Abstract
Pseudomonas aeruginosa OprF is a largely monomeric outer membrane protein that allows the slow, nonspecific transmembrane diffusion of solutes. This protein folds into two different conformers, with the majority conformer folding into a two-domain conformation that has no porin activity and the minority conformer into a one-domain conformation with high porin activity and presumably consisting of a large β barrel. We examined the factors that control the divergent folding pathways of OprF. OprF contains four Cys residues in the linker region connecting the N-terminal β-barrel domain and the C-terminal globular domain in the majority conformer. Prevention of disulfide bond formation either by expression of OprF in an Escherichia coli dsbA strain grown with dithiothreitol or by replacement of all Cys residues with serine (CS OprF) increased the specific pore-forming activity of OprF significantly. Replacement of Phe160 with Ile at the end of the β-barrel termination signal as well as replacement of Lys164 in the linker region with Gly, Cys, or Glu increased porin activity 2-fold. Improving a potential β-barrel termination signal in the periplasmic domain by replacement of Asp211 with asparagine also increased porin activity. The porin activity could be improved about 5-fold by the combination of these replacements. OprF mutants with higher porin activity were shown to contain more one-domain conformers by surface labeling of the A312C residue in intact cells, as this residue is located in the periplasmic domain in the two-domain conformers. Finally, when the OprF protein was expressed in an E. coli strain lacking the periplasmic chaperone Skp, the CS OprF protein exhibited increased pore-forming activity.
IMPORTANCE
High intrinsic levels of resistance to many antimicrobial agents, seen in Gram-negative bacterial species such as Pseudomonas aeruginosa and Acinetobacter species, are largely due to the extremely low permeability of their major porin OprF and OmpA. Because this low permeability is caused by the fact that these proteins mostly fold into a two-domain conformer without pores, knowledge as to what conditions increase the production of the pore-forming minority conformer may lead to dramatic improvements in the treatment of infections by these bacteria. We have found several factors that increase the proportion of the pore-forming conformer up to 5-fold. Although these studies were done with Escherichia coli, they may serve as the starting point for the design of strategies for improvement of antimicrobial therapy for these difficult-to-treat pathogens, some strains of which have now attained the “pan-resistant” status.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that is now sometimes pan-resistant (1) and plays a critical role in nosocomial infections as well as in the later phases of cystic fibrosis. One main reason for these troublesome characteristics of this organism is its intrinsic resistance to many antibiotics, which in turn is mainly due to its very low-permeability outer membrane (OM) (2). We have shown that the low permeability is due to the mostly closed OM porin OprF of this organism and that the closure of this porin occurs because most of the protein folds as a conformer with a closed channel (3, 4). The influx of antimicrobial compounds is thus very slow in P. aeruginosa, and this low permeability of the OM enhances, in a synergistic manner, the contribution of broad-substrate-range efflux pumps coded by the chromosomal genes, creating the very effective intrinsic resistance (5).
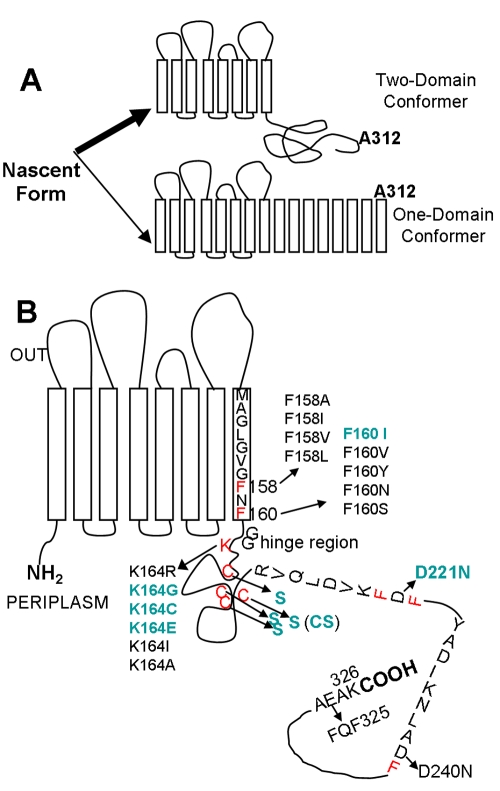
A majority of OprF molecules (96%) fold into closed-channel conformers, and only 4% fold into open-channel conformers (4). Once folded, each of these conformers appears to be stable at least in the OprF homolog OmpA of Escherichia coli, and we have found no evidence so far for a facile interconversion between them (6). (There are, however, reports of interconversion of these conformers in OmpA, and these reports will be examined in Discussion.) The closed-channel conformer presumably folds as a two-domain protein with the N-terminal eight-stranded β-barrel domain and the C-terminal, periplasmic, globular domain, as seen with OmpA (7). We did not observe the diffusion of the smallest organic solutes that we could test (e.g., glycine) in the liposome-swelling assay through the N-terminal domain of OprF (positions 1 to 160) (3), an observation suggesting that the N-terminal eight-stranded β-barrel domain is an essentially closed channel. The C-terminal globular domain shares considerable similarity with the periplasmic C-terminal domain of OmpA, and these domains have been suggested to associate noncovalently with peptidoglycan (8, 9). Thus, the majority conformer of OprF plays a structural role by anchoring the OM to the underlying peptidoglycan (10, 11). In contrast, the rare open-channel conformer of OprF is expected to contain a larger number (perhaps 16, as in the always open E. coli OmpF/C porin) of β strands, which will produce a single domain from the entire sequence (4). OprF preparations enriched in the open-channel conformers were obtained (4) either by utilizing the tendency of these conformers to form oligomers (thus through high-resolution gel filtration) or by taking advantage of the fact that the A312C residue, close to the C terminus, is exposed on the cell surface only in these single-domain conformers (thus through biotin labeling of A312C mutant OprF in intact cells) (Fig. 1A). These preparations were shown to have much higher specific porin activity than unfractionated OprF both by the liposome-swelling assay (4) and by the planar bilayer assay (3). The open-channel conformer was shown to be resistant to protease digestion, as expected for an all-β-barrel protein, but the closed majority conformer was very sensitive to proteases that apparently attack the linker region between two domains (4).
FIG 1 .
Presumed folding patterns of OprF and the residues altered by site-directed mutagenesis. (A) OprF appears to fold into two stable conformers. The majority population (top) folds into a two-domain conformer, with an N-terminal, outer-membrane-spanning, 8-stranded β barrel and a C-terminal, periplasmic, globular domain that binds to the underlying peptidoglycan. A minority population (bottom) folds into a one-domain conformer, presumably containing many transmembrane β strands in view of the large channel size. The position of the A312C residue that was used to examine the proportion of each conformer is also indicated. (B) The residues mutated by site-directed mutagenesis are shown, with those mutants yielding higher permeability shown in bold, green fonts. Phe residues (potentially) forming the C-terminal end of the β-barrel recognition sequence are shown in red. Alterations involving the presumed peptidoglycan-binding sites (see text) as well as those involving the addition of factor X/TEV cleavage site at the C terminus (Fig. 6) are not shown to avoid cluttering of the figure.
We thus believe that the two different conformers of OprF are generated by a divergent folding pathway from the nascent protein. However, it is currently unclear how the entry of the nascent protein into one of the divergent branches is controlled. OM proteins (OMPs) are secreted into the periplasm, where several chaperones have been shown to play a role in OMP biogenesis (12–14). Furthermore, disulfide bond formation and isomerization are catalyzed in the periplasm by the DsbA and DsbC proteins (15), a process that possibly precedes OM insertion (16) (although a protein in which disulfide bonds are formed late in the process has recently been reported [17]). Recently, the machinery involved in OM insertion and β-barrel assembly of OMPs in E. coli was found to consist of at least five interacting components, i.e., four lipoproteins (YfgL, NlpB, YfiO, and SmpA [recently renamed as BamB, BamC, BamD, and BamE, respectively]) and the conserved integral OM protein YaeT (BamA) (18, 19). E. coli BamA binds C-terminal signature sequences of β-barrel OMPs, which contain the C-terminal Phe (or Tyr) residue (20). In E. coli OmpA, the OMP signature sequence is found at the end of the N-terminal β-barrel domain (20), and a similar situation is also found for P. aeruginosa OprF (Fig. 1B).
In this study, we investigated how the divergent folding of this protein is controlled and found several factors that increase the population of the one-domain open-channel conformers.
RESULTS
Initial studies.
Our initial strategy was to use P. aeruginosa and to select for higher-permeability mutants of OprF after random mutagenesis of the cloned oprF gene. For selection, we used a chemostat selection technique to find P. aeruginosa growing faster on larger substrates. However, such efforts were unsuccessful because of the strong tendency of this organism to form biofilms on the surface of the chemostat apparatus, even though strains severely compromised in biofilm formation through the deletion of pslAB genes (21) were used.
We therefore decided to rely on site-directed mutagenesis of chosen residues in OprF and to examine the activity of the purified mutant OprF proteins. For this approach, we decided to use an E. coli host expressing P. aeruginosa OprF from plasmids. In addition to the convenience in mutagenesis and protein purification, this approach was also advantageous because a great deal is known about the folding and assembly of OM proteins in E. coli (19, 20) and because the system will not suffer from the possible inability of the mutated single-domain OprF protein to function as an anchor of the OM to the underlying peptidoglycan (22). We have already shown (4) that a modified OprF protein with the N-terminal OmpA signal sequence and a decahistidine tag was expressed and apparently folded correctly in an E. coli host when the gene was cloned on a medium-copy-number vector, pKY9790. Here, we used a similar plasmid (pKY-His6OprF) expressing a hexahistidine-tagged OprF protein under the Ptac promoter (see Materials and Methods). When the protein was expressed in E. coli DH5α with isopropyl-1-thio-β-d-galactopyranoside (IPTG) (0.1 mM) induction at 30°C, a prominent OprF protein band of about 40 kDa was seen in the Sarkosyl-insoluble OM fraction on SDS-PAGE gel (not shown). To confirm that the expression of the OprF protein from the plasmid is not overloading the pathway for OM protein folding and assembly, we examined the possible accumulation of folding intermediates of this protein in the Sarkosyl-soluble fraction (representing the periplasmic and inner membrane [IM] proteins). After 1 to 3 h of induction with 0.1 mM IPTG at 30°C, no OprF protein was found in this fraction, although a large amount of a protein of about 30 kDa was seen. This corresponds to chloramphenicol transacetylase (219 amino acid residues), the selection marker of the vector, which was also expressed in the control sample from the host containing only the vector.
For the functional assay of OprF protein, hexahistidine-tagged OprF protein was expressed from pKY-His6OprF in an E. coli strain lacking the endogenous trimeric porins OmpF and OmpC (HN705), because the trimeric porins have 50- to 100-fold-higher specific pore-forming activities than OprF (23), and even a minor contamination by these porins would interfere with the OprF assay. The protein was usually expressed at 30°C without IPTG, and this baseline expression led to the recovery of nearly 2 mg of purified OprF from a liter of culture harvested at an A600 of 1.0. When the isolated OprF protein was treated with trypsin at a weight ratio of 200 (OprF) to 1 (trypsin) for 2 h at 37°C, the majority conformer of OprF was converted to a smaller fragment of about 25 kDa by the cleavage within the linker region connecting the N-terminal and C-terminal domains, as with the OprF protein purified from P. aeruginosa PAO1 (4). The value for the pore-forming activity of OprF recovered from E. coli was 1.6 ± 0.2 10−3 × optical density (mOD)/min/µg protein, as measured by the liposome-swelling assay with arabinose as a substrate (4). This value was similar to that for the C-terminally His-tagged OprF protein expressed from a plasmid in P. aeruginosa (1.0 ± 0.15 mOD/min/µg protein) (unpublished results). These results suggest that hexahistidine-tagged recombinant OprF protein was expressed and folded correctly in E. coli, as shown by the trypsin sensitivity and pore-forming activity of this protein.
Prevention of disulfide bond formation in OprF increases its pore-forming activity.
The mature form of the P. aeruginosa OprF protein has four cysteines at the positions 176, 185, 191, and 205 (Fig. 1B). As expected, these cysteines form two disulfide bridges, as the free sulfhydryl groups measured by the colorimetric assay with 5,5′-dithiobis(2-nitrobenzoate) (24) amounted to less than 0.2 residues in the OprF protein from P. aeruginosa (unpublished results). Because the formation of disulfide bonds is thought to accelerate and direct the protein folding pathways (25), because the disulfide bond formation seems to precede the OM insertion of at least some OMPs (16), and because these Cys residues are located just after the end of the N-terminal β barrel in the majority conformer (Fig. 1B), we examined if the inhibition of disulfide bond formation would affect the divergent folding pathways of OprF.
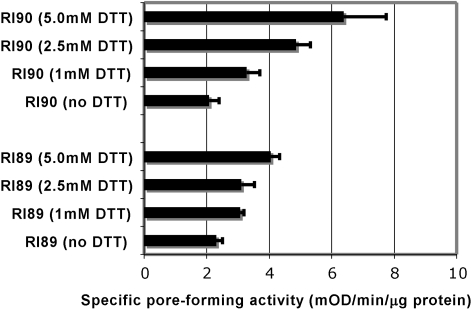
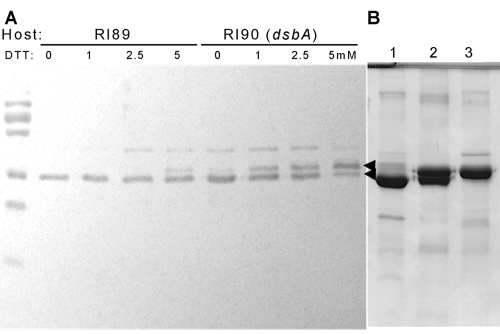
OprF protein was expressed in an E. coli strain deficient in DsbA (15), the major periplasmic disulfide oxidoreductase, and OprF maturation was studied by a functional assay and SDS-PAGE. The absence of the functional DsbA protein alone, however, did not affect the pore-forming activity (Fig. 2). We thought that the Cys residues are very close to each other and thus could form disulfide bonds even without DsbA, perhaps with the assistance of a minor disulfide oxidoreductase(s). Thus, to further inhibit the formation of disulfide bonds, we grew the dsbA strain and its parent in the presence of several concentrations of dithiothreitol (DTT). In the presence of DTT, both strains produced more-permeable OprF proteins, and the increase was more pronounced in the dsbA mutant (Fig. 2). The OprF protein was located in the OM (Fig. 3A), and the periplasmic and IM proteins found in the Sarkosyl-soluble fraction contained no folding intermediates detectable with anti-OprF antibody (not shown). Interestingly, DTT also had a strong effect on the mobility of OprF bands. OprF usually produced two bands in SDS-PAGE (a major, faster-migrating band and a very minor, slower-migrating one). The presence of DTT during growth produced a clear increase in the slower-migrating band, especially in the RI90 (dsbA) host (Fig. 3A). The samples were here treated with SDS at room temperature, a condition causing only the periplasmic domain of the majority conformer to become denatured (26). Under the nonreducing conditions in which the SDS-PAGE was run, most probably the small region containing the disulfide bonds in the periplasmic domain remained undenatured in the oxidized OprF protein, whereas this region became fully denatured in the reduced OprF protein, thus decreasing the migration rate of this protein in SDS-PAGE (Fig. 3A).
FIG 2 .
Porin activity of OprF protein purified from E. coli R189 (wt) and R190 (dsbA::kan1) grown at different concentrations of DTT. Hexahistidine-tagged OprF protein from a recombinant plasmid (pKY-His6OprF) was expressed in E. coli strains RI89 and RI90 in the LB broth containing different concentrations of DTT at 30°C overnight. The purified protein (20 to 40 µg) was reconstituted into proteoliposomes, and the specific pore-forming activity of each OprF protein was examined by determining the rate of proteoliposome swelling with l-arabinose.
FIG 3 .
Effect of disulfide bond formation on the electrophoretic pattern of OprF. (A) OprF was expressed in wt (RI89) and dsbA (RI90) E. coli in the presence of different concentrations of dithiothreitol (DTT) at 30°C. Cells were sonicated in the presence of iodoacetamide as described in Materials and Methods, and the OM fraction was solubilized at room temperature in SDS sample buffer without β-mercaptoethanol (4% SDS, 0.125 M Tris-HCl, pH 6.8, 20% glycerol, 0.01% Bromophenol Blue). Samples each containing 5 µg protein were analyzed by SDS-PAGE in the absence of reducing agents, and OprF was detected by Western blotting with an anti-OprF antibody. The two arrowheads denote the positions of the two bands seen. (B) Electrophoretic patterns of various purified OprF preparations, stained with Coomassie blue. Cells were disintegrated by a French pressure cell. OM proteins were solubilized with a solution containing 1.5% dodecyl-β-d-maltoside, 0.25 mg/ml lysozyme, 1 mM PMSF, and 20 mM HEPES buffer, pH 7.5. Histidine-tagged OprF protein was purified by using a Ni-NTA Superflow column (Qiagen). The purified proteins were analyzed by SDS-PAGE as indicated for panel A. Lane 1, OprF (wt) expressed in RI89 in the absence of DTT; lane 2, OprF (wt) expressed in RI89 in the presence of 5 mM DTT; lane 3, CS OprF expressed in RI89.
In view of these results, we constructed a cysteineless OprF protein. All four cysteines in OprF (all present in the periplasmic domain in the majority conformer [Fig. 1B]) were changed to serines (CS). Hexahistidine-tagged CS OprF and wild-type (wt) OprF were expressed in E. coli strains. As expected, the specific pore-forming activity of the CS OprF protein was 2.4 times higher (3.84 ± 0.25 mOD/min/µg protein) than that of the wild-type protein (1.61 ± 0.17 mOD/min/μg protein). Because a faster solute penetration may be produced by an increase in pore size rather than by an increase in the fraction of the open-channel conformer, the pore sizes of both proteins were examined by determining the dependence of penetration rate on solute size, ranging from 150 to 342 Da (see Materials and Methods). The CS OprF protein and the wt protein showed indistinguishable behavior patterns in this test, indicating that there is no difference in pore size between the two proteins (not shown). Finally, analysis by SDS-PAGE showed that the CS OprF band behaved exactly like the slower-migrating wt OprF band obtained by growth under reducing conditions (Fig. 3B).
β-Barrel termination signals in the middle of the OprF sequence.
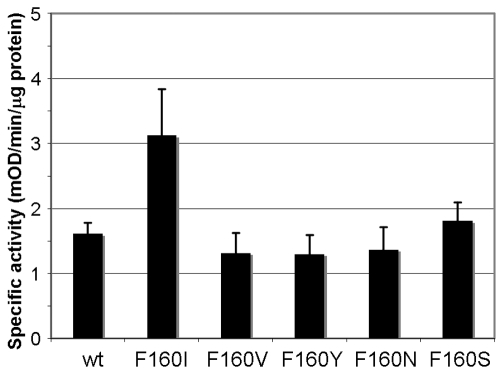
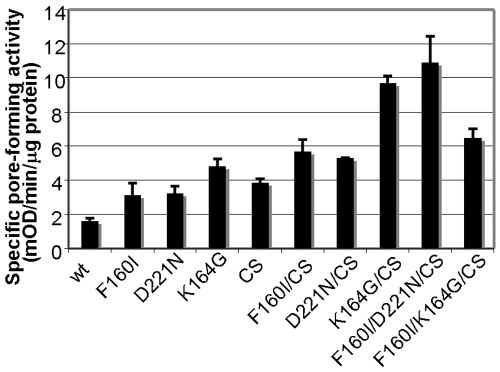
Past studies revealed that the C-terminal OMP signature sequence, ending in Phe, is the sorting signal for β-barreled OM proteins, recognized by the OM assembly factor YaeT (BamA) (20). In P. aeruginosa OprF, this sequence is found in the middle of the whole sequence between Met151 and Phe160 (MAGLGVGFNF) and appears to form the end of the β-barrel domain in the two-domain majority conformer in a situation similar to that of E. coli OmpA (27, 28). We expected that disrupting this signal may favor the production of the one-domain minority conformer by disfavoring the termination of the eight-stranded β barrel. Since carboxyl-terminal phenylalanine is known to be most important within this signal sequence (28), we replaced Phe160 with other residues, including Ile, Val, Tyr, Asn, and Ser. These hexahistidine-tagged mutant proteins were expressed and assembled in the OM at the same level as the wt OprF protein when expressed in E. coli strain HN705 at 30°C (not shown). The replacement of Phe160 with Ile increased specific pore-forming activity nearly 2-fold, but the rest of the mutant proteins had unchanged specific porin activity (Fig. 4). The F160I protein also showed higher pore-forming activity in the CS background (Fig. 5, F160I/CS), increasing the already high activity of the CS protein even further. When Phe158 was replaced with Ala, Ile, Val, or Leu without Phe160 being changed, all mutant proteins produced pore-forming activities similar to that of the wt protein (not shown).
FIG 4 .
Effect of site-directed mutagenesis at Phe160 on porin activity. The last residue of the β-barrel termination sequence, Phe160, was changed to various other amino acids by site-directed mutagenesis with plasmid pKY-His6OprF as a template as described in Materials and Methods. All mutant proteins were expressed in E. coli HN705 in LB broth at 30°C overnight and purified, and the specific pore-forming activity of each protein was analyzed by a proteoliposome assay with l-arabinose.
FIG 5 .
Porin activity of OprF containing D221N and K164G mutations and their combinations with other mutations. Mutant porins were expressed and purified as described in the legend for Fig. 4, and the specific pore-forming activity was tested by a liposome-swelling assay with l-arabinose.
During our cysteine-scanning mutagenesis experiment, we found that replacement of Lys164 in the linker region only a few residues after Phe160 with Gly, Cys, or Glu (but not with Ile, Ala, or Arg) increased the specific pore-forming activity about 2-fold or slightly more in the wt or CS background (the results obtained with K164G are shown in Fig. 5).
In the periplasmic domain of OprF, there exist additional short sequences that might function as possible β-barrel termination sequences, consisting of Phe at the C-terminal position and hydrophobic residues at positions −2, −4, −6, and −8. One such sequence covers Arg213 to Phe222 (RVQLDVKFDF) and another Tyr232 to Phe241 (YADIKNLADF) (Fig. 1B). Interestingly, in these potential signature sequences, there is a negatively charged Asp at position −1 relative to Phe, but this position is never occupied by an acidic residue in the known, functional C-terminal β-barrel termination sequences (20). We thought that perhaps strengthening these potential sequences might lead to competition with the real signature sequence at Met151 to Phe160 and thereby increase the proportion of open-channel conformers. When Asp at position 221 was replaced with Asn, the D221N protein indeed showed a 2-fold increase in pore-forming activity (Fig. 5), and one of the highest specific activities was achieved by combining this mutation with the CS and F160I mutations (Fig. 5, F160I/D221N/CS). In contrast, the D240N protein showed unaltered activity (not shown).
Another sequence in the C-terminal domain that may function as a possible last transmembrane β strand in the one-domain open-channel conformer is that between amino acids 315 and 326 (315RRVEAEVEAEAK326), which is predicted as a transmembrane β strand in one folding model of OprF (29). We wondered if this putative β-strand sequence could be converted into a sequence resembling the canonical C-terminal β-barrel termination signal. Because this sequence lacks terminal aromatic amino acids, we examined if the introduction of an Aro-X-Aro motif at the end of this putative β strand might promote the production of the one-domain conformer. We introduced the Aro-X-Aro motif by converting Ala323-Glu324-Ala325 into Phe323-Gln324-Phe325 (FQF325) in various backgrounds (the wild-type, F160I, CS, and F160I/CS proteins). The porin activities of these mutant proteins, however, did not increase in any combination (not shown).
Other modifications of the C-terminal half of OprF.
The C-terminal domain of the OprF two-domain conformer is known to bind to peptidoglycan (11). We considered the possibility that this interaction with peptidoglycan may prevent the insertion of the C-terminal domain into the OM, which is required for the one-domain minority conformation. Based on a computer docking experiment with the OmpA-like domain of RmpM from Neisseria meningitidis and a peptidoglycan fragment, four conserved residues in RmpM, D120, Y127, R135, and R197, were implicated as the sites of interaction with peptidoglycan (10). Therefore, we tested if the replacement of two of these conserved amino acids in the putative peptidoglycan-binding sites of OprF (D257 and Y264, corresponding to D120 and Y127 of RmpM, respectively) with Ala, in combination with the FQF325 mutation, could promote the formation of one-domain open-channel conformers and thereby increase the porin activity. We did not observe any positive effect of D257A, Y264A, or their combination in this background (not shown).
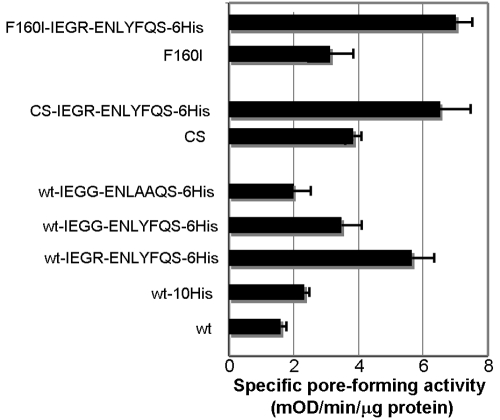
Finally, we accidentally found that the introduction of a short amino acid sequence (IEGRENLYFQS) encoding factor X and tobacco etch virus (TEV) cleavage sites, between the OprF C-terminal end and a hexahistidine tag, significantly increased the porin activity in several different backgrounds (Fig. 6). Although this short peptide is unlikely by itself to form a transmembrane β strand, it contains two aromatic amino acids, Tyr and Phe, and the conversion of these two aromatic amino acids to Ala reduced the porin activity (Fig. 6). This result supports the idea that these aromatic amino acids in the protease cleavage site may function as the C-terminal β-barrel termination sequence for the folding of the one-domain conformer. Although combining this alteration with other beneficial mutations, such as F160I or CS, did increase the specific activity of mutant OprF, the extents of increase were modest (Fig. 6).
FIG 6 .
Introduction of the TEV cleavage site to the C terminus increased pore-forming activity. OprF (wt, F160I, or CS) was modified by the addition, at the C terminus, of a factor X/TEV cleavage site (IEGRNLYFQS), followed by a hexahistidine tag. These derivatives of pKY plasmids were expressed in HN705 without IPTG induction, and the Opr proteins were purified and assayed by a liposome-swelling assay with l-arabinose. In some plasmids, the cleavage site sequence was altered.
Specific labeling of the one-domain conformer in intact cells.
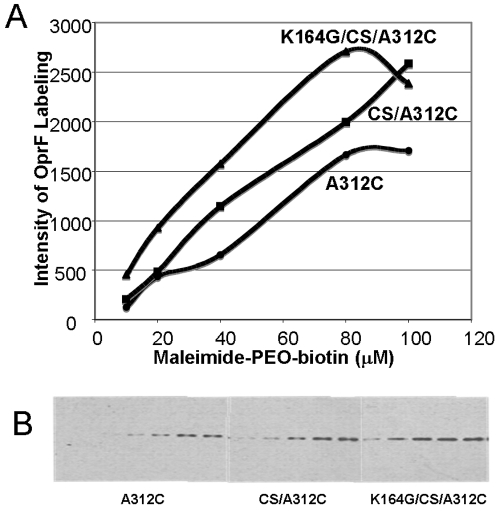
In our previous study (4), we were able to distinguish between the one-domain conformer and the two-domain conformer by introducing a single cysteine at the position of Ala312 (Fig. 1A). Because this residue is located in the C-terminal periplasmic domain in the predominant two-domain conformer, it is shielded, in intact cells, from external agents by the OM. Thus, intact cells of E. coli expressing OprF-A312C were not labeled by biotin-maleimide significantly, whereas this protein was extensively modified by the same reagent when the OM barrier was breached (4). We used a similar approach to assess the fraction of the one-domain conformer in various constructs with a membrane-impermeable cysteine-labeling reagent, maleimide-PEO2-biotin. OprF mutants were expressed in E. coli strain DH5α without IPTG at 30°C. Cells expressing the K164G-CS OprF protein containing the A312C mutation were labeled with 10 µM maleimide-PEO2-biotin to follow the time course of labeling. The labeling was more or less linear until 8 min at room temperature (not shown). Thus, the extents of labeling of OprF were compared for cells expressing wt and mutant proteins by carrying out the labeling reaction for 5 min using different concentrations of maleimide-PEO2-biotin. At every concentration of the reagent tested, the extent of labeling was highest in the cells expressing K164G/CS OprF (with very high porin activity), followed by cells expressing CS OprF (with modest porin activity) and finally by cells expressing wt OprF (with the lowest porin activity) (Fig. 7). These results demonstrate that increased specific pore-forming activity correlates well with increased population of one-domain open-channel conformers of modified OprF proteins.
FIG 7 .
OprF mutants with higher porin activity contain larger fractions of one-domain conformers. The fraction of the one-domain conformer was assessed by determining the extent of maleimide-PEO2-biotin labeling of the A312C residue of OprF in intact cells. Intact cells were labeled with maleimide-PEO2-biotin (10 to 100 µM) for 5 min at room temperature. After the reaction was stopped by the addition of 2-mercaptoethanol, proteins from identical numbers of cells were separated by SDS-PAGE, and the blot was stained with streptavidin reagent for biotin as described in Materials and Methods. The enhanced chemiluminescence (ECL) profile was recorded (B), and the intensity of maleimide-PEO-biotin bound to OprF was quantified after each band was scanned by using NIH Image software (A). Panel B shows the labeling with (from left to right) 20, 40, 60, 80, and 100 µM reagent for the indicated strains.
Influence of Skp, SurA, and YfgL in the folding of OprF.
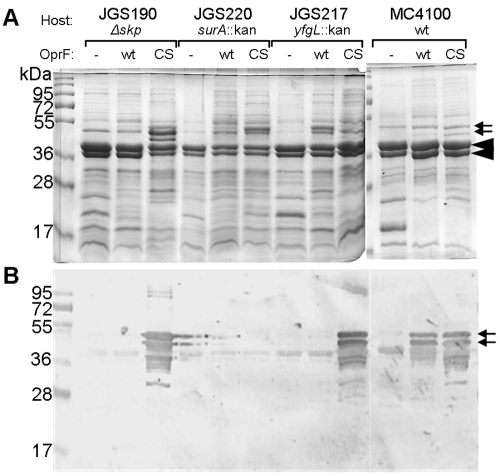
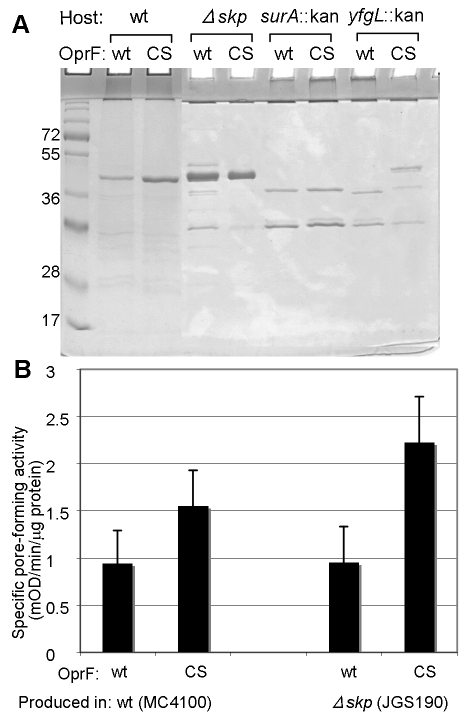
Several chaperones and assembly factors, followed by the YaeT (BamA)-based complex, participate in the folding and insertion of OMPs, which are first secreted into the periplasm (20, 30–32). We asked if the divergent folding pathway of OprF is affected by the absence of the Skp or SurA chaperone or the OM lipoprotein YfgL (BamB), a component of the YaeT-containing OMP assembly complex. OprF proteins (wt and CS) were expressed in E. coli strains lacking Skp (JGS190), SurA (JGS220), or YfgL (JGS217). When the amounts of OprF proteins inserted into the OM were assessed by Western blotting (Fig. 8B), CS OprF (but surprisingly not wild-type OprF) was seen to be overproduced in Δskp and yfgL::kan mutants. Although the Coomassie-stained gels (Fig. 8A) were difficult to interpret because of the presence of many bands, the results were consistent with the observation for Western blots. The levels of the trimeric porins and OmpA were much reduced in the surA-null mutant, and we detected neither wt nor CS OprF protein in this mutant (Fig. 8A and B). Interestingly, we also noticed decreased levels of classical porins and OmpA protein in the Δskp strain (Fig. 8A), which expressed CS OprF protein strongly (Fig. 8B), perhaps suggesting that these proteins may be competing in the same folding and/or insertion pathway.
FIG 8 .
Effect of mutations in periplasmic chaperones and the OMP assembly complex in the folding and insertion of OprF. Hexahistidine-tagged OprF proteins (wt and CS) were expressed in E. coli strains MC4100 (wt), JGS190 (∆skp), JGS220 (surA::kan), and JGS217 (yfgL::kan) at 30°C overnight. The outer membrane proteins (20 µg) were analyzed by SDS-PAGE (the gel was stained with Coomassie blue) (A) and by immunoblotting with anti-OprF antibody (B). Small arrows point to the OprF bands, and large arrowheads point to porins and OmpA.
OprF proteins expressed in the mutant strains were isolated by using Ni-nitrilotriacetic acid (NTA) columns, and the porin functions of these proteins were determined by the proteoliposome-swelling assay. Although the yfgL::kan mutant gave an enhanced level of production of CS OprF (Fig. 8), the CS OprF protein was mostly lost during purification (Fig. 9A), presumably due to degradation. However, OprF proteins (both wt and CS) produced in the JGS190 strain (Δskp) were stable (Fig. 9A), and the CS OprF protein expressed in this mutant showed a somewhat higher level of activity than that expressed in the parent strain MC4100 (Fig. 9B).
FIG 9 .
CS OprF produced in the ∆skp strain has a higher level of pore-forming activity than OprF produced in the parent strain. His-tagged OprF proteins (wt and CS) were expressed in E. coli strains MC4100 (wt), JGS190 (∆skp), JGS220 (surA::kan), and JGS217 (yfgL::kan) at 30°C overnight. OprF proteins were purified from these strains and analyzed by SDS-PAGE followed by immunoblotting (A). Proteins (20 to 40 µg) purified from the outer membrane of MC4100 (wt) and JGS190 (∆skp) were reconstituted into proteoliposomes, and their specific pore-forming activities were determined with l-arabinose (B).
DISCUSSION
We showed earlier that the exceptionally low permeability of the major porin OprF of P. aeruginosa, which makes this organism intrinsically less susceptible to a number of antimicrobial agents and difficult to eradicate in human infection, is due to the fact that only a small percentage of the population folds into a one-domain conformer that contains open channels (3, 4). In view of the clinical relevance of this phenomenon, it would be important to find conditions for influencing this bifurcate folding pathway of OprF, so that a larger fraction of this protein is led to produce an open-channel conformer. In this study, we examined both the roles of individual residues of OprF and the effects of periplasmic chaperones and the YaeT OMP assembly complex in this process.
One of the most effective methods for increasing the specific pore-forming activity of OprF, produced in E. coli, was to prevent the formation of disulfide bonds in the linker region of the protein. The presence of 5 mM DTT in the growth medium caused a modest increase in OprF specific activity in the wt host, but the same treatment in the dsbA mutant strain lacking the major periplasmic sulfhydryl oxidoreductase produced a solid 3-fold increase in activity (Fig. 2). Furthermore, the conversion of all four cysteine residues into serine in the CS mutant OprF protein produced a 2.4-fold increase in activity (Fig. 5).
The majority conformer of OprF has an N-terminal eight-stranded β-barrel structure that ends with the typical β-barrel termination sequence (20), ending with Phe160. Weakening this sequence was expected to favor the continuation of the β-barrel structure toward the C terminus, creating larger barrels with open channels inside. Indeed, the F160I mutation increased the specific porin activity 2-fold (Fig. 4). However, other residues tried at this position had little effect (Fig. 4): perhaps Asn and Ser were too hydrophilic, and Tyr could substitute for Phe as the termination signal. But it was surprising to see that Val, which is so similar to Ile, had little effect. Val, however, does not seem to occur among the terminal three residues of E. coli β-barrel proteins, in contrast to Ile (20).
“Improvement” of one of the downstream potential β-barrel termination signals (D221N) (Fig. 5) and an addition of the factor X/TEV cleavage site, apparently sharing a feature of the β-barrel termination signal, to the C terminus (Fig. 6) also increased the specific pore-forming activity. However, some modifications that were accidentally found to increase this activity, for example the replacement of Lys164 (4 residues after the end of the N-terminal β barrel in the majority conformer) with Gly, Cys, or Glu, are difficult to explain. This is especially so because replacement with Ala, which is expected to behave similarly to Cys, was not effective.
Combination of various mutations in most cases resulted in increased pore-forming activity. We had the ultimate goal of producing a protein completely composed of open conformers. Since the fractionation of unilamellar vesicles each containing only a few wild-type OprF proteins showed that about 5% of OprF proteins folded into an open-channel conformer (4), a preparation consisting of all open conformers is expected to have a specific pore-forming activity 20 times higher than that of wild-type OprF. However, the effect of adding each mutation was not completely multiplicative: for example, Fig. 6 shows that CS has a 2.2-fold-higher activity level than the wt, and the addition of the factor X/TEV cleavage site increased the activity 3.4-fold, but the combination of the two resulted in an activity level 3.9-fold higher than that of the wt, not the 7.5-fold (2.2 × 3.4 = 7.48) effect expected for multiplicative interaction. Thus, our best combination had only 5-fold-higher activity than the wild type, and we estimate that only about 25% of the OprF population was in the open-channel conformation under such conditions, although this leaves us some hope of obtaining an essentially pure preparation of open-channel conformers through purification steps that enrich for such conformers, as described earlier (4).
The high pore-forming activity of various OprF mutant proteins indeed appeared to be due to the production of higher fractions of one-domain open-channel conformers, because a Cys residue inserted at position 312 was more effectively labeled by a membrane-impermeable biotinylation reagent in intact cells in such mutants (Fig. 7). Since this residue is only 15 residues away from the C terminus, it is inaccessible for such a reagent in the classical two-domain conformer of OprF (Fig. 1A).
When we expressed OprF and CS OprF in mutants defective in the periplasmic chaperones and YfgL, a component of the YaeT-based OMP assembly complex, more CS OprF was found in Δskp and yfgL::kan mutants than in the wt (Fig. 8). However, CS OprF produced in the yfgL::kan mutant was apparently degraded during the purification process (Fig. 9A), presumably because a periplasmic protease(s) such as DegP was also expressed more strongly here as a part of the stress response. CS OprF produced in the Δskp mutant, however, could be purified and showed a somewhat higher specific activity than the same protein expressed in the wild-type parent (Fig. 9). Skp, which has a cup-like shape (33), apparently binds the β-barrel domain of E. coli OmpA, an OprF homolog, within its cavity and prevents the complete folding of this domain (34), whereas the periplasmic domain becomes folded into its native globular conformation outside the cavity (35, 36). Our results are consistent with this notion, as the absence of Skp may prevent the early folding of the C-terminal portion of CS OprF into a separate periplasmic domain and thus may increase the chances of the folding of the entire protein into the OM as a single-domain protein with open channels.
We have been assuming that each of the conformers of OprF is reasonably stable, and that there is no rapid interconversion between these two conformations, once the folding is complete. However, we note that there is a claim that the closed conformers of the OprF homolog OmpA become open at high temperatures (37). We have not found a similar phenomenon with our OprF preparations inserted into proteoliposomes (unpublished results). However, these authors (37) used OmpA preparations made by extraction in the denaturing detergent dodecyl sulfate; such preparations contain fully denatured C-terminal domains in the majority conformer (26). When they added this preparation to a planar bilayer of diphytanoylphosphatidylcholine, the N-terminal portion of OmpA left the detergent and became inserted into the bilayer. However, the C-terminal half stayed in the aqueous phase in a denatured state. Perhaps the fact that this half had not been stabilized by the folding into a globular conformation might have led, with the help of the loose structure of the unnatural isoprenoid bilayer, to the facile insertion of also the C-terminal domain into the bilayer to produce a one-domain open conformer. Thus, it seems probable that what these authors observed was the transition from an N-terminal β barrel plus the C-terminal denatured conformation into a single-domain open-channel conformation. (Although another study [38] suggests facile interconversion between the open and closed states of OmpA, the open-channel conductance has two peaks of about equal heights at 270 and 320 pS that differ by exactly the size of the “closed” channel conductance, 50 pS. Thus, it is quite possible that the authors observed independent opening events of “closed” and open channels, not the conversion of the former into the latter.)
MATERIALS AND METHODS
Bacterial strains.
OprF was most often expressed in the E. coli strain HN705 (ΔompC ompF::Tn5) (39). Other strains used for this purpose included DH5α, RI90 (dsbA::kan1) and its parent RI89 (both obtained from J. R. Beckwith) (40), JGS190 (MC4100 Δskp zae-502::Tn10), JGS220 (MC4100 surA::kan) (41), and JGS217 (MC4100 yfgL::kan) (42) (the JGS strains were obtained from N. Ruiz and T. J. Silhavy).
Bacteria were grown at 30°C in LB medium with aeration by shaking. Chloramphenicol (30 µg/ml) was added when needed for plasmid maintenance. pHSG-oprF, which contains the entire oprF gene and its upstream sequence in a low-copy-number vector, pHSG576, has previously been described (4).
Expression plasmids for histidine-tagged OprF.
The plasmid for expression of N-terminally hexahistidine-tagged OprF was made as described for the cloning of the N-terminal half of OprF (4), except that the amplicon contained the entire mature OprF sequence and that a medium-copy-number vector, pKY9790 (4), was used, generating the plasmid pKY-His6OprF.
The plasmid for the expression of C-terminally decahistidine-tagged OprF was constructed similarly by using a PCR amplicon covering the entire mature OprF sequence. This sequence was inserted in between the PstI and BamHI sites of the vector pBCKS (+), previously modified by inserting a sequence coding for the signal sequence of the E. coli OmpA protein just in front of the PstI site. After the correctness of the sequence was confirmed, an EcoRI-NotI fragment was excised from the recombinant plasmid and was inserted into pKY9790. The plasmid for the expression of OprF containing at its C terminus a factor X and TEV cleavage site (IEGRENLYFQS) followed by a hexahistidine tag was constructed similarly, except that the backward primer contained a sequence coding for IEGRENLYFQS, hexahistidine, a stop codon, and a BamHI site.
The reagents required for DNA manipulations such as restriction enzymes were obtained from New England Biolabs. Plasmid DNA purification from E. coli DH5α was performed using a GeneJET plasmid miniprep kit (Fermentas Life Science).
Site-directed mutagenesis.
Site-directed mutagenesis of OprF was performed by a modified QuikChange mutagenesis protocol (Stratagene) with two synthetic oligonucleotide primers containing the desired mutation. PCR was performed using Pfu ultrahigh-fidelity DNA polymerase (Stratagene) according to the instructions of the manufacturer.
Subcellular distribution of the expressed OprF protein.
For examination of cellular distribution of OprF expressed from plasmid pKY-His6OprF in E. coli DH5α and HN705, 0.5 ml of overnight culture (in LB containing 1% glucose and 30 µg/ml chloramphenicol) at 30°C was diluted into 50 ml of the fresh LB broth containing 30 µg/ml chloramphenicol, and the culture was incubated at 30°C with aeration until the A600 reached 0.6. Then, 0.1 mM IPTG was added. After 0, 1, 2, and 3 h of induction, cells from 5 ml of culture were washed and resuspended in 0.5 ml of 20 mM HEPES-NaOH buffer, pH 7.5, and were disintegrated with a Gallenkamp Soniprep 150 sonicator. After the removal of unbroken cells by low-speed centrifugation, Sarkosyl (final concentration, 0.5%) was added to the supernatant, and the Sarkosyl-insoluble OM fraction was separated by centrifugation for 45 min at 60,000 rpm with a Beckman TLA100.2 ultracentrifuge from the Sarkosyl-soluble fraction containing IM, periplasmic, and cytosolic proteins. The Sarkosyl-insoluble fraction was washed with 1 ml of the same buffer and was resuspended in 0.2 ml of the same buffer.
Fractionation of OprF expressed in E. coli RI90 and RI89 was done similarly, except that plasmid pHSG-OprF (4) was used and that the cells were grown in the presence of 0 to 5 mM DTT and 30 µg/ml chloramphenicol. The harvested cells were washed in a buffer containing 1 mM phenylmethanesulfonyl fluoride (PMSF) and 10 mM iodoacetamide and were suspended in 0.5 ml of the same buffer containing DNase (50 µg/ml). After sonication and low-speed centrifugation, the supernatant (the soluble fraction containing cytoplasmic and periplasmic proteins) was separated from the crude envelope fraction by centrifugation at 60,000 rpm for 60 min in the TLA100.2 ultracentrifuge. The pellet was resolved by specifically solubilizing the inner membrane proteins with 400 µl of 0.5% Sarkosyl in 20 mM HEPES buffer, pH 7.5. The insoluble OM fraction was pelleted by centrifugation under the same conditions, washed with 1 ml of the same buffer, and resuspended into 200 µl of the same buffer. The OM was dissolved in a lysis buffer without mercaptoethanol and analyzed by SDS-PAGE in the absence of reducing agents.
His-tagged OprF protein for examination of pore-forming activity.
Derivatives of pKY-OprF were usually transformed into E. coli HN705, which is deficient in both porin OmpF and porin OmpC. A 10-ml portion of overnight culture at 30°C (in LB containing 1% glucose and 30 µg/ml chloramphenicol) was diluted into 1 liter of fresh LB broth containing 30 µg/ml chloramphenicol, and the suspension was incubated at 30°C with aeration overnight without IPTG. The crude envelope fraction was prepared with a French pressure cell disruption and was then extracted with 0.5% Sarkosyl in 20 mM HEPES-NaOH buffer, pH 7.5, 1 mM PMSF to remove inner membrane proteins. OM proteins were solubilized with a solution containing 1.5% dodecyl-β-d-maltoside, 0.25 mg/ml lysozyme, 1 mM PMSF, and 20 mM HEPES buffer, pH 7.5. His-tagged OprF was then adsorbed to a Ni-NTA Superflow column (Qiagen). After two washings with 5 ml of the binding buffer (10 mM imidazole, 0.15 M NaCl, 0.05% dodecylmaltoside, 10% glycerol in 20 mM HEPES buffer, pH 7.5) and two washings with 5 ml of the binding buffer containing 20 mM imidazole, OprF protein was eluted with 5 ml of the binding buffer containing 250 mM imidazole. The solution was concentrated by using centrifugal filtration with Vivaspin20 (molecular weight cutoff [MWCO], 10,000; Vivascience, Inc.). In order to remove imidazole, a 10-fold excess of binding buffer was added to the concentrated protein solution, and the solution was concentrated again by a similar centrifugal filtration step.
Determination of pore-forming activity.
Pore-forming activity was routinely assayed by determining the osmotic swelling rates of proteoliposomes containing different amounts of OprF (23). As a measure of pore-forming activity, rates of swelling in iso-osmotic l-arabinose were used, calculated in units of mOD/min/µg protein. The pore size was inferred from the dependence of the swelling rates on the sizes of the permeating solutes, as described earlier (23).
Labeling of intact cells with maleimide-PEO2-biotin.
E. coli DH5α expressing the A312C mutant OprF protein was grown overnight in LB broth containing 30 µg/ml chloramphenicol at 30°C. A 0.5-ml volume of this culture was diluted into 50 ml of the same but fresh medium, and the culture was shaken at 30°C up to the mid-exponential phase (A600 = 1.0). Ten-milliliter portions were harvested, and the cells were washed twice with M63 medium by centrifugation at room temperature. Cells were resuspended in 1 ml of M63, and optical density (OD) was determined after 10-fold dilution. The amount of cells corresponding to 1 ml at an OD600 of 1.0 was added to 1 ml M63 medium containing maleimide-PEO2-biotin [(+)-biotinyl-3-maleimidepropionamidyl-3, 6-dioxaoctanediamine] (Pierce) and was incubated for 5 min at room temperature. The reaction was stopped with 1% (vol/vol) 2-mercaptoethanol. The labeled OprF protein was visualized by SDS-PAGE followed by streptavidin staining exactly as was specified earlier (4).
ACKNOWLEDGMENTS
We thank J. Beckwith, N. Ruiz, and T. J. Silhavy for strains and T. J. Silhavy for insightful discussions.
This research was supported by a grant from the U.S. Public Health Service (AI-009644).
Footnotes
Citation Sugawara, E., K. Nagano, and H. Nikaido. 2010. Factors affecting the folding of Pseudomonas aeruginosa OprF porin into the one-domain open conformer. mBio 1(4):e00228-10. doi:10.1128/mBio.00228-10.
REFERENCES
- 1. Hsueh P. R., Tseng S. P., Teng L., Ho S. W. 2005. Pan-drug-resistant Pseudomonas aeruginosa causing nosocomial infection at a university hospital in Taiwan. Clin. Microbiol. Infect. 11:670–673 [DOI] [PubMed] [Google Scholar]
- 2. Yoshimura F., Nikaido H. 1982. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J. Bacteriol. 152:636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nestorovich E. M., Sugawara E., Nikaido H., Bezrukov S. M. 2006. Pseudomonas aeruginosa porin OprF: properties of the channel. J. Biol. Chem. 281:16230–16237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugawara E., Nestorovich E. M., Bezrukov S. M., Nikaido H. 2006. Pseudomonas aeruginosa porin OprF exists in two different conformations. J. Biol. Chem. 281:16220–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nikaido H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell Dev. Biol. 12:215–223 [DOI] [PubMed] [Google Scholar]
- 6. Sugawara E., Nikaido H. 1994. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269:17981–17987 [PubMed] [Google Scholar]
- 7. Pautsch A., Schultz G. E. 1998. Structure of the outer membrane protein OmpA transmembrane domain. Nat. Struct. Biol. 5:1013–1017 [DOI] [PubMed] [Google Scholar]
- 8. De Mot R., Vanderleyden J. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333–334 [DOI] [PubMed] [Google Scholar]
- 9. Koebnik R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269–1270 [DOI] [PubMed] [Google Scholar]
- 10. Grizot S., Buchanan S. K. 2004. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 51:1027–1037 [DOI] [PubMed] [Google Scholar]
- 11. Rawling E. G., Brinkman F. S. L., Hancock R. E. W. 1998. Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 180:3556–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bos M. P., Tommassen J. 2004. Biogenesis of the gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7:610–616 [DOI] [PubMed] [Google Scholar]
- 13. Duguay A. R., Silhavy T. J. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694:121–134 [DOI] [PubMed] [Google Scholar]
- 14. Mogensen J. E., Otzen D. E. 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 57:326–346 [DOI] [PubMed] [Google Scholar]
- 15. Kadokura H., Katzen F., Beckwith J. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111–135 [DOI] [PubMed] [Google Scholar]
- 16. Eppens E. F., Nouwen N., Tommassen J. 1997. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO J. 16:4295–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruiz N., Chng S.-S., Hiniker A., Kahne D., Silhavy T. J. 2010. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl. Acad. Sci. U. S. A. 107:12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sklar J. G., Wu T., Gronenberg L. S., Malinverni J. C., Kahne D., Silhavy T. J. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245 [DOI] [PubMed] [Google Scholar]
- 20. Robert V., Volokhina E. B., Senf F., Bos M. P., van Gelder P., Tommassen J. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:1984–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma L., Jackson K. D., Landry R. M., Parsek M. R., Wozniak D. J. 2006. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188:8213–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Woodruff W. A., Hancock R. E. W. 1988. Construction and characterization of Pseudomonas aeruginosa outer membrane protein F-deficient mutants after in vitro and in vivo insertion mutagenesis of the cloned gene. J. Bacteriol. 170:2592–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nikaido H., Nikaido K., Harayama S. 1991. Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem. 266:770–779 [PubMed] [Google Scholar]
- 24. Jocelyn P. C. 1987. Spectrophotometric assay of thiols. Methods Enzymol. 143:44–57 [DOI] [PubMed] [Google Scholar]
- 25. Creighton T. E. 1997. Protein folding coupled to disulphide bond formation. Biol. Chem. 378:731–744 [DOI] [PubMed] [Google Scholar]
- 26. Sugawara E., Steiert M., Rouhani S., Nikaido H. 1996. Secondary structures of outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J. Bacteriol. 178:6067–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klose M., Schwarz H., MacIntyre S., Freudl R., Eschback M. L., Henning U. 1988. Internal deletions in the gene for an Escherichia coli outer membrane protein define an area possibly important for recognition of the outer membrane by this polypeptide. J. Biol. Chem. 263:13291–13296 [PubMed] [Google Scholar]
- 28. Struyve M., Moons M., Tommassen J. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of bacterial outer membrane protein. J. Mol. Biol. 218:141–148 [DOI] [PubMed] [Google Scholar]
- 29. Rawling E. G., Martin N. L., Hancock R. E. W. 1995. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect. Immun. 63:38–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lazar S. W., Kolter R. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178:1770–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruiz N., Kahne D., Silhavy T. J. 2006. Advances in understanding bacterial outer membrane biogenesis. Nat. Rev. Microbiol. 4:57–66 [DOI] [PubMed] [Google Scholar]
- 32. Ureta A. R., Endres R. G., Wingreen N. S., Silhavy T. J. 2007. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J. Bacteriol. 189:446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Korndorfer I. P., Dommel M. K., Skerra A. 2004. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite different architecture. Nat. Struct. Mol. Biol. 11:1015–1020 [DOI] [PubMed] [Google Scholar]
- 34. Qu J., Behrens-Kneip S., Holst O., Kleinschmidt J. H. 2009. Binding regions of outer membrane protein A in complexes with the periplasmic chaperone Skp. A site-directed fluorescence study. Biochemistry 48:4926–4936 [DOI] [PubMed] [Google Scholar]
- 35. Walton T. A., Sousa M. C. 2004. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol. Cell 15:367–374 [DOI] [PubMed] [Google Scholar]
- 36. Walton T. A., Sandoval C. M., Fowler C. A., Pardi A., Sousa M. C. 2009. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc. Natl. Acad. Sci. U. S. A. 106:1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zakharian E., Reusch R. N. 2005. Kinetics of folding of Escherichia coli OmpA from narrow to large pore conformation in a planar bilayer. Biochemistry 44:6701–6707 [DOI] [PubMed] [Google Scholar]
- 38. Arora A., Rinehart D., Szabo G., Tamm L. K. 2000. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 275:1594–1600 [DOI] [PubMed] [Google Scholar]
- 39. Sugawara E., Nikaido H. 1992. Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267:2507–2511 [PubMed] [Google Scholar]
- 40. Rietsch A., Belin D., Martin N., Beckwith J. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:13048–13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rizzitello A. E., Harper J. R., Silhavy T. J. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruiz N., Falcone B., Kahne D., Silhavy T. J. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317 [DOI] [PubMed] [Google Scholar]