Abstract
Coronaviruses efficiently inhibit interferon (IFN) induction in nonhematopoietic cells and conventional dendritic cells (cDC). However, IFN is produced in infected macrophages, microglia, and plasmacytoid dendritic cells (pDC). To begin to understand why IFN is produced in infected macrophages, we infected bone marrow-derived macrophages (BMM) and as a control, bone marrow-derived DC (BMDC) with the coronavirus mouse hepatitis virus (MHV). As expected, BMM but not BMDC expressed type I IFN. IFN production in infected BMM was nearly completely dependent on signaling through the alpha/beta interferon (IFN-α/β) receptor (IFNAR). Several IFN-dependent cytokines and chemokines showed the same expression pattern, with enhanced production in BMM compared to BMDC and dependence upon signaling through the IFNAR. Exogenous IFN enhanced IFN-dependent gene expression in BMM at early times after infection and in BMDC at all times after infection but did not stimulate expression of molecules that signal through myeloid differentiation factor 88 (MyD88), such as tumor necrosis factor (TNF). Collectively, our results show that IFN is produced at early times postinfection (p.i.) in MHV-infected BMM, but not in BMDC, and primes expression of IFN and IFN-responsive genes. Further, our results also show that BMM are generally more responsive to MHV infection, since MyD88-dependent pathways are also activated to a greater extent in these cells than in BMDC.
IMPORTANCE
Coronaviruses cause diseases with various degrees of severity in humans, including severe acute respiratory syndrome (SARS). In domestic and companion animals, coronaviruses induce interferon (IFN) in only a few cell types. In particular, macrophages, which are known to have both protective and pathogenic roles in coronavirus infections, express IFN while dendritic cells do not. Little is known about the basis of these cell-specific differences in IFN induction. Here, we show that an animal coronavirus, mouse hepatitis virus, induces IFN and other IFN-responsive molecules in macrophages, but not in dendritic cells, via a feedback loop that is dependent upon low-level IFN expression at early times after infection. This pathway of cellular activation may be a useful target for modulating macrophage function in order to selectively enhance the antivirus immune response and diminish the pathogenic role of these cells in SARS and other coronavirus infections.
Introduction
Type I interferon (IFN), alpha/beta interferon (IFN-α/β), plays a critical role during the initial antivirus immune response. Type I IFNs directly inhibit viral replication by creating an “antiviral state” by upregulating interferon-responsive genes, such as the genes encoding 2′,5′-oligoadenylate synthetase, RNase L, and Mx protein, and they are also critical for activating and regulating the adaptive immune response (1, 2). The importance of the type I IFN response during viral infection is reflected by the fact that most viruses express IFN antagonists that are able to suppress IFN pathways at the level of IFN production or signaling or that directly interact with antiviral IFN-responsive genes (reviewed in references 3 to 5). Type I IFN production is initiated after recognition of pathogen-associated molecular patterns (PAMPs), such as viral double-stranded RNA (dsRNA), by host cell pathogen recognition receptors (PRRs). There are two major types of PRRs that induce IFN production, cytoplasmic RNA helicases, including retinoic acid-inducible gene I product (RIG-I) and melanoma differentiation-associated gene 5 product (MDA5), and Toll-like receptors (TLRs), including Toll-like receptor 2 (TLR2), TLR3, TLR4, TLR7, TLR8, and TLR9 (6). Almost all cells express type I IFN receptor, which allows them to respond to IFN, and are able to produce various amounts of type I IFN during viral infections. In particular, plasmacytoid dendritic cells (pDC) produce large amounts of alpha interferon (IFN-α) during early stages of the inflammatory response. Conventional dendritic cells (cDC) and macrophages also express type I IFN, although not to the same extent as pDC do (7).
Coronaviruses are large, positive-sense, single-stranded RNA viruses that cause clinically significant diseases in humans and domestic and companion animals (8). All coronaviruses encode four or five structural proteins, approximately 16 replicase-associated proteins, and one to eight accessory proteins (proteins interspersed between or within structural proteins encoded at the 3′ end of the genome). SARS-CoV (severe acute respiratory syndrome coronavirus) accessory proteins ORF3b and ORF6, mouse hepatitis virus (MHV) accessory protein ORF5a, MHV and SARS-CoV nucleocapsid (N) proteins and replicase-associated nonstructural protein 1 (nsp1) and nsp3 are all able to inhibit IFN induction and/or signaling (9–15) (reviewed in reference 16). In addition, MHV delays IFN-stimulated gene (ISG) induction by IFN or Sendai virus (SenV) (17). Further, in some cell types, including fibroblasts and cDC, neither SARS-CoV nor MHV induces IFN expression. However, neither virus is able to prevent IFN induction by other viruses or by poly(I ⋅ C), which suggests that these viruses are “invisible” to intracellular IFN sensors (18, 19). This “passive” method of immune evasion may occur because coronaviruses replicate on membranous structures, including double-membrane vesicles, thereby limiting exposure of sequestered RNA to innate immune sensors (18–20). On first glance, these results appeared to contradict the observation that type I IFNs are detected in MHV-infected mice and in patients with SARS (21, 22). However, IFN is induced in pDC by a TLR7- and interferon regulatory factor 7 (IRF7)-dependent pathway, in macrophages and microglia in an MDA5-dependent pathway, and in oligodendrocyte cell lines via MDA5- and RIG-I-dependent pathways (23–25). Collectively, these results show that coronaviruses use multiple approaches to evade the innate immune response and that the extent and nature of evasion are cell type dependent. The basis of these cell type-specific differences in IFN expression after coronavirus infection is not well understood.
Both macrophages, in which IFN is induced, and DC, in which IFN is not induced, are important target cells for several coronaviruses. DC in particular are critical for initiation of the antivirus T- and B-cell responses, which are ultimately critical for virus clearance. Infection of DC and macrophages likely contributes to the development of effective innate and adaptive immune responses, but macrophage infection, in the case of some coronaviruses, also contributes to immunopathological disease and enhanced lethality (26, 27). Macrophage infection has been demonstrated in infected humans or animals (28–30), and both types of cells are readily infected with coronaviruses after culture in vitro (24, 31, 32).
Here, we cultured primary bone marrow (BM) cells in vitro in the presence of cytokines and growth factors, resulting in differentiation into cells with the characteristics of macrophages (BMM, cultured in the presence of macrophage colony-stimulating factor [M-CSF]; CD11b+ CD11c−) or dendritic cells (BMDC, cultured in the presence of granulocyte-macrophage colony-stimulating factor [GM-CSF] and interleukin-4 [IL-4]; CD11b+ CD11c+). Using these cells, we showed that most of the IFN produced after MHV infection was dependent on signaling through the IFN-α/β receptor (IFNAR). This IFN was critical for virus-induced upregulation of several IFN-related molecules, including MDA5, RIG-I, and CXCL10 (chemokine [C-X-C motif] ligand 10). Further, levels of type I IFN produced in MHV-infected BMM were sufficient to induce maximal activation, since exogenous type I IFN treatment did not further stimulate IFN-related gene upregulation at late times postinfection (p.i.). Our results also showed that the expression of some inflammation-associated molecules, most notably tumor necrosis factor (TNF), IL-12, and IL-6, was mediated, in large part, via signaling through myeloid differentiation factor 88 (MyD88)-dependent pathways.
RESULTS
MHV-infected BMM produce type I IFN at late times p.i.
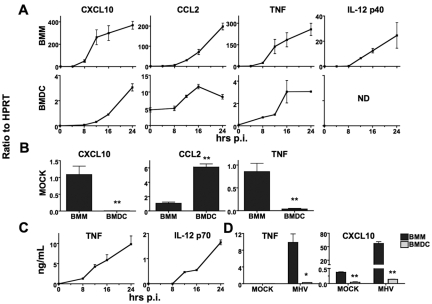
We previously showed that neither BMDC nor fibroblasts produce type I IFN after infection with the JHM strain of MHV (JHMV), and others have found similar results using the A59 strain (MHV-A59) (18, 19, 23, 33). For the experiments described here, we used only MHV-A59 (MHV) because this virus grows to high titer and forms syncytia at later times postinfection (p.i.) than JHMV, allowing prolonged cell survival. Roth-Cross et al. showed that BMM produced type I IFN after MHV infection, but only late times p.i. (24 h p.i.) were examined in that initial study (24). To determine the kinetics of IFN expression, we infected BMM or BMDC with MHV and then measured IFN-β mRNA and IFN protein levels at different times p.i. by real-time quantitative PCR (qPCR) and by an IFN bioassay, respectively. Sendai virus (SenV) was included as a positive control, since it is known to induce IFN. SenV-infected BMM rapidly upregulated IFN expression with protein detected as early as 2 h p.i., and peak levels observed at approximately 8 h p.i. (Fig. 1A). In contrast, MHV-infected BMM produced IFN much more slowly with levels of IFN-β mRNA or IFN protein barely detectable at 8 h p.i. Peak levels of IFN-β mRNA and protein were reached at 16 to 24 h p.i. (Fig. 1A to C). Maximal IFN protein levels were approximately 20- to 25-fold lower in MHV-infected cells than in SenV-infected cells (Fig. 1A and B). Although the IFN protein levels were much lower in MHV-infected BMM than in SenV-infected cells, IFN-β mRNA levels were higher in MHV-infected cells (Fig. 1A to C; see also Fig. 3C and E). This likely resulted from MHV-mediated inhibition of host cell translation, observed in MHV-infected fibroblasts (11, 14, 34, 35). Consistent with previous findings (23, 32, 33), we detected less than 3 U/ml of IFN in MHV-infected DC at 24 h p.i. (Fig. 1B). These results did not reflect differences in the ability of the two cell types to produce IFN, because DC rapidly produced IFN after SenV infection and peak IFN protein levels were only 2-fold less in SenV-infected BMDC than in BMM (Fig. 1A). Of note, both BMM and BMDC were readily infected by MHV, although virus replication proceeded more rapidly in BMM, with syncytium formation detected as early as 6 h p.i. and peak viral titers observed at 8 to 12 h p.i. MHV reached maximal virus titers in BMDC that were approximately 1 log unit higher than in BMM (Fig. 1D).
FIG 1 .
Mouse hepatitis virus (MHV)-infected bone marrow-derived macrophages (BMM) produce type I IFN at late times p.i. (A) BMM or bone marrow-derived dendritic cells (BMDC) were infected with MHV or Sendai virus (SenV), and IFN protein levels were determined using a VSV-based IFN bioassay at the indicated times (in hours) postinfection (p.i. ). BMM-MHV, BMM infected with MHV; BMDC-MHV, BMDC infected with MHV. (B) MHV-induced IFN production in panel A is shown using an enlarged scale. (C) BMM were infected with MHV or SenV, and IFN-β mRNA levels were measured by real-time qPCR. Cycle threshold (CT) values were calculated as a ratio to hypoxanthine phosphoribosyltransferase (HPRT) as described in Materials and Methods. (D) MHV titers in supernatants were determined as previously described (52). Two or three replicates were performed in each experiment, and one of three independent experiments is shown.
FIG 3 .
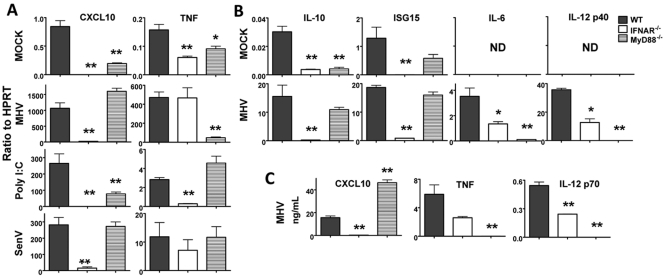
MHV-induced upregulation of IFN-β, MDA5, and RIG-I in BMM is dependent on IFN-α/β receptor (IFNAR). BMM isolated from WT, IFNAR−/−, or MyD88−/− mice were infected with MHV (A and C) or SenV (E) or transfected with poly(I ⋅ C) (D). (A) MHV titers were determined in cell supernatants at 16 hours postinfection as described in Materials and Methods. (B) mRNA was harvested from mock-infected BMM, and basal levels of IFN-β, MDA5, and RIG-I were measured by real-time qPCR. (C to E) mRNA was harvested 16 h after infection or transfection. mRNA levels were measured by real-time qPCR. CT ratios to HPRT are shown. MHV induced upregulation to a greater extent than poly(I ⋅ C) or SenV did (note the differences in scale in panels B to E). Two or three replicates were performed in each experiment, and one of three independent experiments is shown. Values for IFNAR−/− or MyD88−/− BMM that were statistically significantly different from the values for wild-type cells are indicated as follows: *, P < 0.05; **, P < 0.01.
MHV-induced upregulation of type I IFN, MDA5, and RIG-I in BMM is dependent on IFNAR.
To begin to investigate why BMM but not BMDC produced IFN after MHV infection, we examined the levels of MDA5, since this intracellular helical sensor is required for IFN induction in BMM (24). Basal levels of MDA5 mRNA in mock-infected BMM were approximately 2-fold higher than in BMDC (Fig. 2A). After infection with MHV, MDA5 levels were upregulated with similar kinetics in BMM and BMDC; however, levels in BMM were about 20 to 30 times higher than in BMDC at 24 h p.i. (Fig. 2B and C). Similar changes in expression of a second intracellular sensor molecule, RIG-I, were also observed. RIG-I is not believed to be critical for IFN production in MHV-infected BMM, although it has an important role in cells such as oligodendrocytes (25).
FIG 2 .
MHV upregulates MDA5 and RIG-I expression to higher levels in BMM than in BMDC. (A) Basal mRNA levels of MDA5 and RIG-I in mock-infected BMM and BMDC were measured by real-time qPCR. Values that are significantly different (P < 0.05) from the value for BMM are indicated by an asterisk. (B and C) mRNA levels of MDA5 and RIG-I in BMM (B) and in BMDC (C) at different times p.i. were measured by real-time qPCR. CT ratios to HPRT were calculated as described in Materials and Methods. MDA5 and RIG-I were upregulated to a greater extent in BMM than in BMDC (note the differences in scale). Two or three replicates were performed in each experiment, and one of three independent experiments is shown.
Type I IFN mediates enhanced expression of MDA5 and RIG-I (36). Thus, elevated baseline levels of MDA5 and RIG-I in MHV-infected BMM (Fig. 2A) might facilitate enhanced IFN production at early times p.i., which could then result in increased MDA5 and RIG-I expression after signaling through IFNAR. To examine this possibility, we cultured BMM from wild-type and IFNAR−/− mice. We included BMM from MyD88−/− mice in these experiments, since MyD88 is involved in signaling through TLRs but not IFNAR. We detected low basal levels of IFN, MDA5, and RIG-I in BMM from all mice, although the levels were lowest in IFNAR−/− BMM (Fig. 3B). The levels of IFN-β, MDA5, and RIG-I mRNA were significantly lower in infected IFNAR−/− BMM than in wild-type BMM (Fig. 3C). However, the mRNA levels of these molecules in infected IFNAR−/− BMM were still higher than in mock-infected IFNAR−/− cells (Fig. 3B and C), indicating that virus infection directly upregulates IFN-β, MDA5, and RIG-I, but optimal induction requires signaling through the IFNAR. These differences did not result from differences in susceptibility to infection because equivalent titers of virus were observed in IFNAR−/−, MyD88−/−, and wild-type (WT) BMM at 16 h p.i. (Fig. 3A). Of note, MHV titers are much higher in tissues of IFNAR−/− mice than in wild-type mice. The lack of difference in titers in Fig. 3A may reflect the high multiplicity of infection that we used in our experiments or the well-described relative insensitivity of MHV to IFN signaling in tissue culture cells (13, 14, 37).
MHV induces IFN production in pDC through a TLR7-dependent pathway, which signals through MyD88. TLR7 is also expressed in BMM (38). However, we found that type I IFN production in MHV-infected BMM was not dependent on MyD88 in agreement with a previous report (24) (Fig. 3C). Further, the levels of MDA5 and RIG-I in MyD88−/− BMM were approximately equal to those found in WT cells, indicating that regulation of these genes in MHV-infected BMM is not dependent on MyD88 (Fig. 3C). These findings are not unique for MHV, since expression of IFNAR, but not MyD88, was also required for expression of IFN-β, MDA5, and RIG-I in BMM that were either transfected with poly(I ⋅ C) (Fig. 3D) or infected with SenV (Fig. 3E).
MHV infection results in upregulation of IFN-dependent and -independent cytokines and chemokines to a greater extent in BMM than in BMDC.
MHV infection induces expression of an array of cytokines and chemokines in the infected murine central nervous system (CNS), including CXCL10, CCL2 (chemokine [C-C motif] ligand 2), TNF, IL-6, IL-10, and IL-12, which play important roles in the anti-MHV immune response (39, 40). We next examined whether BMM produced these cytokines and chemokines after MHV infection and, if so, whether their expression depended upon type I IFN production. We found that these genes were upregulated with similar kinetics in BMM and BMDC; however, BMM expressed much higher levels than BMDC did (Fig. 4A) (data for IL-6 and IL-10 are not shown). These results were confirmed for TNF, CXCL10, and IL-12 p70 at the protein level by performing an enzyme-linked immunosorbent assay (ELISA) (Fig. 4C and D). No IL-12 p70 mRNA or protein was detected in BMDC. Of note, basal mRNA levels of CXCL10 and TNF were higher in BMM than in BMDC, while the reverse was true for CCL2 (Fig. 4B). Nevertheless, all three were expressed at higher levels in BMM during the peak time of infection (Fig. 4A), indicating that the differences in basal levels did not explain the ability of BMM to produce larger amounts of inflammatory cytokines and chemokines in response to MHV infection.
FIG 4 .
MHV infection results in greater cytokine and chemokine upregulation in BMM than in BMDC. BMM and BMDC were infected with MHV. (A) mRNA levels of CXCL10, CCL2, TNF, and IL-12 p40 were measured by real-time qPCR in BMM and BMDC at the indicated times p.i. ND, not detectable. (B) Basal levels of CXCL10, CCL2, and TNF were measured by real-time qPCR in mock-infected BMM and BMDC. CT ratios to HPRT are shown in panels A and B (note the differences in scale). In panels A and B, three or four replicates were performed in each experiment, and one of three independent experiments is shown. (C) Protein levels of TNF and IL-12 p70 in BMM were measured by ELISA at different times p.i. (D) Protein levels of TNF and CXCL10 in BMM and BMDC were assayed at 24 h p.i. In panels C and D, one of two independent experiments is shown. Values for BMDC that are significantly different from the value for BMM are indicated as follows: *, P < 0.05; **, P < 0.01.
To assess whether the upregulation of these cytokines and chemokines was IFN dependent, we analyzed only BMM, since BMDC produced very low levels of these molecules and did not express type I IFN. We also included an additional IFN-responsive gene ISG15 (IFN-stimulated gene 15) in these assays. Consistent with a previous study of lipopolysaccharide (LPS)-treated BMM and BMDC (41), we found that CXCL10, IL-10, and ISG15 expression was dependent on IFNAR, but not MyD88, signaling (Fig. 5). TNF, IL-6, and IL-12 p40 mRNA upregulation was abolished in MHV-infected MyD88−/− cells and was variably dependent upon IFNAR signaling: the levels of IL-6 and IL-12 p40 were decreased by about 50 to 80% in IFNAR−/− BMM (Fig. 5A and B). These mRNA data were confirmed by measuring the levels of CXCL10, TNF, and IL-12 p70 protein in an ELISA (Fig. 5C). As in MHV-infected cells, CXCL10 expression in SenV-infected or poly(I ⋅ C)-transfected BMM was also dependent on IFNAR, but not MyD88. However, TNF expression in these samples was not dependent on MyD88, consistent with a previous report showing that TNF expression was mediated by direct MDA5 recognition in SenV-infected cells (Fig. 5A) (42). IL-6 and IL-12 p40 were expressed at very low levels in poly(I ⋅ C)-treated or SenV-infected BMM, so expression of these cytokines was not analyzed further.
FIG 5 .
MHV induces IFNAR-dependent and MyD88-dependent upregulation of cytokines and chemokines. (A) BMM isolated from WT, IFNAR−/−, or MyD88−/− mice were mock infected, infected with MHV or SenV, or transfected with poly(I ⋅ C) for 16 h. mRNA levels of CXCL10 and TNF were measured by real-time qPCR. (B) mRNA levels of IL-10, ISG15, IL-6, and IL-12 p40 in mock-infected or MHV-infected BMM isolated from WT, IFNAR−/−, or MyD88−/− mice were measured by real-time qPCR at 16 h p.i. ND, not detectable. CT ratios to HPRT are shown in panels A and B (note the differences in scale). (C) Protein levels of CXCL10, TNF, and IL-12 p70 were measured by ELISA. Two or three replicates were performed in each experiment, and one of two independent experiments (for panel C) or one of three independent experiments (for panels A and B) is shown. Values that are statistically significantly different from the value for infected WT BMM are indicated as follows: *, P < 0.05; **, P < 0.01.
Exogenous IFN enhances expression of cytokines and other IFN-dependent molecules in MHV-infected BMM at early but not late times p.i.
Our results suggested that BMM expression of several inflammation-related molecules, including IFN-α/β, MDA5, RIG-I, and CXCL10, required signaling through the IFNAR. However, from these results, it could not be determined whether upregulated expression of these molecules occurred solely as a consequence of MHV-induced IFN induction or if some aspect of virus replication synergistically enhanced expression. Further, the amount of IFN produced in MHV-infected BMM was much lower than in SenV-infected cells, raising questions as to whether this amount was sufficient to fully upregulate these IFN-induced genes. To address these questions, we treated mock-infected or MHV-infected BMM with IFN-β and then measured mRNA levels of inflammatory molecules by real-time qPCR. The cells were treated with IFN-β at 4 h p.i. to minimize any effect that IFN treatment might have on virus replication; we could not detect any MHV-induced IFN-β RNA or IFN protein at this time p.i. (Fig. 1C). We used a high dose of IFN (800 U of murine IFN-β) so that any IFN-mediated effects would be readily apparent. For comparison, we also treated mock- and MHV-infected BMDC and 17Cl-1 cells with IFN, since IFN was barely detected at any time p.i. in these cells. IFN treatment did not reduce virus titers in any of these cells, when measured at 16 h p.i. (Fig. 6A), consistent with previous reports (13, 32, 33).
FIG 6 .
Exogenously added IFN is required for maximal levels of cytokine and chemokine expression in BMM at 8 h p.i. and in BMDC and fibroblasts at 16 h p.i. BMM, BMDC, and 17Cl-1 cells were infected with MHV. Some samples were treated with 800 U of IFN-β at 4 h p.i. (A) Supernatants were harvested at 16 h p.i., and virus titers were measured. (B to E) RNA was prepared from mock-infected or MHV-infected BMM, BMDC, or 17Cl-1 cells at 8 or 16 h p.i. mRNA levels of type I IFN (IFN-β and IFN-α4) (B), RNA helicases (MDA5 and RIG-I) (C), IFN-dependent genes (CXCL10 and ISG15) (D), and an IFN-independent gene (TNF) (E) were measured by real-time qPCR. To determine whether upregulation required active virus replication, BMM were exposed to UV-inactivated virus prior to IFN treatment, and the levels of select genes were determined (shown in the boxes to the right of the graph for BMM 16-h sample graphs). CT ratios to HPRT are shown. All genes were induced to higher levels in BMM than in BMDC or 17Cl-1 cells (note the differences in scale). Three or four replicates were performed in each experiment, and one of three independent experiments is shown. Values that were statistically significantly different from the values for cells infected with MHV and treated with IFN are indicated as follows: *, P < 0.05; **, P < 0.01.
To determine whether cytokine production by BMM required virus replication, we exposed BMM to UV-inactivated MHV prior to treatment with IFN-β. IFN-β treatment did not stimulate expression of IFN-β or IFN-α4 in mock-infected cells or in cells incubated with UV-inactivated virus, demonstrating that only active virus replication could initiate IFN-α/β production (Fig. 6B). Treatment with IFN-β enhanced IFN production in MHV-infected BMM at 8 h p.i. (Fig. 6B), which suggests that the levels of MHV-induced IFN were insufficient to maximally increase IFN-α/β expression at this time p.i. However, by 16 h p.i., MHV induced sufficient amounts of IFN so that exogenous IFN had no additional effects (Fig. 6B). No IFN protein was detected in either MHV-infected BMDC or 17Cl-1 cells (32, 43), but low levels of IFN mRNA were detected at late times p.i. in these cells (23). Treatment of MHV-infected BMDC with IFN induced significantly (P < 0.01) higher levels of IFN-α/β mRNA than did IFN-β treatment or MHV infection alone, when measured at 16 h p.i., although the levels were only about 2% of those observed in infected BMM. In 17Cl-1 cells, IFN-α/β was expressed at very low levels after infection, even after IFN-β treatment (Fig. 6B). Combined with data generated from IFNAR−/− BMM, these results suggest that virus replication is required to initiate production of type I IFN, which functions by autocrine signaling to reach maximal levels.
IFN-β treatment upregulated expression of IFNAR-dependent molecules MDA5, RIG-I, CXCL10, and ISG15 in mock-infected BMM and BMDC to maximal levels as early as 4 h posttreatment, and the level remained constant over the next 8 hours (Fig. 6C and D). In contrast, expression of these molecules was minimal in MHV-infected BMM at 8 h p.i. but increased over the following 12 to 16 h p.i. (Fig. 2B and C, 4A, and 6C and D). Exogenous IFN-β treatment enhanced expression of these molecules compared to MHV infection alone at 8 but not 16 h p.i., again suggesting that the amount of IFN produced by infected BMM was sufficient for maximal expression at later times p.i. (Fig. 6C and D). We detected much higher mRNA levels of MDA5 and RIG-I in MHV-infected BMM at 16 h p.i. than in IFN-β-treated mock-infected cells, although the IFN level induced by MHV was actually lower than 800 U (Fig. 1A and B). This likely occurred because MDA5 and RIG-I recognize viral PAMPs and directly upregulate expression of these IFN-dependent molecules via a positive-feedback loop. Additionally, these results also suggest that the effects of IFN on neighboring uninfected cells do not account for the elevated levels of IFN, MDA5, and RIG-I detected in these cultures after MHV infection, since the levels were substantially greater than those observed after IFN treatment of mock-infected cells.
In contrast, mRNA expression of MDA5, RIG-I, CXCL10, and ISG15 was lower at 16 h p.i. in BMDC than after IFN treatment alone (Fig. 6C and D). No IFN protein was produced at any time after MHV infection in BMDC or 17Cl-1 cells, so exogenous IFN greatly enhanced expression of these genes in both cell types. The lower levels observed in infected BMDC resulted only in part from an inability to produce IFN because even when we treated these cells with IFN-β, cytokine expression was still much lower than detected in MHV-infected BMM (Fig. 6C and D). Furthermore, virus replication and IFN acted synergistically only in enhancing CXCL10 expression, with maximal levels obtained when MHV-infected BMDC or 17Cl-1 cells were treated with IFN. Consistent with analyses of IFNAR−/− cells (Fig. 5A), TNF expression was not upregulated by IFN treatment in any cell type in the presence or absence of MHV infection (Fig. 6E).
Finally, we measured CXCL10 protein levels by an ELISA and, as expected, observed increased production of CXCL10 in infected cells. However, CXCL10 protein levels in MHV-infected BMM were comparable to those in IFN-treated mock-infected cells, even though mRNA amounts in MHV-infected cells were appreciably higher at 16 h p.i. (compare Fig. 6D with Fig. 7A). IFN treatment of MHV-infected BMM, BMDC, and 17Cl-1 cells did not increase CXCL10 protein production compared to IFN treatment alone. These results likely reflect MHV-induced inhibition of host cell protein synthesis, mediated in part by nsp1 (11, 14, 34, 35). Of note, expression of the MyD88-dependent molecule TNF was not upregulated by IFN treatment in mock-infected or MHV-infected cells (Fig. 7B), consistent with the real-time qPCR data shown in Fig. 6E.
FIG 7 .
Exogenously added IFN enhances expression of CXCL10 but not TNF. BMM, BMDC, and 17Cl-1 cells were infected with MHV with (+) or without (−) IFN-β treatment at 4 h p.i. The levels of CXCL10 (A) and TNF (B) protein were determined by an ELISA. One of two independent experiments is shown. Values that were significantly different from the values for MHV-infected, IFN-treated cells (P < 0.01) are indicated (**).
DISCUSSION
The ability of coronaviruses to induce an interferon response in some cells and not others is not well understood. BMM and BMDC are derived from the same BM progenitor cells in the presence of different cytokines, but they show substantial differences in type I IFN production and in cytokine and chemokine profiles (41, 44). Here, we show that maximal IFN production in murine coronavirus-infected BMM requires signaling through the IFNAR (Fig. 3C). We could detect small amounts of IFN mRNA but not protein in MHV-infected BMM at 8 h p.i. (Fig. 1). However, IFN may have been produced at low levels at this early time p.i. but immediately depleted by binding to the IFNAR. In support of this, in a prior study, basal expression of IFN by BMM was detected when IFNAR−/− but not WT BMM were examined by an ELISA (41). Together, these results suggest that a very small amount of IFN produced initially following infection effected a large increase in the levels of IFN and other inflammatory molecules via autocrine signaling. However, treatment of uninfected BMM with exogenous IFN did not induce the same level of expression of some of these molecules as occurred in MHV-infected cells at 16 h p.i. (Fig. 6). Thus, active virus replication induced cytokine and chemokine expression via additional pathways in BMM. Furthermore, these molecules were largely produced by infected cells and not neighboring uninfected cells.
It is likely that MDA5 is involved in one or both steps of this amplification process, because MDA5 is expressed in mock-infected BMM (Fig. 2), and previous work has shown that IFN expression was significantly decreased in MHV-infected MDA5−/− BMM (24). The levels of MDA5 and RIG-I increased greatly after infection via an IFN-dependent pathway, providing additional pathogen recognition receptors (PRRs) for detecting virus replication products. By late times p.i., sufficient levels of IFN were synthesized so that exogenously added IFN had no effect on the levels of any molecules that were examined. Similar effects were observed in BMDC and 17Cl-1 fibroblasts, but in these cells, no or very small amounts of IFN mRNA (Fig. 6B) and no protein (18, 23, 32, 33) were produced endogenously. Consequently, in both cell types, exogenous IFN treatment significantly upregulated expression of MDA5, RIG-I, CXCL10, and ISG15. These results are consistent with recent reports showing that IFN priming prior to tissue culture cell infection with SARS-CoV resulted in augmented expression of several molecules involved in IFN induction and signaling pathways, including CXCL10, RIG-I, MDA5, IRF-7, IFN-β, ISG15, and 2′,5′-oligoadenylate synthetase 1 (OAS1) (45). Analogous results were obtained when human conventional DC were primed with IFN prior to infection with influenza A virus (IAV) (46). Collectively, our results suggest that IFN is critical for maximal expression of these proinflammatory mediators, but unlike MHV-infected BMDC or 17Cl-1 cells, sufficient amounts of IFN are expressed endogenously via an MDA5-dependent pathway in infected BMM to initiate the process.
The enhanced responsiveness of BMM compared to BMDC or fibroblasts to MHV infection also occurs after exposure to other stimuli. The levels of IFN-dependent proinflammatory cytokines and chemokines, such as CCL2, CCL5, and CXCL10, were higher in BMM than in BMDC after stimulation with LPS (44). This increased sensitivity to MHV infection is not likely to be a function of differences in basal levels because baseline levels of MDA5 and RIG-I are only 2-fold higher in BMM than in BMDC (Fig. 2A); both cell types should have responded nearly equivalently to infection, if basal levels of MDA5 were the only factor. On the other hand, MDA5 levels are extremely low in 17Cl-1 cells (Fig. 6C) (24), and this may have contributed to the inability of these cells to produce IFN and other IFN-dependent proteins. Another possible explanation for the difference in IFN expression in infected BMM compared to BMDC or fibroblasts is that virus replication could occur on different types of membranous structures in the different cell types. MHV is known to replicate on double-membrane vesicles (DMV) and other intracellular membranes (20). DMV may shield viral dsRNA from detection by MDA5 and RIG-I, so differences in the numbers of DMV or in the types of membranous structures used in replication could potentially contribute to differences in PAMP recognition and subsequent IFN induction. However, we detected no obvious differences in the types of virus-specific membranous structures observed in BMM compared to other cells when analyzed by electron microscopy (data not shown); if anything, the numbers of DMV in infected BMM were increased more than in other cells, probably reflecting the more rapid course of infection observed in these cells (Fig. 1D). Thus, an outstanding question is how the milieu of resting macrophages differs from those of BMDC and fibroblasts, enabling rapid and extensive upregulation of IFN-α/β and other proinflammatory molecules after infection with coronaviruses.
While a type I IFN amplification loop is critical for maximal production of IFN and other molecules such as CXCL10, some aspects of the innate response in MHV-infected BMM are not IFN dependent. Thus, expression of TNF, IL-12, and IL-6 (Fig. 5) is completely dependent upon signaling through MyD88. MyD88 is a key adaptor molecule in signaling pathways used by IL-1, IL-18, and all of the TLRs except TLR3. We examined the expression of TLR2, -3, -4, -6, and -7 and found that they were expressed at low levels in uninfected BMM and upregulated after MHV infection (data not shown). Therefore, signaling through one of these molecules could initiate the expression of these MyD88-dependent cytokines. In fact, IL-6 and TNF production in peritoneal macrophages infected with the hepatotropic MHV-3 strain is TLR2 dependent (47). However, in preliminary experiments, we detected the same levels of TNF and IL-6 in MHV-A59-infected B6 and TLR2−/− BMM, suggesting that TLR2 does not have the same critical role in BMM as observed in MHV-3-infected peritoneal macrophages. Consistent with this putative cell-specific difference in expression, splenic macrophages but not BMM produce TNF in response to TLR2 ligands (48).
The relative importance of IFN production by macrophages compared to pDC is not known in MHV-infected mice. Cervantes-Barragan et al. showed that pDC depletion diminished IFN-α production by approximately 10-fold in mice with MHV-induced hepatitis (23). Type I IFN-mediated protection of MHV-infected mice occurred via effects on macrophages and conventional dendritic cells (cDC) because deletion of IFNAR from macrophages and cDC but not nonhematopoietic or other hematopoietic cells resulted in severe liver disease (49). Thus, it is possible that IFN produced by infected macrophages at sites of infection is critical in protecting neighboring macrophages and DC from infection and in priming all cells in the CNS and other sites of infection for subsequent IFN and cytokine production.
Taken together, our results show that macrophages, long considered a major target for infection with MHV, use both IFNAR-dependent and -independent pathways to enhance the inflammatory milieu at sites of infection and that these pathways are preferentially activated in these cells compared to DC or fibroblasts. Combined with previous studies (50), they suggest that macrophages are an attractive target for modulating the immune response to enhance virus clearance while minimizing immunopathological disease.
MATERIALS AND METHODS
Viruses and cell cultures.
MHV-A59 (MHV) was grown on 17Cl-1 cells, and the titers of the virus on HeLa cells expressing CEACAM1 (carcinoembryonic antigen-related cell adhesion molecule 1), the MHV receptor (51), were determined as previously described (52). The cells were infected with MHV at a multiplicity of infection (MOI) of 5 or, as a control, with 100 hemagglutination activity units of Sendai virus (SenV) (Cantell strain; Charles River Laboratories, Wilmington, MA). 17Cl-1 cells were cultured in medium consisting of Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal calf serum (FBS), 5% tryptose phosphate, and 1% penicillin-streptomycin.
In some experiments, bone marrow-derived macrophages (BMM) were transfected with 20 µg/ml poly(I ⋅ C) (InvivoGen, San Diego, CA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). In some experiments, mock-infected or MHV-infected BMM, bone marrow-derived dendritic cells (BMDC), or 17Cl-1 cells were treated with 800 U of murine IFN-β (PBL Biomedical Laboratories, Piscataway, NJ) at 4 h p.i., and either cells (mRNA) or supernatants (protein) were harvested at 8 h or 16 h p.i.
Mice.
Pathogen-free C57BL/6 mice were purchased from the National Cancer Institute (Frederick, MD). Mice deficient in IFN-α/β receptor expression (IFNAR−/−) or in MyD88 expression (MyD88−/−) were kindly provided by John Harty (University of Iowa, Iowa City, IA) or by Shizuo Akira (Osaka University, Osaka, Japan) and Madhu Singh (University of Iowa, Iowa City, IA), respectively.
Generation of BMM and BMDC.
Primary bone marrow cells were isolated from the hind limbs of WT, IFNAR−/−, or MyD88−/− C57BL/6 mice as previously described (32). BMM were cultured in DMEM supplemented with 10% FBS, 10% L929 cell-conditioned medium (as a source for macrophage colony-stimulating factor [M-CSF]), 1 mM sodium pyruvate, and 1% penicillin-streptomycin. The cells were harvested 7 days after plating and were >90% pure (CD11b+ CD11c–), as determined by flow cytometry. BMDC were grown in RPMI 1640 medium supplemented with 10% FBS, 1.0 mM HEPES, 0.2 mM l-glutamine, 1% penicillin-streptomycin, 1 mM sodium pyruvate, 0.02 mM 2-mercaptoethanol, 1,000 U/ml recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) (BD Pharmingen, San Diego, CA), and 25 U/ml recombinant interleukin-4 (IL-4) (PeproTech, Rocky Hill, NJ). The cells were used after 7 days of culture and were >80% pure (CD11b+ CD11c+) as determined by flow cytometry.
Real-time quantitative PCR (qPCR) analysis.
RNA was isolated from BMM, BMDC, or 17Cl-1 cells using Trizol (Invitrogen). The levels of mRNA of genes of interest were determined by real-time qPCR as previously described (43). Primers are listed in Table 1. Cycle threshold (CT) values were normalized to the values for the housekeeping gene hypoxanthine phosphoribosyltransferase (HPRT) by the following equation: CT = CT (gene of interest) − CT (HPRT). All results are shown as a ratio to HPRT calculated as −2CT.
TABLE 1 .
Primers for quantitative reverse transcription-PCR
| Gene | Sequence for: |
|
|---|---|---|
| Forward 5′ primer | Reverse 3′ primer | |
| HPRT | GCGTCGTGATTAGCGATGATG | CTCGAGCAAGTCTTTCAGTCC |
| IFN-β | CCCTATGGAGATGACGGAGA | TCCCACGTCAATCTTTCCTC |
| IFN-α4 | TCCATCAGCAGCTCAATGAC | AGGAAGAGAGGGCTCTCCAG |
| MDA5 | CGATCCGAATGATTGATGCA | AGTTGGTCATTGCAACTGCT |
| RIG-I | CAGACAGATCCGAGACACTA | TGCAAGACCTTTGGCCAGTT |
| IL-12 p40 | GAAGTTCAACATCAAGAGCAGTAG | AGGGAGAAGTAGGAATGGGG |
| CCL2 | AGCACCAGCCAACTCTCACT | TCATTGGGATCATCTTGCTG |
| CXCL10 | AAGTGCTGCCGTCATTTTCT | TTCATCGTGGCAATGATCTC |
| ISG15 | GGCCACAGCAACATCTATGA | CGCAAATGCTTGATCACTGT |
| TNF | GAACTGGCAGAAGAGGCACT | AGGGTCTGGGCCATAGAACT |
| IL-6 | ACAACGATGATGCACTTGCAGA | GATGAATTGGATGGTCTTGGTC |
| IL-10 | TGGCCCAGAAATCAAGGAGC | CAGCAGACTCAATACACACT |
IFN bioassay.
The levels of IFN were measured using a bioassay based on the inhibition of vesicular stomatitis virus (VSV) growth in L929 cells. Supernatants were harvested from infected BMM or BMDC, and infectious virus was inactivated by exposure to UV light. L929 cells infected with 10,000 PFU VSV were treated with dilutions of supernatants or defined amounts of recombinant murine IFN-β at 30 min p.i. The titers of VSV were determined on Vero cells. IFN levels were calculated on the basis of standard curves generated with recombinant IFN-β.
ELISA.
To measure the protein levels of cytokines of interest, supernatants were harvested from mock-infected or MHV-infected BMM or BMDC and analyzed for CXCL10 (PBL Biomedical Laboratory) or IL-12 p70 or TNF (eBioscience, San Diego, CA) by an ELISA per the manufacturer’s instructions.
Statistics.
A Student’s t test was used to analyze differences in mean values between groups. All results are expressed as means ± standard errors of the means (SEM). P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant NS36592 from the National Institute of Neurological Disorders and Stroke.
We thank Steven Varga and Taeg S. Kim for careful review of the manuscript.
Footnotes
Citation Zhou, H., J. Zhao, and S. Perlman. 2010. Autocrine interferon priming in macrophages but not dendritic cells results in enhanced cytokine and chemokine production after coronavirus infection. mBio 1(4):e00219-10. doi:10.1128/mBio.00219-10.
REFERENCES
- 1. Garcia-Sastre A., Biron C. A. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879–882 [DOI] [PubMed] [Google Scholar]
- 2. Iwasaki A., Medzhitov R. 2010. Regulation of adaptive immunity by the innate immune system. Science 327:291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hengel H., Koszinowski U. H., Conzelmann K. K. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26:396–401 [DOI] [PubMed] [Google Scholar]
- 4. Perry A. K., Chen G., Zheng D., Tang H., Cheng G. 2005. The host type I interferon response to viral and bacterial infections. Cell Res. 15:407–422 [DOI] [PubMed] [Google Scholar]
- 5. Sarkar S. N., Sen G. C. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245–259 [DOI] [PubMed] [Google Scholar]
- 6. Rehwinkel J., Reis e Sousa C. 2010. RIGorous detection: exposing virus through RNA sensing. Science 327:284–286 [DOI] [PubMed] [Google Scholar]
- 7. Asselin-Paturel C., Trinchieri G. 2005. Production of type I interferons: plasmacytoid dendritic cells and beyond. J. Exp. Med. 202:461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perlman S., Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Devaraj S. G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C. J., Tseng C. T., Baker S. C., Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208–32221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kopecky-Bromberg S. A., Martinez-Sobrido L., Frieman M., Baric R. A., Palese P. 2006. SARS coronavirus proteins Orf 3b, Orf 6, and nucleocapsid function as interferon antagonists. J. Virol. 81:548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C. T., Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 83:4471–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wathelet M. G., Orr M., Frieman M. B., Baric R. S. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81:11620–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye Y., Hauns K., Langland J. O., Jacobs B. L., Hogue B. G. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 81:2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koetzner C. A., Kuo L., Goebel S. J., Dean A. B., Parker M. M., Masters P. S. 2010. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J. Virol. 84:8262–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rose K. M., Weiss S. R. 2009. Murine coronavirus cell type dependent interaction with the type I interferon response. Viruses 1:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rose K. M., Elliott R., Martinez-Sobrido L., Garcia-Sastre A., Weiss S. R. 2010. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J. Virol. 84:5656–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Versteeg G. A., Bredenbeek P. J., van den Worm S. H., Spaan W. J. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou H., Perlman S. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knoops K., Kikkert M., van den Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cameron M. J., Ran L., Xu L., Danesh A., Bermejo-Martin J. F., Cameron C. M., Muller M. P., Gold W. L., Richardson S. E., Poutanen S. M., Willey B. M., Devries M. E., Fang Y., Seneviratne C., Bosinger S. E., Persad D., Wilkinson P., Greller L. D., Somogyi R., Humar A., Keshavjee S., Louie M., Loeb M. B., Brunton J., McGeer A. J., Kelvin D. J. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81:8692–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rempel J. D., Murray S. J., Meisner J., Buchmeier M. J. 2004. Differential regulation of innate and adaptive immune responses in viral encephalitis. Virology 318:381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cervantes-Barragan L., Zust R., Weber F., Spiegel M., Lang K. S., Akira S., Thiel V., Ludewig B. 2006. Control of coronavirus infection through plasmacytoid dendritic cell-derived type I interferon. Blood 109:1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roth-Cross J. K., Bender S. J., Weiss S. R. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82:9829–9838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J., Liu Y., Zhang X. 2010. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 84:6472–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marsden P. A., Ning Q., Fung L. S., Luo X., Chen Y., Mendicino M., Ghanekar A., Scott J. A., Miller T., Chan C. W., Chan M. W., He W., Gorczynski R. M., Grant D. R., Clark D. A., Phillips M. J., Levy G. A. 2003. The Fgl2/fibroleukin prothrombinase contributes to immunologically mediated thrombosis in experimental and human viral hepatitis. J. Clin. Invest. 112:58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim T. S., Perlman S. 2005. Viral expression of CCL2 is sufficient to induce demyelination in RAG1−/− mice infected with a neurotropic coronavirus. J. Virol. 79:7113–7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nicholls J. M., Butany J., Poon L. L., Chan K. H., Beh S. L., Poutanen S., Peiris J. S., Wong M. 2006. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 3:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Templeton S., Kim T. H., O’Malley K., Perlman S. 2007. Maturation and localization of macrophages and microglia during infection with a neurotropic murine coronavirus. Brain Pathol. 18:40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye J., Zhang B., Xu J., Chang Q., McNutt M. A., Korteweg C., Gong E., Gu J. 2007. Molecular pathology in the lungs of severe acute respiratory syndrome patients. Am. J. Pathol. 170:538–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Turner B. C., Hemmila E. M., Beauchemin N., Holmes K. V. 2004. Receptor-dependent coronavirus infection of dendritic cells. J. Virol. 78:5486–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou H., Perlman S. 2006. Preferential infection of mature dendritic cells by mouse hepatitis virus strain JHM. J. Virol. 80:2506–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roth-Cross J. K., Martinez-Sobrido L., Scott E. P., Garcia-Sastre A., Weiss S. R. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 81:7189–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hilton A., Mizzen L., MacIntyre G., Cheley S., Anderson R. 1986. Translational control in murine hepatitis virus infection. J. Gen. Virol. 67(Pt. 5):923–932 [DOI] [PubMed] [Google Scholar]
- 35. Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 103:12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Honda K., Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658 [DOI] [PubMed] [Google Scholar]
- 37. Ireland D. D., Stohlman S. A., Hinton D. R., Atkinson R., Bergmann C. C. 2008. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J. Virol. 82:300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 [DOI] [PubMed] [Google Scholar]
- 39. Bergmann C. C., Lane T. E., Stohlman S. A. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lane T. E., Asensio V., Yu N., Paoletti A. D., Campbell I., Buchmeier M. J. 1998. Dynamic regulation of α- and β-chemokine expression in the central nervous system during mouse hepatitis virus-induced demyelinating disease. J. Immunol. 160:970–978 [PubMed] [Google Scholar]
- 41. Fleetwood A. J., Dinh H., Cook A. D., Hertzog P. J., Hamilton J. A. 2009. GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J. Leuk. Biol. 86:411–421 [DOI] [PubMed] [Google Scholar]
- 42. Gitlin L., Benoit L., Song C., Cella M., Gilfillan S., Holtzman M. J., Colonna M. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pewe L., Zhou H., Netland J., Tangadu C., Olivares H., Shi L., Look D., Gallagher T. M., Perlman S. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fleetwood A. J., Lawrence T., Hamilton J. A., Cook A. D. 2007. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178:5245–5252 [DOI] [PubMed] [Google Scholar]
- 45. Kuri T., Zhang X., Habjan M., Martinez-Sobrido L., Garcia-Sastre A., Yuan Z., Weber F. 2009. Interferon priming enables cells to partially overturn the SARS coronavirus-induced block in innate immune activation. J. Gen. Virol. 90:2686–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phipps-Yonas H., Seto J., Sealfon S. C., Moran T. M., Fernandez-Sesma A. 2008. Interferon-beta pretreatment of conventional and plasmacytoid human dendritic cells enhances their activation by influenza virus. PLoS Pathog. 4:e1000193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jacques A., Bleau C., Turbide C., Beauchemin N., Lamontagne L. 2009. Macrophage interleukin-6 and tumour necrosis factor-alpha are induced by coronavirus fixation to Toll-like receptor 2/heparan sulphate receptors but not carcinoembryonic cell adhesion antigen 1a. Immunology 128:e181–e192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berghaus L. J., Moore J. N., Hurley D. J., Vandenplas M. L., Fortes B. P., Wolfert M. A., Boons G. J. 2010. Innate immune responses of primary murine macrophage-lineage cells and RAW 264.7 cells to ligands of Toll-like receptors 2, 3, and 4. Comp. Immunol. Microbiol. Infect. Dis. 33:443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cervantes-Barragan L., Kalinke U., Zust R., Konig M., Reizis B., Lopez-Macias C., Thiel V., Ludewig B. 2009. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 182:1099–1106 [DOI] [PubMed] [Google Scholar]
- 50. Levy G. A., Liu M., Ding J., Yuwaraj S., Leibowitz J., Marsden P. A., Ning Q., Kovalinka A., Phillips M. J. 2000. Molecular and functional analysis of the human prothrombinase gene (HFGL2) and its role in viral hepatitis. Am. J. Pathol. 156:1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williams R. K., Jiang G., Holmes K. V. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88:5533–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perlman S., Schelper R., Bolger E., Ries D. 1987. Late onset, symptomatic, demyelinating encephalomyelitis in mice infected with MHV-JHM in the presence of maternal antibody. Microb. Pathog. 2:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]