a substrate for further Cas6 cleavage (Fig. 5B), consistent with the role of Cas6 in psiRNA biogenesis (see Fig. 1). The Cas6 binding site defined in this work (Fig. 2) remains intact in the end product of Cas6 cleavage of the CRISPR transcript, the 1× intermediate. We found that Cas6 remains associated with the 1× intermediate RNA (Fig. 5B), suggesting that Cas6 may also influence downstream events such as the 3′-end processing that generates mature psiRNAs (Hale et al. 2008, 2009).
The overall protein structures of Thermus thermophilus Cse3 and P. furiosus Cas6 are very similar (Ebihara et al. 2006; Carte et al. 2008). Both Cse3 and Cas6 are composed of duplicated ferrodoxin folds separated by a central cleft that contains a conserved glycine-rich loop. In both enzymes, a highly conserved histidine residue is positioned in a spatially comparable location adjacent to the glycine-rich loop. For both Cse3 and Cas6, mutational analysis supports the hypothesis that this histidine is an active-site residue critical for catalyzing cleavage of CRISPR repeat RNA (Fig. 3A; Ebihara et al. 2006; Brouns et al. 2008). We found that a trio of clustered, conserved amino acids (P. furiosus Tyr31, His46, Lys52) was important for cleavage activity and appear to define the Cas6 active site. Additional residues may also cooperate with the invariant histidine in Cse3 to catalyze CRISPR RNA cleavage.
Suggested Cas6 CRISPR RNA cleavage mechanism
Like other metal-independent ribonucleases, Cas6 likely cleaves RNAs via a trans-esterification reaction employing general acid–base chemistry (Steyaert 1997; Raines 1998; Xue et al. 2006; Yajima et al. 2006; Keppetipola et al. 2009). The available evidence suggests that Cas6 shares similar active-site architecture and likely reaction characteristics with the archaeal tRNA splicing endonuclease (Xue et al. 2006; Calvin and Li 2008). In the proposed mechanism for the archaeal tRNA splicing endonuclease, the active-site tyrosine abstracts a proton from the 2′-hydroxyl of the ribose ring (Calvin and Li 2008). This leads to nucleophilic attack of the 2′-oxygen on the phosphate backbone, resulting in the formation of a pentavalent transition state, whose charge is stabilized by a conserved lysine residue located in the active site (Calvin and Li 2008). Cleavage of the scissile phosphate is then facilitated by proton donation from the imidazole ring of the active-site histidine residue, producing cleavage products with 5′-hydroxyl and 2′–3′ cyclic phosphate ends (Calvin and Li 2008). This predicted cleavage mechanism is supported by extensive structural, kinetic, mutational, and computational analyses (Xue et al. 2006; Kim et al. 2007; Min et al. 2007; Calvin et al. 2008). The present mutational analysis of Cas6 suggests a similar acid–base cleavage mechanism in which Tyr31 may act as a general base, His46 may act as a general acid, and Lys52 may stabilize the transition state. Alternatively, His46 may act as a general acid and Tyr31 as a general base for cleavage of the repeat RNA.
A more detailed understanding of the Cas6 cleavage mechanism and substrate recognition will require further investigation. For example, a high-resolution structure of the Cas6 enzyme bound to the CRISPR RNA could help elucidate the specific roles of each of the active-site residues in catalysis as well as provide a greater understanding how Cas6 recognizes the 5′ end of the substrate RNA and positions the scissile bond near the 3′ end of the RNA within the active site of the enzyme.
MATERIALS AND METHODS
Expression and purification of PfCas6 and mutants
Primers to generate site-specific mutants were designed and ordered from Eurofins MWG Operon (listed in Tables 1 and 2). Mutant cas6 genes were generated from a pET24d plasmid containing the PF1131 gene (cas6 from P. furiosus) using QuikChange site-directed mutatagenesis (Stratagene). The DNA sequences of the mutated constructs were confirmed by nucleotide sequencing analysis. N-terminal, 6x histidine-tagged proteins were expressed in E. coli BL21 codon + (DE3, Invitrogen) and purified to homogeneity as described previously (Carte et al. 2008).
TABLE 1.
Oligonucleotides used in this study
TABLE 2.
Combinations of oligonucleotides used in this study
Generation of radiolabeled RNAs
The synthetic RNA (see Table 1 for sequence) and RNA size standards (Decade markers) used in this study were purchased from Integrated DNA Technologies (IDT) and Applied Biosystems, respectively. The Northern deoxyoligonucleotide probes used in this study (see Hale et al. 2008 for sequences) were purchased from Eurofins MWG Operon. RNAs were 5′-end-labeled with T4 polynucleotide kinase (Applied Biosystems) and [γ-32P]ATP (7000 Ci/mmol; MP Biomedicals) as described previously (Carte et al. 2008). End-labeling at the 3′ end was performed with T4 RNA ligase (Promega) and [α-32P]pCp (2500 Ci/mmol; MP Biomedicals). A typical reaction contained 10 pmol of RNA, 20 U of T4 RNA ligase, 10 U of SUPERase-IN RNase inhibitor (Applied Biosystems), 1× T4 RNA ligase buffer (Promega), 20% polyethylene glycol M.W. 3350, and ∼12 pmol of [α-32P]pCp. The uniformly labeled CRISPR repeat RNA substrate was generated by in vitro transcription by T7 polymerase using annealed DNA oligonucleotides containing the T7 promoter sequence as a template (see Tables 1 and 2) in the presence of [α-32P]UTP (MP Biomedicals) and purified as described previously (Carte et al. 2008). All radiolabeled RNAs were extracted with phenol/chloroform/isoamyl alcohol (PCI), precipitated with ethanol, and gel-purified as described previously (Carte et al. 2008).
RNA footprinting
Lead (II)–induced and RNase A cleavage were carried out essentially as described previously (Youssef et al. 2007). Briefly, 0.1 pmol of 32P end-labeled RNA (either 5′ or 3′) was incubated in the absence (free RNA) or presence of increasing concentrations of Cas6 for 30 min at 65°C–70°C in buffer A (20 mM HEPES-KOH at pH [7.0], 500 mM KCl). Lead (II)–induced cleavage was initiated by the addition of 15 mM Pb(II) acetate prepared fresh in sterile water. Reactions were carried out for 10 min at room temperature and were stopped by the addition of EDTA to a final concentration of 20 mM, followed by PCI extraction and ethanol precipitation. RNase A cleavage was initiated by the addition of 10 pg of RNase A (Applied Biosystems) and incubated for 15 min at 37°C. Reactions were stopped by PCI extraction followed by ethanol precipitation. Alkaline hydrolysis ladders (cleavage after each nucleotide) were generated as described previously (Youssef et al. 2007). In each case, precipitated RNAs were resuspended in RNA loading dye (10 M urea, 2 mM EDTA, 0.5% SDS, and 0.02% [w/v] each bromophenol blue and xylene cyanol) and separated on 38 × 30 cm 15% polyacrylamide (acrylamide:bis ratio 19:1) 7 M urea-containing gels. The gels were dried, and the radiolabeled RNAs were visualized by PhosphorImaging.
RNA binding and cleavage reactions
RNA binding and cleavage reactions were carried out as described previously (Carte et al. 2008). Briefly, 50 fmol of uniformly 32P-labeled RNA was incubated in the absence (free RNA) or presence of increasing concentrations of Cas6 (as indicated in figure legends) in buffer A containing 20 mM EDTA for 30 min at 65°C–70°C. Half of each reaction was electrophoresed on an 8% native polyacrylamide gel to assess RNA binding by gel mobility shift analysis. RNA cleavage was assessed by separation of the RNAs on denaturing, 7 M urea-containing, 15% polyacrylamide gels following PCI extraction and ethanol precipitation. For analysis of native Cas6 cleavage activity, either ∼40 μg of protein from whole-cell extract (WCE) or supernatant from an immunoprecipitation reaction (see below) was incubated with 50 fmol of uniformly 32P-labeled RNA for 30 min at 70°C. Alternatively, 10 μL of resin from an immunoprecipitation reaction (see below) was added. In this case, samples were mixed every 5 min by pipetting up and down during the 30-min incubation at 70°C. The gels were dried and radiolabeled RNAs were visualized by PhosphorImaging. Quantitation of the cleavage products was performed using ImageQuant TL software (GE Life Sciences).
Preparation of P. furiosus cell extract
Four grams of P. furiosus cells was lysed in 10 mL of 50 mM Tris (pH 8.0) in the presence of 100 U of RQ1 DNase (Promega) and 0.1 mM phenylmethanesulfonylfluoride (PMSF). The extract was then subjected to ultracentrifugation at 100,000g for 90 min. The resulting S100 was stored at −80°C until use.
Preparation of polyclonal antibodies against PfCas6 in Gallus gallus
Specific antibodies against PfCas6 were raised in leghorn hens (Gallus gallus). For immunization, each hen received three injections of 200 μg of 6x histidine-tagged PfCas6 in buffer B (20 mM sodium phosphate at pH 7.0, 500 mM NaCl). Each injection was separated by 2 wk. For the initial injection, 500 μL of antigen (0.4 mg/mL) was emulsified in 500 μL of Freund's complete adjuvant prior to bilateral injection in the breast muscle. For the two booster injections, 500 μL of antigen (0.4 mg/mL) was emulsified in 500 μL of Freund's incomplete adjuvant prior to injection. Beginning 1 wk after the final injection, immune eggs were collected daily for 3 mo.
IgY was purified from the egg yolks by polyethylene glycol (PEG) precipitation as described previously (Polson et al. 1980). Briefly, egg yolks were separated from the egg whites and washed with dH2O. In a typical purification, three egg yolks were combined, and resuspended in ∼250 mL of lysis buffer (10 mM Tris at pH 7.5, 100 mM NaCl, 0.02% NaN3). Polyethylene glycol (PEG) 8000 (Fisher Scientific) was added to 3.5% (w/v). The sample was mixed by shaking and centrifuged at 10,000g for 10 min. The supernatant was filtered through 100% cotton cheesecloth, and PEG 8000 was added to 9% (w/v). The sample was mixed by shaking and centrifuged at 10,000g for 10 min. The supernatant was removed and discarded. The pellet was resuspended in ∼35 mL of lysis buffer by incubation overnight at 4°C. The PEG precipitation was repeated, and the pellet from the 9% (w/v) PEG step, containing the purified antibodies, was resuspended in ∼7 mL of lysis buffer and stored at either 4°C or −80°C until use. The protein concentration was determined by the BCA assay (Pierce).
Immunoprecipitation of Cas6 from P. furiosus extract
Immunoprecipitations (IP) were performed using anti-Cas6 IgY antibodies conjugated to CarboLink coupling gel (Pierce). Coupling was performed according to the manufacturer's protocol and was verified by A260 absorbance readings.
P. furiosus S100 cell extract was pre-cleared in a reaction containing ∼8 mg of total protein, ∼550 μg of non-immune IgY-coupled CarboLink resin, 1× Complete Mini protease inhibitor (Roche), and 50 U of SUPERase-IN RNase inhibitor, and brought up to a total volume of 1 mL with IPP-300 (10 mM Tris at pH 8.0, 300 mM NaCl, 0.05% Igepal). The pre-clearing reaction was incubated at room temperature for 2 h with end-over-end rotation. The sample was centrifuged at 3000g for 2 min, and the supernatant was split between preimmune and immune IP reactions. A typical IP reaction contained 500 μL of pre-cleared cell extract (∼4 mg of total protein), 1× Complete Mini protease inhibitor, 50 U of SUPERasin RNase inhibitor, and 270–550 μg of antibody (preimmune or immune)–coupled resin brought up to a total volume of 1 mL with IPP-300. The reactions were incubated for 2 h at room temperature with end-over-end rotation and washed four times with IPP-300. The pellets were resuspended in an equal volume of buffer A.
Northern analysis
For Northern blot analysis, RNAs were extracted from immunoprecipitation samples (immune or preimmune) and whole-cell extract using TRIzol LS Reagent (Invitrogen) according to the manufacturer's recommendations. Northern blots were performed essentially as described previously (Hale et al. 2008). Briefly, RNAs were separated on a 15% polyacrylamide, 7 M urea-containing gel (Criterion, Bio-Rad) and transferred onto Zeta-Probe nylon membranes (Bio-Rad) using a Trans-Blot SD Semi-Dry Cell (Bio-Rad). The membranes were baked for 1 h at 80°C before prehybridization in a ProBlot hybridization oven (LabNet) for 1 h at 42°C. Prehybridization and hybridization were performed in Oligo-UltraHyb (Applied Biosystems). Hybridization was initiated by addition of 5′-end-labeled probe (see Hale et al. 2008 for sequences) to the prehybridization buffer, and hybridization was carried out overnight at 42°C. Following hybridization, the membranes were washed twice with 2× SSC (30 mM sodium citrate at pH 7.0, 300 mM NaCl) with 0.5% SDS for 30 min at 42°C. Radiolabeled RNAs were visualized by PhosphorImaging.
ACKNOWLEDGMENTS
We thank members of the Terns lab and Claiborne Glover for helpful discussions throughout the course of this work. This study was supported by NIH grant R01GM54682 (including American Recovery and Reinvestment Act [ARRA] funds) from the National Institute of General Medical Sciences to M.P.T. and R.M.T.
Footnotes
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; Cas, CRISPR-associated; psiRNA, prokaryotic silencing RNA.
Article published online ahead of print. Article and publication data are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2230110.
REFERENCES
- Andersson AF, Banfield JF 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel C, Romby P 2000. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol 318: 3–21 [DOI] [PubMed] [Google Scholar]
- Calvin K, Li H 2008. RNA-splicing endonuclease structure and function. Cell Mol Life Sci 65: 1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvin K, Xue S, Ellis C, Mitchell MH, Li H 2008. Probing the catalytic triad of an archaeal RNA splicing endonuclease. Biochemistry 47: 13659–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara A, Yao M, Masui R, Tanaka I, Yokoyama S, Kuramitsu S 2006. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci 15: 1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60 doi: 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP 2008. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14: 2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP 2009. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ 2010. The CRISPR system: Small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Jain R, Meineke B, Diver M, Shuman S 2009. Structure–activity relationships in Kluyveromyces lactis γ-toxin, a eukaryal tRNA anticodon nuclease. RNA 15: 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Mizutani K, Rhee KH, Nam KH, Lee WH, Lee EH, Kim EE, Park SY, Hwang KY 2007. Structural and mutational analysis of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic mechanism of tRNA intron-splicing endonucleases. J Bacteriol 189: 8339–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8: R61 doi: 10.1186/gb-2007-8-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Redder P, Garrett RA, Brugger K 2006. A putative viral defence mechanism in archaeal cells. Archaea 2: 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell M, Romby P, Wagner EG 2002. Lead(II) as a probe for investigating RNA structure in vivo. RNA 8: 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV 2006. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1: 7 doi: 10.1186/1745-6150-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2010a. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2010b. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463: 568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S, Griffin C, Coffey A, Meijer WC, Hafkamp B, Ross RP 2009. CRISPR analysis of bacteriophage-insensitive mutants (BIMs) of industrial Streptococcus thermophilus— implications for starter design. J Appl Microbiol 108:945–955 [DOI] [PubMed] [Google Scholar]
- Min D, Xue S, Li H, Yang W 2007. ‘In-line attack’ conformational effect plays a modest role in an enzyme-catalyzed RNA cleavage: A free energy simulation study. Nucleic Acids Res 35: 4001–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60: 174–182 [DOI] [PubMed] [Google Scholar]
- Polson A, von Wechmar MB, van Regenmortel MH 1980. Isolation of viral IgY antibodies from yolks of immunized hens. Immunol Commun 9: 475–493 [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151: 653–663 [DOI] [PubMed] [Google Scholar]
- Raines RT 1998. Ribonuclease A. Chem Rev 98: 1045–1066 [DOI] [PubMed] [Google Scholar]
- Shah SA, Hansen NR, Garrett RA 2009. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans 37: 23–28 [DOI] [PubMed] [Google Scholar]
- Steyaert J 1997. A decade of protein engineering on ribonuclease T1—atomic dissection of the enzyme–substrate interactions. Eur J Biochem 247: 1–11 [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol 55: 469–481 [DOI] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10: 200–207 [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34: 401–407 [DOI] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H 2006. RNA recognition and cleavage by a splicing endonuclease. Science 312: 906–910 [DOI] [PubMed] [Google Scholar]
- Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H 2006. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA–tRNA interaction. Nucleic Acids Res 34: 6074–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef OA, Terns RM, Terns MP 2007. Dynamic interactions within sub-complexes of the H/ACA pseudouridylation guide RNP. Nucleic Acids Res 35: 6196–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]