Abstract
Intracellular mRNA targeting and localized translation are potential determinants for protein localization. To facilitate targeting, mRNAs possess specific cis-acting sequence motifs that are recognized by trans-acting RNA-binding proteins (RBPs). While many mRNAs are trafficked, our knowledge of the RBPs involved and presence of additional transcripts within these ribonucleoprotein (RNP) complexes is limited. To facilitate the identification of RBPs and transcripts that bind to specific mRNAs, we developed RNA-binding protein purification and identification (RaPID), a novel technique that allows for the affinity purification of MS2 aptamer-tagged mRNAs and subsequent detection of bound RBPs and transcripts using mass-spectometry and RT–PCR, respectively. RaPID effectively isolated specific mRNAs from both yeast and mammalian cells, and identified known mRNA–RBP interactions (e.g., ASH1-She2; β-Actin-IMP1). By isolating tagged OXA1 mRNA using RaPID, we could identify a yeast COPI subunit (i.e., Sec27) as a candidate interacting protein. This finding was strengthened by the observation that a portion of OXA1 mRNA was delocalized in a sec27-1 temperature-sensitive mutant at restrictive temperatures. Finally, RaPID could also be used to show biochemically the coexistence of different RNA species within the same RNP complex (e.g., coprecipitation of the yeast SRO7, WSC2, SEC3, and IST2 mRNAs with ASH1 mRNA) for the first time.
Keywords: RNA affinity purification, MS2 aptamer, RNA-binding proteins, She2, IMP1, ASH1, OXA1
INTRODUCTION
Eukaryotic cells distribute most of their newly synthesized proteins in an asymmetric fashion in order to create organelles and to establish subcellular domains necessary for a wide variety of activities. Yet, only a fraction of proteins possess targeting sequences and signal peptides embedded in their primary structure; thus, it is unclear how the others target to their sites of action. Targeted mRNA localization and localized translation has been proposed to account for the site-specific deposition of protein and assembly into complexes/domains/organelles and is thought to be important for cellular processes such as cell division and fate determination (Du et al. 2007), polarity and motility (Condeelis and Singer 2005; Mili and Macara 2009), and responses to external cues (Du et al. 2007; Elson et al. 2009; Mili and Macara 2009; Yoon et al. 2009). This process is widespread and has been demonstrated in yeast (Chartrand et al. 2001), flies (Lecuyer et al. 2007), plants (Okita and Choi 2002), fungi (Zarnack and Feldbrugge 2007), and mammals (Rodriguez et al. 2008).
mRNA localization is a complex process that begins with the recognition of cis-acting elements in nascent transcripts by trans-acting RBPs (Chabanon et al. 2004; Pan et al. 2007; Shen et al. 2009). After the RNP complex is established, it exits the nucleus and is joined by additional cytoplasmic factors to create a RNP granule that associates either with the cytoskeleton (Bohl et al. 2000; Farina et al. 2003) or with specific membranes (e.g., endoplasmic reticulum [ER] [Gerst 2008], mitochondria [Saint-Georges et al. 2008], and peroxisomes [Zipor et al. 2009]) to facilitate mRNA delivery and, possibly, localized translation. While mRNAs localize to distinct sites within eukaryotic cells, little is known of the nature and types of the different RNP complexes, especially in regard to their protein and transcript content, as well as the RBPs that contribute to mRNA targeting. Different strategies have been used to help identify the cis- and trans-acting factors involved in mRNA transport and localization. One strategy involves immunoprecipitation (IP) of an RBP known to reside within a specific RNP particle, and subsequent identification of the bound proteins (Jonson et al. 2007) or interacting RNAs (Niranjanakumari et al. 2002; Gilbert and Svejstrup 2006; Keene et al. 2006). Other approaches isolate specific exogenously expressed RNAs that bear short sequence tags (called aptamers) embedded in the RNA, such as D8 and S1 (Srisawat and Engelke 2002), MS2 (Beach and Keene 2008), or StreptoTag (Bachler et al. 1999) by affinity purification and subsequent analysis of the bound factors. However, these existing procedures have been put to little use, probably due to problems stemming from the nonspecific binding of either protein or RNA.
In this study, we report an improved method for RNA-binding protein purification and identification, named RaPID. This method advances the existing state-of-the-art by using a fluorescent reporter (e.g., MS2-CP-GFP), which is able to visualize mRNAs bearing the MS2 aptamer (Bertrand et al. 1998; Beach et al. 1999; Haim et al. 2007), fused to a streptavidin-binding protein (SBP) tag, which allows for mRNA–protein complex purification using streptavidin-conjugated beads. This procedure takes advantage of the high-affinity interaction between the MS2 aptamer and the MS2-CP RBP (to bind mRNA), as well as that between SBP and streptavidin (to isolate RNP particles containing MS2 aptamer-tagged mRNAs). By adding well-defined preclearance and elution steps to reduce nonspecific binding, we successfully precipitated multiple MS2 aptamer-tagged mRNAs from both yeast and mammalian cells, and identified both known and novel interacting proteins. We were also able to identify endogenous nontagged transcripts contained within the same RNP particle for the first time. Thus, RaPID is a simple, but elegant, procedure to identify proteins and RNAs arrayed in trans with a given mRNA.
RESULTS
General description of RaPID
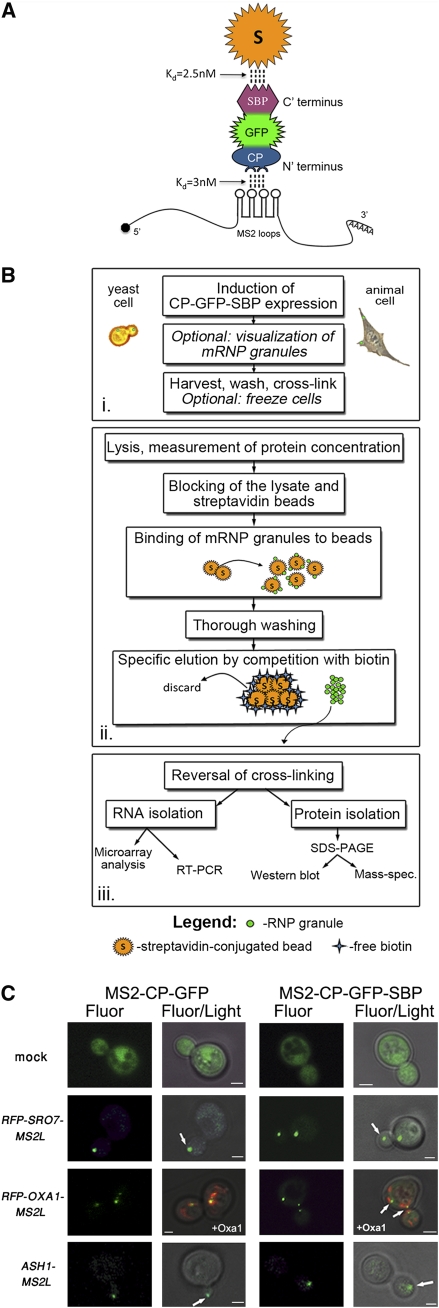
The bacteriophage MS2 coat protein (MS2-CP) has been used to visualize mRNAs bearing the MS2 aptamer loop sequence (MS2L) when fused to a fluorescent reporter molecule (e.g., GFP; to give MS2-CP-GFP) in yeast and mammalian cells (Bertrand et al. 1998; Beach et al. 1999; Fusco et al. 2003; Haim et al. 2007). Thus, we exploited the high-affinity MS2-CP::MS2L aptamer interaction (Kd = 3 x 10-9 M) (Lim and Peabody 1994) as the basis for RaPID to isolate MS2L-tagged mRNAs from cells by affinity purification. To do this, MS2-CP-GFP was fused with a streptavidin-binding protein (SBP) tag to yield MS2-CP-GFP-SBP (Fig. 1A), which allows for mRNA visualization (using fluorescence microscopy) as well as affinity purification via the interaction with a streptavidin-conjugated matrix (Kd = 2.5 x 10-9 M) (Keefe et al. 2001). The RaPID pull-down procedure consists of three principle steps (Fig. 1B). First, cells expressing a MS2L aptamer-tagged mRNA are grown in culture, and the expression of the MS2-CP-GFP-SBP is induced in a regulated manner. This step allows for the visualization of mRNP granules in live cells by fluorescence microscopy. The cells are then harvested, treated with formaldehyde (to cross-link protein–RNA interactions), and either processed directly or frozen for later use. Second, the cells are lysed and the extract is incubated with free avidin to block biotinylated moieties contained therein. In parallel, streptavidin-conjugated beads are blocked with yeast tRNA to eliminate nonspecific RNA binding. RNP granules containing MS2-CP-GFP-SBP bound to the tagged mRNA are isolated using the beads, washed thoroughly, and eluted by competition with free biotin. Third, cross-linking is reversed and both bound RNA and proteins are isolated from the eluate to enable identification and further analysis.
FIGURE 1.
RaPID and its components. (A) Functional moieties of MS2-CP-GFP-SBP. MS2-CP-GFP-SBP binds to MS2 aptamer-tagged mRNAs via the MS2-CP moiety located at the N-terminal and interacts with immobilized streptavidin via the SBP moiety at the C-terminal. The affinities of both interactions are indicated. The GFP moiety located in the central region allows for mRNA visualization via fluorescence microscopy and is recognized by anti-GFP antibodies in Westerns. (B) A flowchart of the RaPID procedure. The schematic is divided into three main steps; the first, (i) involves cell growth, induction of the MS2-CP-GFP-SBP expression, and cross-linking after harvesting. The second step (ii) includes cell lysis, the pull-down reaction, and gentle elution of the bound material with biotin. In the last step (iii), the cross-links are reversed and the RNA and protein fractions are isolated and subjected to further analysis. (C) Addition of the SBP tag does not alter mRNA visualization using the MS2 system. Yeast expressing either MS2-CP-GFP (CP-GFP) or MS2-CP-GFP-SBP (CP-GFP-SBP) were transformed with plasmids expressing MS2L-tagged RFP-SRO7 and RFP-OXA1, or ASH13′UTR. Yeast were grown to mid-log phase, transferred to medium lacking methionine for 1 h, fixed, and visualized by confocal microscopy. In cells expressing RFP-OXA1-MS2L, the RFP-tagged Oxa1 protein acts as a mitochondrial marker. White arrows indicate localization of the mRNA granules, i.e., bud-tip in the case of ASH1 and SRO7 mRNAs, and mitochondria in the case of OXA1 mRNA. (Fluor) The localization of GFP-labeled mRNA or GFP-labeled mRNA and RFP-labeled mitochondria; (Fluor/Light) the merge between the fluorescence and light microscopy (differential interference contrast) windows. Size bars, 1 μm.
MS2-CP-GFP-SBP: A fusion protein for visualizing and isolating MS2 aptamer-tagged mRNAs
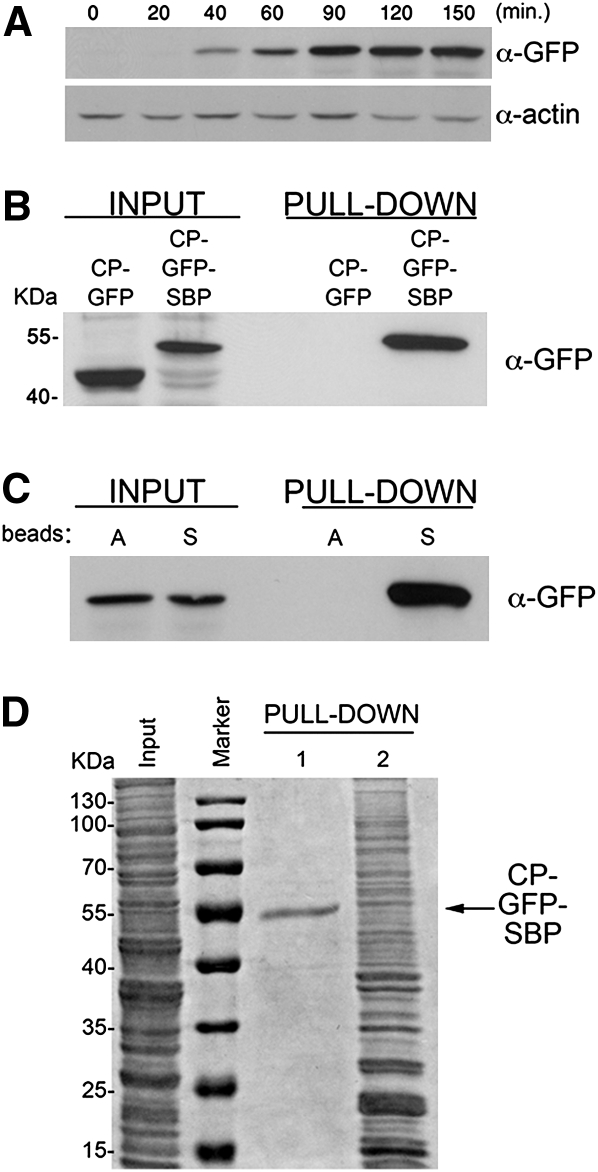
After the construction of MS2-CP-GFP-SBP, we first tested whether addition of the SBP moiety alters the ability of MS2-CP-GFP to visualize RNP granules in yeast cells. We examined in vivo the localization of three MS2L-tagged mRNAs that were visualized in previous studies: (1) ASH13′UTR mRNA, which localizes to the bud tip in a cell cycle-dependent manner (Bertrand et al. 1998; Beach et al. 1999; Aronov and Gerst 2004); (2) SRO7 mRNA, which is also polarized to the bud tip (Aronov et al. 2007; Haim et al. 2007); and (3) OXA1 mRNA, which localizes to mitochondria (Aronov et al. 2007; Haim et al. 2007). By using either conventional MS2-CP-GFP or MS2-CP-GFP-SBP, we found that addition of the SBP epitope did not affect either the visualization or localization of the fluorescent granules for any of these mRNAs (Fig. 1C). Since MS2-CP may potentially oligomerize upon high levels of expression, we placed the gene fusion encoding MS2-CP-GFP-SBP under a methionine starvation-inducible MET25 promoter in order to allow for regulated expression. We examined expression after growth in methionine-depleted medium (Fig. 2A) and found that a starvation period of 60–75 min yielded the optimal signal-to-background ratio (i.e., no aggregation of MS2-CP-GFP-SBP in the absence of a tagged mRNA and typically little to no degradation products were observed in Westerns; data not shown) and allowed for clear visualization of the mRNA granules, as seen with MS2-CP-GFP. Next, we examined whether addition of the SBP epitope allows for the isolation of MS2-CP-GFP-SBP from cell extracts using streptavidin-coated beads. Indeed, the MS2-CP-GFP-SBP protein was efficiently pulled down in contrast to MS2-CP-GFP, which lacks SBP (Fig. 2B).
FIGURE 2.
Characterization of the MS2-CP-GFP-SBP fusion protein. (A) Starvation-induced expression of MS2-CP-GFP-SBP. Yeast transformed with pMS2-CP-GFP-SBP plasmid were grown to mid-log phase (O.D.600 ≅ 1) in synthetic medium, shifted to medium lacking methionine, grown for the indicated times (minutes), and then collected. Cells were lysed and 50 μg of protein samples were analyzed by Western blotting using anti-GFP antibodies to detect MS2-CP-GFP-SBP or anti-actin antibodies to detect actin, as a loading control. (B) MS2-CP-GFP-SBP, but not MS2-CP-GFP, is efficiently pulled down with streptavidin-conjugated beads. Yeast transformed with plasmids expressing MS2-CP-GFP or MS2-CP-GFP-SBP were grown in 200-mL cultures to mid-log phase, starved for methionine for 75 min, and harvested. Following lysis, 5 mg of total protein extract derived from each transformant was incubated with streptavidin beads, washed with lysis buffer, and eluted using free biotin. Eluates (Pull-down) were resolved by SDS-PAGE along with 50-μg samples of total protein (Input) and probed with anti-GFP antibodies. Molecular mass is given in kilodaltons (kDa). (C) MS2-CP-GFP-SBP does not bind to immobilized avidin. Yeast transformed with pMS2-CP-GFP-SBP were grown to mid-log phase in 400-mL cultures, incubated in medium lacking methionine for 75 min, and harvested. After lysis, separate aliquots of 8 mg of total protein were incubated overnight with beads conjugated to either avidin (A beads) or streptavidin (S beads). Following washing, proteins were eluted with biotin and both the eluates (Pull-down) and 50 μg of samples of total protein (Input) were analyzed by Western blotting using anti-GFP antibodies. (D) Avidin blocking and biotin-mediated elution greatly improve the signal-to-noise ratio. Yeast transformed with pMS2-CP-GFP-SBP were grown in 400-mL cultures, starved for methionine for 75 min, lysed, and two aliquots each of 2 mg of total protein were incubated overnight with streptavidin-conjugated beads. One aliquot was blocked for 1 h with free avidin prior to the pull-down, and the bound material was eluted by competition with free biotin (PULL-DOWN 1), while the second aliquot (PULL-DOWN 2) was not avidin blocked and was eluted by boiling in sample buffer containing 0.1 M dithiothreitol for 5 min. Both eluates were resolved by SDS-PAGE along with a 50-μg aliquot of total protein (Input) and were stained with a general protein stain (Imperial; Sigma).
Before performing the affinity purification of MS2-CP-GFP-SBP using the streptavidin matrix, several measures were used to improve specificity. First, since streptavidin is an oligomeric version of avidin, it may interact with biotinylated moieties in the cell extract and, thus, create streptavidin–biotin complexes that bind nonspecifically to the beads (and potentially lead to false-positive results). However, since MS2-CP-GFP-SBP bound only to streptavidin-conjugated beads, and not to avidin-conjugated beads (Fig. 2C), we used free avidin to block intracellular biotin and biotinylated moieties in the lysate prior to the pull-down and, thus, decreased nonspecific binding to the immobilized streptavidin. Second, SBP is efficiently eluted from immobilized streptavidin by competition with free biotin (Keefe et al. 2001), which binds to avidin or streptavidin with very high affinity (Kd = 10-15 M). Biotin-mediated elution from the streptavidin matrix is preferable, since it does not necessitate elution at elevated temperatures nor involve reducing agents, proteases, or RNases as used by others (Niranjanakumari et al. 2002; Beach and Keene 2008). To examine the consequences of blocking by avidin and elution by biotin, we used two equal aliquots of a cell lysate containing MS2-CP-GFP-SBP; one aliquot was preincubated with free avidin prior to precipitation with streptavidin-conjugated beads and elution by biotin competition (Fig. 2D, PULL-DOWN, lane 1); while the other was precipitated directly and eluted by boiling in the presence of reducing agent (Fig. 2D, PULL-DOWN, lane 2). Importantly, we noted striking differences in the signal-to-background ratio of the two eluted samples after SDS-PAGE and nonspecific protein staining. Only MS2-CP-GFP-SBP was clearly detected after blocking with avidin and upon elution with biotin, whereas denaturation led to the appearance of a large number of additional proteins in the eluate (i.e., background). Therefore, we conclude that use of the SBP epitope does not alter the ability of MS2-CP-GFP to visualize mRNAs in vivo, allows for efficient pull-down of MS2-CP-GFP-SBP fusion protein, and confers specific elution from the streptavidin matrix.
RaPID allows for the isolation of specific mRNAs
As MS2-CP-GFP-SBP interacts with MS2L-tagged mRNAs via its amino terminus and is captured by streptavidin via its carboxyl terminus (Fig. 1A), we determined whether aptamer-tagged mRNAs can be isolated from cell lysates using RaPID. To do this, red fluorescent protein (RFP) gene- and MS2L-tagged OXA1 mRNA (i.e., RFP-OXA1-MS2L mRNA) was exogenously expressed along with MS2-CP-GFP-SBP in yeast and subjected to RaPID (Fig. 3A, lane 1). As controls, we examined the pull-down of RFP-OXA1-MS2L mRNA using beads coupled to avidin, instead of streptavidin (Fig. 3A, lane 2); with MS2-CP-GFP instead of MS2-CP-GFP-SBP (Fig. 3A, lane 3); or in the absence of the MS2L tag (i.e., untagged RFP-OXA1; Fig. 3A, lane 4). Importantly, RFP-OXA1 mRNA was identified in the eluate only when all components of the system (i.e., the MS2L tag, MS2-CP-GFP-SBP, and immobilized streptavidin) were present (Fig. 3A, lane 1). Moreover, a control mRNA (e.g., HOM2) was not recovered in the eluate, indicating that precipitation of the MS2L-tagged message was specific. Next, we precipitated exogenously expressed MS2L-tagged RFP-SRO7 and ASH13′UTR mRNAs using RaPID (Fig. 3B), and found that, like tagged RFP-OXA1-MS2L mRNA (Fig. 3A, lane 1), they could only be isolated when MS2-CP-GFP-SBP was present.
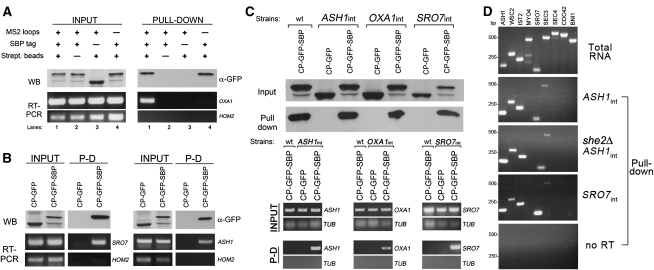
FIGURE 3.
RaPID allows for the specific isolation of MS2L-tagged mRNAs. (A) MS2 aptamer-tagged OXA1 mRNA is specifically precipitated using RaPID. Wild-type yeast cultures (300 mL each) expressing RFP-OXA1-MS2L mRNA and either pMS2-CP-GFP or pMS2-CP-GFP-SBP, and yeast expressing RFP-OXA1 mRNA and pMS2-CP-GFP-SBP were grown to mid-log phase and processed as described in the Materials and Methods. Following lysis, 10 mg of total protein from each cell type was subjected to RaPID to yield the respective eluates. For Western analysis (WB), 30% of each eluate (PULL-DOWN) and 40 μg from samples of total protein (INPUT) were resolved by SDS-PAGE and detected using anti-GFP antibodies. The remaining 70% of each eluate and 40-μg samples of total extract from each cell type were taken for RNA isolation and subsequent RT–PCR analysis (RT–PCR) with OXA1 or HOM2 primers. (Lane 1) Input from cells expressing RFP-OXA1-MS2L mRNA (+ MS2 loops) and MS2-CP-GFP-SBP (+ SBP tag) pulled down with immobilized streptavidin (+ Strept. beads). For controls, we performed the same experiment using avidin beads (lane 2), MS2-CP-GFP protein lacking the SBP tag (lane 3), or the RFP-OXA1 message lacking the MS2 loops (lane 4). HOM2 primers were used to detect HOM2 as a control mRNA. (B) MS2 aptamer-tagged SRO7 and ASH1 mRNAs can be specifically precipitated using RaPID. Wild-type yeast expressing MS2L-tagged RFP-SRO7 mRNA (left) or MS2L-tagged ASH13′UTR (right), along with either MS2-CP-GFP-SBP or MS2-CP-GFP were grown, lysed, subjected to RaPID, and analyzed as in A. (C) Endogenous MS2L-tagged ASH1, OXA1, and SRO7 transcripts can be specifically isolated using RaPID. Wild-type (wt) cells and yeast strains bearing MS2L-tagged ASH1, OXA1, and SRO7 loci (i.e., ASH1int, OXA1int, and SRO7int) were transformed with pMS2-CP-GFP-SBP or pMS2-CP-GFP plasmids and were grown in 400-mL cultures, treated, and collected. After lysis and RaPID, 25% of the eluate (Pull-down) and 40 μg of the total protein (INPUT) were resolved by SDS-PAGE and detected using anti-GFP antibodies (top). For RNA analysis (bottom), RNA was isolated from the remaining 75% of the eluate (P-D) and from 25 μL of the total extract (INPUT), and analyzed by RT–PCR using the indicated primer pairs (bottom). TUB1 primers (TUB) were used to detect TUB1 mRNA as control. (D) Additional mRNAs coprecipitate with endogenous MS2L-tagged ASH1 mRNA. Yeast (i.e., ASH1int, she2Δ ASH1int, and SRO7int) expressing the indicated endogenous MS2L-tagged mRNAs and pMS2-CP-GFP-SBP were grown in 400-mL cultures and 20 mg of total extract from each lysate was subjected to RaPID. RNAs derived from the total extract (Total RNA) or the eluates (Pull-down) were analyzed by RT–PCR using primer pairs corresponding to known or suspected bud-localized messages (e.g., ASH1 [control], WSC2, IST2, MYO4, SRO7, SEC3, SEC4, CDC42, and BNI1). mRNA coprecipitation was examined in precipitates derived from ASH1int cells (ASH1int), she2Δ ASH1int cells (she2Δ ASH1int), and SRO7int cells (SRO7int). RNA samples derived from the eluate of ASH1int cells were also subjected to PCR without reverse transcription, as controls (no RT). PCR products were resolved on 1.5% agarose gels.
Next, we examined the ability of RaPID to precipitate endogenously expressed mRNAs. We recently developed a method, called m-TAG, that integrates the MS2L sequence between the open reading frame and 3′UTR of any gene of interest in the yeast genome (Haim et al. 2007; Haim-Vilmovsky and Gerst 2009). Thus, we performed RaPID on yeast that express MS2L-tagged ASH1, OXA1, or SRO7 (ASH1int, OXA1int, or SRO7int, respectively) from their chromosomal loci. As seen (Fig. 3C), each tagged mRNA could be identified in the eluate following pull-down using MS2-CP-GFP-SBP, indicating that RaPID allows for the isolation of transcripts expressed at endogenous levels.
We then examined whether the ability of RaPID to precipitate endogenous mRNAs could help identify additional mRNA species in the isolated RNP complexes. Previously, it was suggested that the ASH1, WSC2, and IST2 mRNAs might undergo trafficking in the same granule to reach the bud tip (Lange et al. 2008); however, direct biochemical evidence for the coexistence of these different mRNA species within the same mRNP complex is lacking. Therefore, we precipitated endogenously expressed ASH1 mRNA and tested the eluate for the presence of other bud-targeted mRNAs (Fig. 3D). Indeed, this resulted in coprecipitation of ASH1, IST2, and WSC2 mRNAs, as well as other messages, such as SRO7 and SEC3 (Fig. 3D, ASH1int panel), strongly indicating that a particular mRNP granule can contain more than one mRNA species. Moreover, deletion of the SHE2 RBP gene, which is involved in the polarized delivery of ASH1 and other mRNAs (Long et al. 2000; Shepard et al. 2003; Aronov et al. 2007), did not affect the coprecipitation of the different messages (Fig. 3D; she2Δ ASH1int), indicating that RNP integrity is sustained, at least in part, upon the loss of this protein.
Importantly, the pull-down of endogenous SRO7 mRNA resulted in the reciprocal isolation of ASH1 mRNA, along with the WSC2, IST2, and SEC3 mRNAs (Fig. 3D; SRO7int). This confirms the coexistence of these messages within the same mRNP complex and the consistency of the RaPID procedure. Interestingly, addition of EDTA to the lysis buffer (5 mM) repeatedly impaired the coprecipitation of SEC3 mRNA with other messages in the RNP complex (data not shown), suggesting that the interaction of this particular transcript with the ASH1 mRNP complex might depend, in part, upon polyribosome integrity. Based on these results, we conclude that RaPID allows for the direct isolation of both exogenous and, for the first time, endogenous MS2L-tagged mRNAs and can serve as a tool to reveal the coexistence of different mRNA species within the same RNP particle.
RaPID allows for isolation of RNA-interacting proteins
As a proof of concept, we determined whether RaPID can identify specific RNA–protein interactions. We first examined the well-known interaction between ASH1 mRNA and the yeast She2 protein that binds to structural motifs present in the 3′UTR and ORF of ASH1 mRNA (Olivier et al. 2005). We used MS2-CP-GFP-SBP to precipitate the MS2L-tagged ASH13′UTR mRNA and probed the protein fraction of the eluate with anti-She2 antibodies in Western blots (Fig. 4A). Indeed, endogenous She2 was detected in the pull-down only when the MS2L-tagged ASH13′UTR was expressed (Fig. 4A, lane 2), and not when either the tagged transcript was absent (Fig. 4A, lane 1) or when SHE2 was deleted (Fig. 4A, lane 3). Next, we asked whether proteins that interact with mRNP granules, but do not directly bind to RNA, might be identified by RaPID. She2 binds to She3, an adaptor that interacts with Myo4, a type-V myosin, and connects the ASH1 mRNP granule to the actin cytoskeleton (Bohl et al. 2000; Long et al. 2000). Therefore, we used RaPID to isolate tagged ASH13′UTR granules and probed blots of the eluate with specific antibodies to detect endogenous Myo4 (Fig. 4B). Myo4 was found to precipitate with the ASH13′UTR, suggesting that RaPID can isolate proteins that reside in an mRNP complex, but do not bind directly to RNA.
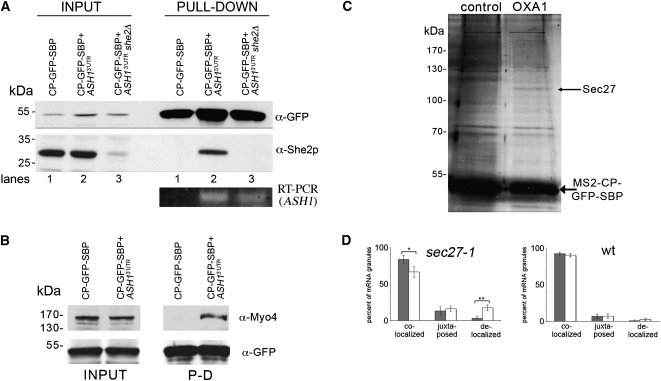
FIGURE 4.
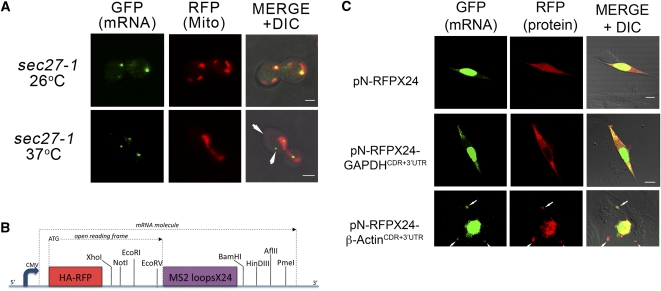
Identification of RNA-binding proteins using RaPID. (A) Endogenous She2 interacts with the 3′UTR of ASH1 mRNA. Wild-type and she2Δ yeast expressing pMS2-CP-GFP-SBP and MS2L-tagged ASH13′UTR mRNA, or wild-type yeast expressing pMS2-CP-GFP-SBP alone were grown in 400-mL cultures, treated, lysed, and 25 mg of the total protein extract from each sample was subjected to RaPID. For Western analysis, 85% of the eluates (PULL-DOWN) and 40-μg samples of the total protein (INPUT) were resolved by SDS-PAGE and analyzed using anti-She2 and anti-GFP antibodies. Note that the faint band detected with anti-She2 antibodies in input lane 3 is probably due to recognition of a nonspecific protein. For RT–PCR analysis, RNA was isolated from the remaining 15% of the eluate and analyzed by RT–PCR with a primer pair that recognizes the ASH13′UTR. (B) Endogenous Myo4 is identified in the precipitated ASH1 mRNP complex. Yeast expressing pMS2-CP-GFP-SBP and MS2L-tagged ASH13′UTR mRNA or cells expressing pMS2-CP-GFP-SBP alone were grown in 400-mL cultures, treated, lysed, and 30 mg of the total protein extract from each sample was subjected to the RaPID procedure. The eluates (PULL-DOWN) and 40-μg samples of the total protein (INPUT) were resolved by SDS-PAGE and analyzed with anti-Myo4 and anti-GFP antibodies. (C) Identification of Sec27 as a candidate OXA1 mRNA interacting protein. Wild-type yeast expressing RFP-OXA1-MS2L (OXA1) or RFP-MS2L (control) were grown in cultures of 800 mL, treated, lysed and aliquots of 50 mg of total protein extract were processed using RaPID. Following the reversal of cross-linking, the eluate was resolved by SDS-PAGE using a 20 x15 cm 9% polyacrylamide gel, silver stained, and select bands were analyzed by mass spectrometry. The thin arrow marks Sec27, while the thick arrow marks the precipitated MS2-CP-GFP-SBP protein. (D) Inactivation of Sec27 increases the proportion of delocalized OXA1 mRNA granules. Wild-type or sec27-1 yeast strains expressing RFP-OXA1-MS2L mRNA and MS2-CP-GFP(x3) were grown to mid-log phase (O.D.600 ≅ 1) at 26°C. Cells were shifted for 1 h to medium lacking methionine at the permissive temperature (26°C) and then either shifted to the restrictive temperature (37°C) for 1 h or maintained at 26°C. The cells were fixed and the localization of OXA1 mRNA granules relative to mitochondrial structures (as labeled with Oxa1-RFP) was analyzed using confocal microscopy. The gray-filled and white (unfilled) columns of the histogram indicate the distribution of granules in cells incubated at 26oC and 37oC, respectively. Statistics show the average percentage (±SE) of colocalized, juxtaposed (within ≤0.2 μm), or delocalized granules relative to the closest mitochondrial structure. (*) P = 0.035; (**) P = 0.006.
As the major aim of this procedure is to allow for the unbiased identification of RNA-interacting proteins, we examined whether RaPID can identify novel proteins that interact with mitochondria-targeted mRNAs. We precipitated either MS2L-tagged RFP-OXA1 mRNA (OXA1) or MS2L-tagged RFP mRNA (control) using MS2-CP-GFP-SBP. After separation of the eluted proteins by SDS-PAGE and silver staining, we identified a band specific for OXA1 mRNA in two separate experiments (Fig. 4C; data not shown). This band was excised, digested with trypsin, and mass-spectrometry (MS) analysis using liquid chromatography-tandem MS identified it as Sec27, the β′ subunit of the COPI coat complex that is involved mainly in retrograde transport in the early secretory pathway. Four unique peptides derived from the protein labeled in Figure 4C (see thin arrow) were identified and gave >5% coverage of Sec27 protein. Importantly, sequencing of the parallel region from the control lane did not result in the identification of Sec27, suggesting that this result is specific to OXA1 mRNA.
To test whether Sec27 has a role in OXA1 mRNA trafficking, we examined the localization of MS2L-tagged OXA1 mRNA granules in wild-type cells and yeast bearing a temperature-sensitive allele of SEC27 (Fig. 4D). Granule localization was evaluated by its proximity to the mitochondria and was divided into three categories: (1) Colocalization, when the signal overlapped with mitochondria; (2) juxtaposition, when the signal is within 0.2 μm but did not overlap; and (3) delocalization, when the signal neither overlapped nor was in close proximity. We noted an approximately sixfold increase in the percentage of delocalized granules in sec27-1 cells upon shifting to the restrictive temperature for 1 h (i.e., from 3.1 ± 2.8% at 26oC to 17.6 ± 3.9% at 37oC; P = 0.006) (Figs. 4D, 5A). In contrast, no significant change was observed in temperature-shifted wild-type cells (i.e., from 1.1 ± 1.0% at 26oC to 2.9 ± 1.7% at 37oC) (Fig. 4D). Examination of the actin cytoskeleton in these cells using rhodamine-conjugated phalloidin (data not shown) indicated that this effect is unlikely to originate from cytoskeletal defects. Thus, Sec27 may play a role in the localization of OXA1 mRNA to mitochondria. Taken together, our results demonstrate that RaPID can identify proteins that interact either directly or indirectly with aptamer-tagged mRNAs.
FIGURE 5.
Effect of SEC27 inactivation upon the localization of OXA1 mRNA in yeast and establishment of a dual mRNA and protein detection system in animal cells. (A) SEC27 inactivation enhances the delocalization of OXA1 mRNA. sec27-1 cells expressing RFP-OXA1-MS2L mRNA and MS2-CP-GFP(x3) were shifted to either restrictive temperatures for 1 h or maintained at permissive temperatures (as detailed in the legend to Fig. 4D) and visualized. White arrows indicate delocalized OXA1 mRNA granules. Size bars, 2 μm. (B) A schematic representation of the pN-RFPX24 expression vector, which is based upon pcDNA3.1(-) (Invitrogen). Downstream of the constitutive CMV promoter is an open reading frame that begins with an encoded HA epitope-tagged RFP gene that lacks a stop codon (HA-RFP), followed by a multiple cloning site (MCS; see indicated sites) for insertion of a gene of interest. Downstream of the MCS are 24 repeats of the MS2 aptamer (MS2 loops x24) and a second MCS (see indicated sites) that serves for the optional insertion of a 3′UTR sequence. (C) Visualization of GAPDH and β-Actin mRNAs and their respective translation products in animal cells. NIH3T3 fibroblasts were seeded onto round glass coverslips (13-mm diameter; in 24-well plates), grown for 24 h, and transfected with a plasmid expressing MS2-CP-GFP bearing nuclear localization sequence (100 ng DNA/well), as well as either the pN-RFPX24 plasmid alone or pN-RFPX24 plasmids that express the GADPH or β-Actin coding regions (CDRs) and corresponding 3′UTRs (500 ng DNA/well), as indicated. Fresh medium was added after 6 h and the cells were grown for an additional 12 h prior to fixation in a 4% formaldehyde solution, mounting on slides, and visualization by confocal microscopy. No asymmetric distribution of either RFP (top) or GAPDH (middle) mRNA and protein was observed. RFP protein was found in the nucleus and cytoplasm, while GAPDH (a cytosolic protein) was restricted to the cytoplasm, as expected. White arrows indicate the colocalization of β-Actin mRNA and protein at polarized extensions of the cell, as seen by others. Size bars, 10 μm.
Use of RaPID in mammalian cells
We next modified RaPID for use in cultured animal cells. First, to express MS2L-tagged mRNAs in animal cells, we created vectors that allow for the simultaneous detection of both mRNA (via the fused MS2 aptamer and MS2-CP-GFP) and its translation product (via the fused RFP moiety) (Fig. 5B), as was done previously in yeast (Aronov et al. 2007). We then cloned the ORF and 3′UTR sequences of either β-Actin or GAPDH genes into the expression vector under the control of a cytomegalovirus (CMV) promoter. Upon expression in NIH3T3 cells, we found that RFP-GAPDH protein showed a diffuse cytoplasmic pattern of localization, while RFP-tagged β-Actin protein localized to the tips of lamellipodia, where it overlapped with the localization of β-Actin mRNA, as was shown by others (Fig. 5C; Hill et al. 1994). Thus, this approach works in animal cells as well as in yeast.
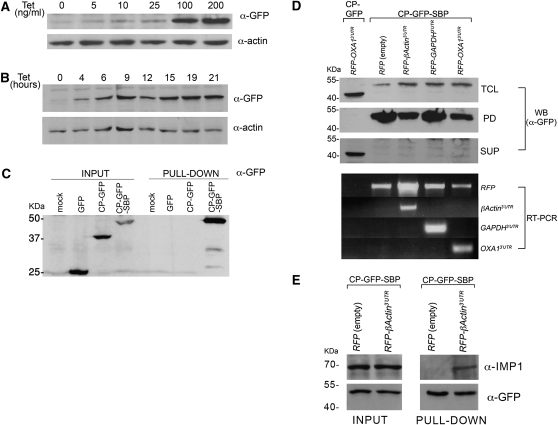
In order to use RaPID to pull-down MS2L-tagged mRNAs in mammalian cells, MS2-CP-GFP-SBP was cloned behind a tetracycline-inducible promoter to allow for regulated expression (Fig. 6A,B). When transiently expressed in HEK293 cells, only MS2-CP-GFP-SBP (and not MS2-CP-GFP or GFP alone) could be precipitated using streptavidin-conjugated beads (Fig. 6C), as described above using the yeast system (Fig. 2B). Next, 293TRex clones stably expressing MS2-CP-GFP or MS2-CP-GFP-SBP from the genome were established (i.e., 293TRex-CP-GFP and 293TRex-CP-GFP-SBP cells, respectively). To test RaPID in these cells, we expressed RFP-MS2L fused to the 3′UTRs of the mammalian GAPDH, OXA1, and β-Actin genes, and used RaPID to isolate their transcripts from cell lysates. While MS2-CP-GFP failed to precipitate the RFP-MS2L-OXA13′UTR message, MS2-CP-GFP-SBP successfully precipitated all of the tagged transcripts, as detected using RT–PCR (Fig. 6D).
FIGURE 6.
Use of RAPID in animal cells. (A) Dose-dependent induction of MS2-CP-GFP-SBP expression. 293TRex cells were transiently transfected with pcDNA4-MS2-CP-GFP-SBP (6 μg of DNA/10-cm dish), harvested after 8 h, and reseeded into 6-well plates. After an additional 12 h, tetracycline was added at the indicated concentration and the cells were grown for an additional 20 h. After harvesting, the cells were lysed and 10% of the total protein extract was resolved by SDS-PAGE and analyzed in Western blots using anti-GFP to detect MS2-CP-GFP-SBP and anti-actin antibodies to detect actin, as a loading control. (B) Time-dependent induction of MS2-CP-GFP-SBP expression. Stable 293TRex-MS2-CP-GFP-SBP cells were grown to ∼50% confluency, and then tetracycline was added to the medium (100 ng/mL) for the indicated times. After harvesting, the cells were lysed and 30 μg of the total protein extract from each time point was resolved by SDS-PAGE and analyzed in blots, as described in A. (C) Mammalian expressed MS2-CP-GFP-SBP precipitates with immobilized streptavidin. HEK293 cells were transiently transfected with pcDNA4/TO plasmids coding for GFP, MS2-CP-GFP, or MS2-CP-GFP-SBP (8 μg of DNA/100-mm dish). Nontransfected HEK293 cells were also included (mock). After 24 h, the cells were harvested, total protein was extracted, and 3.5 mg from each sample was taken for pull-down with streptavidin-conjugated beads. Both the eluates (PULL-DOWN) and 40 μL of the total protein extract (INPUT) samples were resolved by SDS-PAGE and analyzed by Western blotting using anti-GFP antibodies. (D) Isolation of mammalian MS2L-tagged mRNAs using RaPID. 293TRex cells stably expressing MS2-CP-GFP-SBP were transfected (6 μg of DNA/100-mm dish) with pN-RFPX24 plasmids expressing HA-RFP alone or as a fusion with the 3′UTRs of the β-Actin, GAPDH, or OXA1 mRNAs, as indicated. In addition, 293TRex cells stably expressing MS2-CP-GFP were transfected (6 μg of DNA/100-mm dish) with a pN-RFPX24 plasmid expressing HA-RFP fused to the 3′UTR of OXA1 mRNA. The cells were treated with tetracycline (100 ng/mL, for 12 h), collected, cross-linked with 0.1% formaldehyde, lysed, and 5 mg of total cellular extract from each sample was taken for RaPID. For Western analysis, 25% of the eluate (PD), 30 μg of total protein (TCL), and 30 μg of the supernatant remaining after incubation with the streptavidin beads (SUP) were resolved by SDS-PAGE and detected in blots using anti-GFP antibodies. For RT–PCR analysis (RT–PCR), RNA was isolated from the remaining 75% of the eluate, subjected to reverse transcription, and analyzed by PCR using the indicated primers. (E) Precipitation of endogenous IMP1 with human β-Actin mRNA. 293TRex cells stably expressing MS2-CP-GFP-SBP were transfected (10 μg of DNA/10-cm dish) with either pN-RFPX24 (empty) or pN-RFPX24 containing the 3′UTR of β-Actin. Cells were grown in the presence of tetracycline (100 ng/mL) for 12 h, collected, cross-linked using 0.01% formaldehyde, and 18 mg of each total extract was processed by RaPID. The eluates and 50 μg of the total input were analyzed by Western blots using the indicated antibodies.
To verify a known mRNA–RBP interaction in animal cells using RaPID, we expressed RFP-MS2L-β-Actin3′UTR mRNA in 293TRex-CP-GFP-SBP cells and examined the blotted eluates for the presence of endogenous IMP1 protein using specific antibodies. Human β-Actin mRNA was shown to bind ZBP1, the chicken ortholog of IMP1 (Farina et al. 2003), and the sequences sufficient for binding are contained in its 3′UTR (Chao et al. 2010). Indeed, following RaPID we could detect IMP1 bound to the RFP-MS2L-β-Actin3′UTR mRNA, but not to the RFP-MS2L mRNA (Fig. 6E). We also verified by using RaPID that RFP-MS2L fused with the 54-nt zipcode of chicken β-Actin mRNA (Chao et al. 2010) could precipitate endogenous IMP1 (data not shown). These results suggest that RaPID can also be used in mammalian cells to precipitate aptamer-tagged mRNAs and reveal specific RNA–protein interactions.
DISCUSSION
Here, we present RaPID, a method to affinity purify specific mRNAs and to identify interacting proteins and additional transcripts. RaPID uses a novel fusion protein (MS2-CP-GFP-SBP) that possesses three relevant functional domains: (1) an amino terminal MS2-CP moiety that shows high affinity to the MS2L RNA aptamer (Bernardi and Spahr 1972; Valegard et al. 1994); (2) a GFP reporter that allows for visualization of its intracellular localization in vivo and for protein detection in blots using anti-GFP antibodies; and (3) a carboxy-terminal SBP epitope that allows for specific high-affinity interactions with streptavidin-conjugated matrices (Keefe et al. 2001). Together, the CP and GFP domains allow for visualization of the intracellular localization of MS2L-tagged mRNAs of interest (Fig. 1C) before proceeding with RaPID. Correspondingly, the SBP epitope allows for affinity purification using immobilized streptavidin (Figs. 2B, 6C), elution with biotin (Fig. 2D), and the precipitation of specific bound proteins, both known (Figs. 4A,B, 6E) and unknown (Fig. 4C), as well as other RNAs present in the same RNP complex (Fig. 3D). This procedure represents an advancement over previous RNA affinity purification procedures, in that it can be used to both visualize mRNA and to reproducibly precipitate specific exogenously and endogenously expressed RNAs and their associated RBPs at a high signal/background ratio. This is due, in part, to the high-affinity interactions used to bind RNA to the novel MS2-CP-GFP-SBP protein, and the protein to the immobilized support. Together, the RaPID method allows for: (1) stringent washing of the bound RNP complexes (due to the high-affinity interaction between SBP and streptavidin); (2) simple and specific elution of the bound MS2-CP-GFP-SBP and associated RNAs using biotin; and (3) rapid detection of coprecipitated RNAs and RNA-interacting proteins using standard techniques.
In addition to the use of a single molecule (i.e., MS2-CP-GFP-SBP) for both mRNA detection (via fluorescence microscopy) and pull-down (via immobilized streptavidin), several improvements to RaPID were made during its development. First, to avoid potential artifacts resulting from MS2-CP aggregation upon overexpression (Pickett and Peabody 1993), we placed MS2-CP-GFP-SBP under an inducible promoter in both yeast (Fig. 2A) and mammalian cells (Figs. 6A,B). This allows for the tight regulation of protein expression. Second, we used formaldehyde cross-linking to stabilize interactions between the RNA and RBPs, which might otherwise be disrupted during the procedure, especially during high-stringency washing of the immobilized MS2-CP-GFP-SBP::streptavidin complex. Moreover, it may help avoid nonspecific RNA::RBP interactions that occur upon cell lysis (Mili and Steitz 2004). Lastly, formaldehyde treatment helps maintain the integrity of at least some mRNAs by preventing degradation from their ends (data not shown). Indeed, RNAse inhibition has been demonstrated at low concentrations of formaldehyde (Jonson and Lagerstedt 1959). An advantage of formaldehyde cross-linking is that it is reversible (Niranjanakumari et al. 2002), which is necessary for the separation of mRNA from protein and subsequent protein identification by MS and RNA identification using RT–PCR or RNA sequencing. Importantly, we show that formaldehyde cross-linking does not prevent the identification of proteins using MS (Fig. 4C). Moreover, to avoid nonspecific signals that excessive cross-linking might cause (Keene et al. 2006), we tested different concentrations of formaldehyde (0.05%–0.5%) with RaPID in yeast and observed no nonspecific precipitation of an unrelated protein (e.g., She2) with MS2-CP-GFP-SBP at the conditions suggested herein (i.e., 0.01%–0.1% formaldehyde) (data not shown). Thus, we find it advisable to use cross-linking, the conditions of which (e.g., formaldehyde concentration, time of cross-linking) may be adjusted in a case-specific manner depending upon the level of nonspecific binding observed. Third, we used immobilized streptavidin as a trap for MS2-CP-GFP-SBP. Streptavidin is advantageous because its near-neutral pI and lack of glycosylation result in low nonspecific interactions with proteins. Moreover, by blocking the cell lysates with free avidin prior to pull-down we reduced the nonspecific binding of immobilized streptavidin to biotin and biotinylated substrates, thus favoring the streptavidin::SBP interaction. Finally, the use of streptavidin allowed for the simple elution of bound material by competition with free biotin. Overall, RaPID allows for the specific elution of isolated tagged RNAs and their interacting factors and, thus, is probably more efficient than coimmunoprecipitation approaches (e.g., using anti-GFP antibodies to precipitate MS2-CP-GFP), which do not necessarily take advantage of specific blocking steps, stringent washing, and mild elution conditions.
RaPID proved useful for the isolation of tagged mRNAs from the eluate (Figs. 3A,B, 6D) and, importantly, demonstrated that mRNAs expressed at endogenous levels can be efficiently isolated (Fig. 3C). This feature is important, since it allows for the isolation of nonabundant messages and avoids possible artifacts originating from conditions of mRNA overexpression. Moreover, it allowed us to provide the first biochemical evidence for a physical association between polarized species of mRNAs in yeast (i.e., ASH1, WSC2, IST2, and SRO7 mRNAs) (Fig. 3D). This indicates that these different mRNAs are probably packaged in the same RNP particle, as was predicted using live-cell imaging approaches (Lange et al. 2008). Importantly, deletion of the gene encoding the She2 RBP, which is involved in mRNA polarization to the bud tip (Long et al. 2000; Shepard et al. 2003; Aronov et al. 2007), did not disrupt the interaction between these mRNAs (Fig. 3D). This indicates that She2 is not required for the integrity of the ASH1 RNP particle. Thus, RaPID can validate the coexistence of distinct mRNA species within the same mRNP granule.
The best example of the importance of RaPID is its ability to isolate RNA-interacting proteins. By using RaPID in yeast, we verified that endogenous She2 binds specifically to the 3′UTR of ASH1 mRNA (Fig. 4A), as shown by others (Bohl et al. 2000; Long et al. 2000; Olivier et al. 2005). Importantly, we also detected Myo4 in the eluate of isolated ASH13′UTR mRNA, suggesting that RaPID can identify other proteins in RNP complexes, even if they do not interact with the mRNA directly. Moreover, when using RaPID with mammalian cells, we demonstrated that endogenous IMP1, which interacts with β-Actin mRNA, could be identified upon the precipitation of MS2L-tagged β-Actin3′UTR mRNA (Fig. 6E) and the MS2L-tagged 54-nt zipcode derived from chicken β-Actin mRNA (data not shown). Finally, an unbiased approach led to the identification of Sec27, a subunit of the COPI coat complex involved in retrograde ER-to-Golgi transport (McMahon and Mills 2004), as a candidate OXA1 mRNA-interacting protein (Fig. 4C). This protein is likely to play a role in the localization of OXA1 mRNA to mitochondria, since use of a temperature-sensitive sec27-1 allele led to partial OXA1 mRNA and Oxa1 protein delocalization at restrictive temperatures (Figs. 4D, 5A; B Slobodin and JE Gerst, in prep.). This limited level of delocalization might reflect the outcome of Sec27 inactivation only upon actively transported (and not already anchored) OXA1 mRNA granules. Importantly, several studies have suggested that coatomer components might be connected to intracellular mRNA trafficking, e.g., mRNA localization to the bud tip in yeast (Trautwein et al. 2004) and to axons of animal neurons (Bi et al. 2007). Our study suggests that COPI components might also be important for mRNA transport to mitochondria; however, further study is required to elucidate this phenomenon.
Taken together, the results show that RaPID allows for the pull-down of specific mRNAs and subsequent identification of their interacting proteins and transcripts. While tested here using yeast and mammalian cells, we predict that RaPID will work efficiently in other systems as well. Thus, RaPID may become a helpful tool and lead to a better understanding of the processes governing mRNA localization.
MATERIALS AND METHODS
Plasmids
Yeast system
Plasmid pCP-GFP (Beach et al. 1999) was a gift from K. Bloom (University of North Carolina). Plasmid pUG34-MS2-CP-GFP-SBP: In separate reactions, MS2-CP-GFP was amplified by PCR from pCP-GFP, while the SBP tag was amplified from a pcDNA1-H2A-SBP plasmid (provided by G. Lederkremer, Tel Aviv University, Israel). In order to generate the MS2-CP-GFP-SBP gene fusion, chimeric reverse and forward oligonucleotides complementary to GFP and SBP, respectively, were used to generate MS2-CP-GFP lacking a stop codon and bearing the 5′ end of SBP, as well as SBP lacking an initiation codon and containing the 3′ end of GFP by PCR. In a second PCR reaction, the purified fragments were amplified by splice overlap extension. The MS2-CP-GFP-SBP fusion was then digested with XbaI and cloned behind the MET25 promoter in plasmid pUG34 (U. Güldener and J. H. Hegemann, Heinrich-Heine-Universität, Düsseldorf, Germany; see http://mips.helmholtz-muenchen.de/proj/yeast/info/tools/hegemann/gfp.html) that had been predigested with XbaI to remove the existing GFP gene. This yielded plasmid pUG34-MS2-CP-GFP-SBP. Plasmids pAD54-RFP-SRO7-MS2-3′UTR and pAD54-RFP-OXA1-MS2-3′UTR were previously described (Aronov et al. 2007). Plasmid pIIIA/ASH1-UTR, which expresses the MS2 tagged ASH13′UTR was a gift from K. Bloom.
Mammalian system
In order to express tagged mRNAs in mammalian cells, we created plasmid pN-RFPX24 that: (1) allows for fusion of a coding region of interest downstream of mRFP and in the same open reading frame (ORF); (2) bears 24 MS2L repeats; and (3) allows for introduction of a 3′UTR downstream of the aptamer. pN-RFPX24 was prepared as follows: HA epitope-tagged monomeric RFP was amplified and ligated into the NheI and ApaI restriction endonuclease sites of the pcDNA3.1(-) expression vector (Invitrogen) to yield pcDNA3.1-RFP. A fragment bearing 12 MS2L repeats was amplified from plasmid pSL1180 (gift of R. Singer; Albert Einstein College of Medicine, Bronx, NY) by PCR and ligated into the EcoRI and BamHI sites of plasmid pcDNA3.1-RFP to yield pN-RFPX12. To increase the number of loops, another set of 12 repeats was amplified, digested with BclI and BamHI, and ligated into pN-RFPX12 predigested with BamHI, to yield pN-RFPX24. Plasmids pN-RFPX24-βActin3′UTR, pN-RFPX24-GAPDH3′UTR, and pN-RFPX24-OXA13′UTR: the 3′ UTRs of human β-actin, GAPDH, and OXA1 genes were amplified from a human cDNA library and ligated into the BamHI and HindIII sites of pN-RFPX24 to yield the corresponding plasmids. The human β-actin ORF was amplified from human cDNA and ligated into the XhoI and EcoRV sites of the pN-RFPX24-βActin3′UTR to yield pN-RFPX24-βActinORF+3′UTR. The GAPDH ORF was amplified from human cDNA and inserted into the XhoI and EcoRI sites of pN-RFPX24-GAPDH3′UTR to yield pN-RFPX24-GAPDHORF+3′UTR. pN-RFPX24-ZIP54: two complementary oligonucleotides encoding the 54-nt chicken β-actin zipcode (Chao et al. 2010), and the last 10 bases of the chicken β-actin ORF were synthesized, annealed, and ligated into the BamHI and HindIII sites of the pN-RFPX24 vector. pcDNA4-MS2-CP-GFP-SBP plasmid: MS2-CP was amplified from pCP-GFP (Beach et al. 1999) and inserted into the KpnI and BamHI sites in pGEM-T Easy (Promega) to yield pGEM-MS2-CP. EGFP was amplified from pE-GFP (Clontech) and ligated into the BamHI and PstI sites of pGEM-MS2-CP to yield pGEM-MS2-CP-GFP. The SBP tag was amplified from the yeast pUG34-MS2-CP-GFP-SBP construct and ligated into the PstI and XbaI sites in pGEM-MS2-CP-GFP. The entire pGEM-MS2-CP-GFP-SBP fusion construct was then sequenced, excised with KpnI and XbaI, and ligated into pcDNA4/TO (Invitrogen) predigested with KpnI and XbaI to yield pcDNA4-MS2-CP-GFP-SBP. pcDNA4-CP-GFP plasmid: the MS2-CP sequence was amplified from pCP-GFP and ligated into the KpnI and BamHI sites of pcDNA4/TO vector (Invitrogen), while the EGFP gene bearing a stop codon was amplified from the pE-GFP plasmid (Clonetech) and ligated into BamHI and PstI sites to yield pcDNA4-CP-GFP. pcDNA4-NLS-CP-GFP plasmid: the nuclear localization sequence of the IL1α protein (i.e., MKVLKKRR) was inserted upon amplification of MS2-CP before ligation into the KpnI and BamHI sites of pcDNA4/TO prior to the subcloning of EGFP. All constructs created for this study were sequenced for verification.
Yeast strain culture and lysis procedure
A haploid derivative of the BY4743 diploid strain (Euroscarf) containing the MET15 gene was created by sporulation and tetrad analysis (BY4743h: MATa his3Δ1 leu2Δ0 ura3Δ0) (Haim et al. 2007) and is referred to as the wild-type (wt) strain for this study, except for the visualization of OXA1 mRNA presented in Figure 4E, wherein BY4743 was used. Strains possessing 12 copies of the MS2L aptamer integrated into genomic loci included: ASH1INT (MATa his3Δ1 leu2Δ0 met15Δ0 ASH1::loxP::MS2L::ASH13′-UTR); SRO7INT (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 SRO7::loxP::MS2L::SRO73′UTR) and OXA1INT (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 OXA1::loxP::MS2L::OXA13′-UTR) (Haim et al. 2007). Other strains included Y04980 (she2Δ in BY4741; MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YKL130c::kanMX4) and RDY146 (sec27-1; MATα leu2-3 112 trp1 ura3-52 sec27-1) from R. Duden (University of Lubeck, Lubeck, Germany). DNA introduction into yeast was performed using standard procedures.
For cell culture, a single yeast colony was inoculated into 5 mL of synthetic selective medium and grown for 7 h at 26°C with constant shaking. Afterward, the culture was transferred into a flask containing 200–400 mL of selective medium and grown overnight at 26°C with constant shaking to an O.D.600 = ∼1. The cells were centrifuged using a Sorvall SLA3000 rotor at 1100g for 5 min and resuspended in 200 mL of complete synthetic medium lacking methionine in order to induce the expression of the MS2-CP-GFP-SBP protein, and grown for an additional 1 h. The cells were collected by centrifugation as described above, washed with PBS buffer (lacking Ca++ and Mg++), and transferred into a 50-mL tube and pelleted as above. Cellular proteins were cross-linked by the addition of 8 mL of PBS containing 0.05% formaldehyde and incubated at 24°C for 10 min with slow shaking. The cross-linking reaction was terminated by adding 1 M glycine buffer (pH 8.0) to a final concentration of 0.125 M and additional shaking for 2 min. The cells were then washed once with ice-cold PBS buffer and the pellet was flash-frozen in liquid nitrogen, and stored at −80°C. For lysis, cell pellets were thawed upon the addition of ice-cold lysis buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1.8 mM MgCl2, and 0.5% NP40 supplemented with Aprotinin [10 μg/mL], PMSF [1 mM], Pepstatin A [10 μg/mL], Leupeptin [10 μg/mL], 1 mM DTT, and 80 U/ml RNAsin [Promega]) at 1 mL per 100 O.D.600 U, and 0.5-mL aliquots were then transferred to separate microcentrifuge tubes containing an equal volume of 0.5 mm glass beads, and vortexed in an IKA/Vibrax shaker at maximum speed for 45 min at 4°C. Glass beads and unbroken cells were sedimented at 4°C by centrifugation at 1700g for 1 min, and the supernatant removed to new microcentrifuge tubes and centrifuged at 15,300g at 4°C for 15 min. The total cell lysate (TCL) was then removed to a fresh tube and protein concentration was determined using the microBCA protein determination kit (Pierce).
Mammalian cell culture and lysis procedure
Cell lines used in this work included HEK293 and NIH3T3 from the ATCC collection. HEK293-TRex cells stably expressing the tetracycline repressor (pcDNA6-TR, Invitrogen) of the TRex system (Invitrogen) were a generous gift from Sara Lavi (Tel-Aviv University, Israel). Stable cell lines expressing either MS2-CP-GFP or MS2-CP-GFP-SBP under the TRex cassette were obtained by transfection of pcDNA4-MS2-CP-GFP or pcDNA4-MS2-CP-GFP-SBP into the HEK293-TRex cells (10 μg of DNA/10-cm dish) followed by selection of the stable transfectants by growing cells in medium supplemented with 300 μg/mL of Zeocin (Invitrogen).
Cells were maintained in 10-cm culture dishes with DMEM medium containing 10% fetal calf serum at 37°C and 5% CO2. The transfection of 3T3 cells was performed using JetPEI reagent (Polyplus transfection), while the transfection of ∼70% confluent 293HEK cultures was performed using calcium phosphate according to standard protocols. Culture medium was changed 6–8 h following transfection, and the cells were grown for an additional 16 h. The cells were washed once and collected in 3 mL of ice-cold PBS per dish using a cell scraper, transferred into 14-mL tubes, centrifuged at 4000g for 3 min, and incubated with a total of 10 mL of PBS-formaldehyde solution at the indicated concentrations for 10 min at 24°C with slow shaking. The cross-linking reaction was terminated by adding 1 M glycine buffer (pH 8.0) to a final concentration of 0.125 M and additional shaking for 2 min. The cells were then washed once with 10 mL of ice-cold PBS, flash-frozen in liquid nitrogen, and stored at −80°C. For lysis, cells were thawed upon the addition of 2 mL of ice-cold lysis buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1.8 mM MgCl2, and 0.5% NP40 supplemented with Protease inhibitor cocktail for mammalian cells [Sigma], 1 mM DTT, and 80 U/mL RNAsin [Promega]), transferred into 5-mL glass tubes, and sonicated on ice using a Microsonic sonicator for three rounds; each round lasted 20 sec at 7–8 Watts, with 2-min pauses on ice in between rounds. Cell debris was pelleted by centrifugation at 15,300g for 15 min at 4°C, and the supernatant removed to a new microcentrifuge tube. Protein concentration in the TCL (supernatant) was determined using the microBCA protein determination kit (Pierce).
RaPID procedure for precipitation of RNP complexes
Protein aliquots used in the RaPID procedure varied and are listed in the legend to each figure. In order to block endogenous biotinylated moieties, the protein aliquot taken for pull-down was incubated with 10 μg of free avidin (Sigma) per 1 mg of protein input at 4°C for 1 h with constant rotation. In parallel, streptavidin-conjugated beads (Streptavidin-sepharose high performance, GE Healthcare) were aliquoted to microcentrifuge tubes according to 5 μL of the slurry per 1 mg of protein (but not <30 μL overall), washed twice in 1 mL of PBS, once in 1 mL of lysis buffer, and blocked with a 1:1 mixture of 1 mL of lysis buffer containing yeast tRNA (Sigma; 0.1 mg/100 μL of beads) and 1 mL of 4% BSA in PBS at 4°C for 1 h with constant rotation. Following the blocking step, the beads were washed twice in 1 mL of lysis buffer. Pull-down was then performed by adding the indicated amount (see figure legends) of avidin-blocked TCL to the beads, followed by incubation at 4°C for 2–15 h with constant rotation. Yeast tRNA was added to the pull-down reaction (0.1 mg/tube) to reduce nonspecific interactions. We used standard 1.7-mL microcentrifuge tubes when working with small volumes of TCL or 15-mL sterile polypropylene centrifuge tubes with larger volumes. Following pull-down, the beads were centrifuged at 660g at 4°C for 2 min; the supernatant was then removed and the beads washed three times with lysis buffer (e.g., 1-mL volume washes for small tubes, 2-mL for large tubes), twice with wash buffer (20 mM Tris at pH 7.5, 300 mM NaCl, and 0.5% Triton X100), all performed at 4°C, with each step lasting for 10 min with rotation. The beads were then equilibrated by a final wash in 1–2 mL of cold PBS, pelleted by centrifugation as above, and excess buffer aspirated. For elution of the cross-linked RNP complexes from the beads, 100 μL of PBS containing 6 mM free biotin (Sigma) was added to the beads, followed by 1 h of incubation at 4°C with rotation. After centrifugation at 660g for 2 min, the eluate was transferred into a fresh microcentrifuge tube, recentrifuged, and transferred into another tube to assure that no beads were carried over. To reverse the cross-link, the eluate was incubated at 70°C for 1–2 h with an equal volume of 2X cross-link reversal buffer (100 mM Tris at pH 7.0, 10 mM EDTA, 20 mM DTT, and 2% SDS) for RNA analysis or with an appropriate volume of 5X protein sample buffer (5X: 0.4 M Tris at pH 6.8, 50% glycerol, 10% SDS, 0.5 M DTT, and 0.25% bromophenol blue) for protein analysis using SDS-PAGE.
Silver staining and mass spectrometry analysis
Following SDS-PAGE, protein gels were stained by silver staining (Silver SNAP Stain Kit II, Pierce). A region containing a ∼110-kDa band specific to RFP-OXA1 mRNA, as well as the corresponding region in the RFP control lane, were excised and analyzed by liquid chromatography/tandem MS.
RNA isolation, reverse transcription, and PCR
RNA was isolated either from the TCL or from the post-RaPID eluate by adding 175 μL of MPC Protein Precipitation Reagent (MasterPure RNA isolation kit, Epicentre Biotechnologies) per 300-μL volume, as described by the manufacturer, followed by centrifugation at 20,000g for 10 min at 4°C. The supernatant was transferred into a fresh microcentrifuge tube and both NaOAc (pH 5.4; 0.3 M final concentration) and glycogen (80 μg/mL final concentration; Fermentas) were added, followed by thorough vortexing. An equal volume of isopropanol was then added to the tube, followed by vortexing and incubation overnight at −20°C. After centrifugation at 20,000g for 10 min at 4°C, the RNA pellet was rinsed once with 70% ethanol, air-dried, and dissolved in 30 μL of Ultra-pure water (Biological Industries, Israel). The recovered RNA was subjected to treatment with RQ1-Rnase-free DNase (Promega) as detailed by the manufacturer, and 10 μL from this reaction (or 1 μg from the total RNA) was taken for reverse transcription (RT) using the M-MLV RNase H Minus, Point Mutant reverse transcriptase (Promega), and a random hexamer mixture (Fermentas) in a final volume of 25 μL. From the RT product, 1 μL was taken for each PCR reaction (15 μL total reaction volume) using the Taq Purple Mix (Lamda Biotech) and the specific primer pairs listed in Supplemental Table I.
Visualization of mRNA granules in yeast
Yeast cells were grown to mid-log phase in synthetic medium; 0.5-mL samples were aliquoted to 1.7-mL microcentrifuge tubes, centrifuged at 960g for 2 min, and fixed by resuspension in 100 μL of fresh 4% paraformaldehyde solution containing 3% sucrose for 15 min at room temperature. The cells were then washed once in KPO4/sorbitol solution (1.2 M sorbitol, 100 mM potassium phosphate buffer at pH 7.5) and kept in 200 μL of this solution at 4°C. The cells were visualized by confocal microscopy (Zeiss LSM 510) using a 100X oil immersion lens. For the statistical analysis of the intracellular distribution of OXA1 mRNA granules, three experiments were performed (n = 3) in which >200 granules were counted in total for each strain at each temperature. The Student's t-test (two-tailed; homoscedastic) was performed on the percentage of mRNA granule distribution.
Antibodies
Monoclonal anti-GFP antibodies (1:3,000) were obtained from Roche Diagnostics and anti-actin antibodies (1:10,000) were obtained from MP Biomedicals. Polyclonal anti-She2 antibodies (1:3,000) were a gift from R. Jansen (University of Tuebingen, Tuebingen, Germany); anti-Myo4 antibodies were a gift from P. Takizawa, (Yale University, New Haven, CT, USA); and anti-IMP1 polyclonal antibodies (1:500) were from I. Ginzburg (Weizmann Institute of Science, Rehovot, Israel).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank K. Bloom, S. Lavi, R. Duden, R. Jansen, G. Lederkremer, P. Takizawa, I. Ginzburg (deceased), and R. Singer for reagents and the Smoler Proteomics Center at the Technion, Israel for mass-spectrometry. This work was supported by grants to J.E.G. from the Yeda CEO Fund, Weizmann Institute of Science, Israel, and Minerva Foundation, Germany. J.E.G. holds the Besen-Brender Chair of Microbiology and Parasitology.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2091710.
REFERENCES
- Aronov S, Gerst JE 2004. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J Biol Chem 279: 36962–36971 [DOI] [PubMed] [Google Scholar]
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE 2007. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol 27: 3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachler M, Schroeder R, von Ahsen U 1999. StreptoTag: A novel method for the isolation of RNA-binding proteins. RNA 5: 1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Keene JD 2008. Ribotrap : Targeted purification of RNA-specific RNPs from cell lysates through immunoaffinity precipitation to identify regulatory proteins and RNAs. Methods Mol Biol 419: 69–91 [DOI] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K 1999. Localization and anchoring of mRNA in budding yeast. Curr Biol 9: 569–578 [DOI] [PubMed] [Google Scholar]
- Bernardi A, Spahr PF 1972. Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc Natl Acad Sci 69: 3033–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Bi J, Tsai NP, Lu HY, Loh HH, Wei LN 2007. Copb1-facilitated axonal transport and translation of κ opioid-receptor mRNA. Proc Natl Acad Sci 104: 13810–13815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl F, Kruse C, Frank A, Ferring D, Jansen RP 2000. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 19: 5514–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabanon H, Mickleburgh I, Hesketh J 2004. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief Funct Genomics Proteomics 3: 240–256 [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM 2001. RNP localization and transport in yeast. Annu Rev Cell Dev Biol 17: 297–310 [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH 2005. How and why does β-actin mRNA target? Biol Cell 97: 97–110 [DOI] [PubMed] [Google Scholar]
- Du TG, Schmid M, Jansen RP 2007. Why cells move messages: The biological functions of mRNA localization. Semin Cell Dev Biol 18: 171–177 [DOI] [PubMed] [Google Scholar]
- Elson SL, Noble SM, Solis NV, Filler SG, Johnson AD 2009. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet 5: e1000664 doi: 10.1371/journal.pgen.1000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE 2008. Message on the web: mRNA and ER co-trafficking. Trends Cell Biol 18: 68–76 [DOI] [PubMed] [Google Scholar]
- Gilbert C, Svejstrup JQ 2006. RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr Protoc Mol Biol 75: 27.4.1–27.4.11 [DOI] [PubMed] [Google Scholar]
- Haim L, Zipor G, Aronov S, Gerst JE 2007. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4: 409–412 [DOI] [PubMed] [Google Scholar]
- Haim-Vilmovsky L, Gerst JE 2009. m-TAG: A PCR-based genomic integration method to visualize the localization of specific endogenous mRNAs in vivo in yeast. Nat Protoc 4: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Hill MA, Schedlich L, Gunning P 1994. Serum-induced signal transduction determines the peripheral location of beta-actin mRNA within the cell. J Cell Biol 126: 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonson N, Lagerstedt S 1959. The effect of formaldehyde containing fixatives on ribonuclease activity. Histochem Cell Biol 1: 251–256 [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics 6: 798–811 [DOI] [PubMed] [Google Scholar]
- Keefe AD, Wilson DS, Seelig B, Szostak JW 2001. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr Purif 23: 440–446 [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB 2006. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307 [DOI] [PubMed] [Google Scholar]
- Lange S, Katayama Y, Schmid M, Burkacky O, Brauchle C, Lamb DC, Jansen RP 2008. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic 9: 1256–1267 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lim F, Peabody DS 1994. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res 22: 3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P 2000. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Mills IG 2004. COP and clathrin-coated vesicle budding: Different pathways, common approaches. Curr Opin Cell Biol 16: 379–391 [DOI] [PubMed] [Google Scholar]
- Mili S, Macara IG 2009. RNA localization and polarity: From A(PC) to Z(BP). Trends Cell Biol 19: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10: 1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Okita TW, Choi SB 2002. mRNA localization in plants: Targeting to the cell's cortical region and beyond. Curr Opin Plant Biol 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P 2005. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol 25: 4752–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Huttelmaier S, Singer RH, Gu W 2007. ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol Cell Biol 27: 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett GG, Peabody DS 1993. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res 21: 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH 2008. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol 20: 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C 2008. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 3: e2293 doi: 10.1371/journal.pone.0002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Paquin N, Forget A, Chartrand P 2009. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell 20: 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD 2003. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci 100: 11429–11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR 2002. RNA affinity tags for purification of RNAs and ribonucleoprotein complexes. Methods 26: 156–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein M, Dengjel J, Schirle M, Spang A 2004. Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol Biol Cell 15: 5021–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L 1994. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature 371: 623–626 [DOI] [PubMed] [Google Scholar]
- Yoon BC, Zivraj KH, Holt CE 2009. Local translation and mRNA trafficking in axon pathfinding. Results Probl Cell Differ 48: 269–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, Feldbrugge M 2007. mRNA trafficking in fungi. Mol Genet Genomics 278: 347–359 [DOI] [PubMed] [Google Scholar]
- Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE 2009. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc Natl Acad Sci 106: 19848–19853 [DOI] [PMC free article] [PubMed] [Google Scholar]