Abstract
We have developed a new approach to screen bacterial artificial chromosome (BAC) libraries by recombination selection. To test this method, we constructed an orangutan BAC library using an E. coli strain (DY380) with temperature inducible homologous recombination (HR) capability. We amplified one library segment, induced HR at 42°C to make it recombination proficient, and prepared electrocompetent cells for transformation with a kanamycin cassette to target sequences in the orangutan genome through terminal recombineering homologies. Kanamycin-resistant colonies were tested for the presence of BACs containing the targeted genes by the use of a PCR-assay to confirm the presence of the kanamycin insertion. The results indicate that this is an effective approach for screening clones. The advantage of recombination screening is that it avoids the high costs associated with the preparation, screening, and archival storage of arrayed BAC libraries. In addition, the screening can be conceivably combined with genetic engineering to create knockout and reporter constructs for functional studies.
1. Introduction
Bacterial Artificial Chromosome (BAC) libraries have been used extensively for constructing physical maps of the human genome as well as for whole-genome sequencing projects. These applications require the BAC clones to be arrayed into the wells of microtiter plates for archiving single unique clones. The location in the array of each BAC clone is used as clone name to connect the library with genome sequence data. Such arrayed libraries have been frequently used as reference libraries for fingerprinting, BAC-end sequencing, large-scale marker screening, cytogenetic mapping, and BAC shotgun sequencing. However, the high costs of arrayed BAC libraries cannot be easily justified for applications that have a short lifespan or focus on small genomic regions or single gene. For instance, BACs are useful to recover a specific haplotype for a small genomic region in order to identify a disease-causing mutation [1]. BAC libraries made for such applications do not necessarily require a format where each clone is identifiable by a name.
Nonarrayed BAC libraries can be conceivably screened by radioactive probe colony hybridization after plating it on Petri dishes and screening a filter replicate. This process is laborious requiring multiple cycles of colony streaking and screening to generate pure colonies. Alternately, libraries can be amplified and aliquots of the library screened through recombination selection, although feasibility of this approach has not yet been demonstrated for BAC libraries. The latter approach is based on phenotypic modification of targeted clones through homologous recombination (HR) and the subsequent selective growth of these clones. This approach has mostly been followed using bacteriophage lambda libraries amplified on a host containing a “probe” plasmid and then tested in a restrictive E. coli strain for selective growth of recombinant lambda clones with an integrated plasmid [2–5].
Newly developed technologies for site-specific engineering of BAC clones have been collectively labeled “recombineering” [6] because they rely on inducible HR to engineer modified BAC clones. Several systems make use of the RecA-RecBCD pathway [7], the RecE,T proteins [8, 9], or the bacteriophage lambda HR genes: exo, bet, and gam [9–12]. Lee et al. [13] described a new E. coli strain with inducible recombination derived from a defective lambda prophage harboring the lambda exo, bet, and gam genes. This strain (DY380) expresses the exo, bet, and gam genes under control of a temperature-sensitive CI-857 repressor. For our studies we chose to use the DY380 strain with the lambda prophage because the recombination proteins are tightly repressed at low temperature, hence likely reducing the toxicity and presumably ensuring optimal stability of BACs. Furthermore, the high recombination efficiency is essential to increase the sensitivity of recombination selection from complex BAC libraries. Finally, transformation with high-molecular weight DNA requires a specific genetic background and, so far, only DH10B and its derivatives have been found to be compatible with artificial chromosome generation.
Zhang et al. have shown that small-insert high-copy number plasmid libraries can be screened by HR [14]. In particular, their plasmid library was transformed with a DNA construct containing an antibiotic-resistance marker and terminal sequences homologous to the target region. We postulated that a somewhat similar approach could also work for screening BACs, which are high-molecular weight low copy number plasmids, provided that representative libraries can be prepared and the recombination process is adequately efficient.
In this study we tested the feasibility of screening by HR complex BAC library constructed using recombination proficient E. coli strain.
2. Methods
2.1. Media and Plates
LB and SOC medium containing 20 μg/mL chloramphenicol (Cm), and 25 μg/mL kanamycin (Kan) were used to grow BAC clones.
2.2. Bacterial Strain and Cultures
All bacterial cultures in this study were maintained (unless specified) at 32°C because of the temperature-inducible prophage in DY380 bacterial strain [15]. This strain was kindly provided by Dr. N.G. Copeland and contains a defective prophage to supply the expression of Exo, Bet, and Gam necessary to execute HR.
2.3. Analysis of BAC DNA
BAC DNA minipreps were purified using the Autogen-940 robot. For fingerprinting, 0.5 μg of DNA was digested with 1-2 units of EcoRI. EcoRI digests were analyzed on a 1% agarose gel in 0.5xTBE at room temperature, 4 V/cm for 8 h. The gels were stained with ethidium bromide for visualization.
2.4. Plasmids and DNA Templates
pTARBAC2.1 [16] digested with EcoRI and dephosphorylated with CIAP was used as a cloning vector to construct the library and pCYPAC1 [17] as a template for kanamycin PCR amplification. We used a human BAC (RP11-622D14) containing the complete human β-globin locus. This BAC spans position chr11:5218918-5406749 hg19.
2.5. Library Construction
The preparation of recombination proficient orangutan genomic library followed was performed following the cloning approach developed in our laboratory [18]. DNA was isolated from a blood sample from a male Sumatran orangutan (“Segundo”) by embedding DNA in agarose. After pre-electrophoresis, high-molecular weight genomic DNA was partially digested with a combination of EcoRI restriction enzyme and EcoRI Methylase and the resultant fragments were subjected to double-size fractionation by pulsed field gel electrophoresis. DNA fragments from the appropriate size fraction were cloned into the EcoRI digested and dephosphorylated pTARBAC2.1 vector. The ligation products were then transformed into DY380 electrocompetent cells; glycerol stocks of nonarrayed library were made before library amplification on agar plates. To perform control experiments 16,000 colonies (0.7x redundant library) were picked and arrayed into 42 × 384-well microtiter dishes. Same 42 × 384-well microtiter dishes (16,000 clones) were gridded onto one 22 × 22 cm nylon high-density filter stamped in duplicate for screening by probe hybridization. To create a supply of nonarrayed library and ensure that each clone is equally represented before amplification, 10 μL aliquots were collected from 16,000 wells into one flask and snap frozen in a dry ice ethanol bath. After the titer was determined (3 × 108 cfu/mL) a 1.6 × 104 cfu aliquot was taken for genomic library amplification experiment.
2.6. Genomic Library Amplification
To amplify the genomic library 16,000 clones were plated at a density of ≈3,000 cfu on 5 Petri dishes (15 mm diameter) containing Cm-agar. Plates were incubated at 32°C for 18–24 hours to assess amplification rates and representation of the primary library. To harvest the small (0,2–0,5 mm) colony growth, 7 mL of LB media was poured on the agar's surface and incubated at room temperature for 10 min on a rocking platform and 30 mL of LB containing lifted colonies were collected. Glycerol was added up to 10%, 30 × 1 mL aliquots were snap frozen and stored at −80°C with titer of 3 × 108 cfu/mL or 9 × 109 cfu in total.
2.7. Preparation and Transformation of Electrocompetent Cells
For 10 electroporations approximately 0.6 mL (≈3 × 108 cells) of an overnight culture of DY380 cells (or 3 × 108 cells from the amplified library) was inoculated into 60 mL of SOB and were grown in 1 L baffled conical flasks shaking at 320 rpm until an OD600 = 0.6 was reached. Cells were collected by centrifugation at 5500 g for 5 min, washed with ice-cold water 3 times and finally resuspended in 200 μl of ice-cold sterile water. Electroporations were performed with a mix of 2 μl of DNA and 20 μl of cells (1.5 × 108 cells/μl or 3 × 109 cells per electroporation) using a Gibco BRL Cell-Porator set at 315 Volts, 350 μF, 2 KΩ with resulting pulse more than 1.88 KV. After electroporation cells were resuspended in 500 μl SOC and incubated at 32°C for 1.5 hour with shaking and spread on selective agar media. The achieved efficiency of transformation for large (180-Kb) BAC molecules was 107 cfu/μg of DNA.
2.8. Induction of λ-Recombination
Electrocompetent cells for HR experiments were made from temperature-induced cultures. Induction of λ-recombination of the culture for a total of 15 min at 42°C was performed following a protocol reported by Yu et al. [19]. About 1 mL (3 × 108 cells) of glycerol stock from amplified library was inoculated into 60 mL of SOB media with chloramphenicol and grown at 32°C with shaking in a baffled flask for approximately 4-5 hours until an O.D.600 = 0.6 was reached. The flask containing the culture was then placed in a water bath shaker at 42°C for 15 min (300 revs/min). Immediately after, the flask was swirled in ice-water slurry for 10 minutes to cool down. Cells were collected and washed following procedure for preparation and transformation of electrocompetent cells hereinabove.
2.9. Preparation of Linear kanR-Cassette for HR
The 1.4-Kb kanamycin cassette was PCR amplified from NotI/EcoRI linearized pCYPAC2N vector using PAGE-purified chimeric oligos. Their structure is shown in Table 3 in Supplementary Material available online at doi:10.1155/2011/560124. Each chimeric primer is 69-70 bases long and contains a portion (shown in upper case characters, 49-50 nt long) homologous to the sequence of the targeted region and a portion homologous to the kanamycin gene (shown in lower case characters, 20 nt long). The PCR products were DpnI digested and purified by washing with water using Centricon spin columns (Millipore). PCR colony screening was performed with 19–21 nt-long screening primers (Table 3). These primers yield PCR products in the range of 300–700 bp from unmodified gene, while insertion of the kanR cassette should yield a PCR product over 1.7-Kb (Figure 6). The same flanking primers were used for verification of library representation after amplification and to generate the probes for library screening by hybridization.
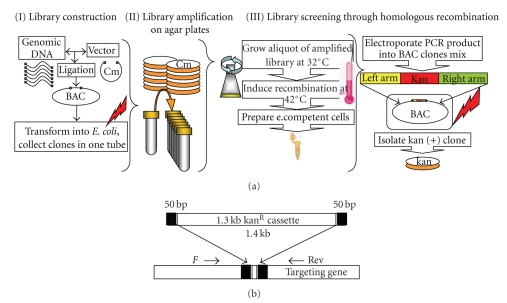
Figure 6.
(a) Strategy for library screening through homologous recombination. There are 3 steps: (I) Library construction—consists of DNA ligation and E. coli transformation; (II) Library amplification—library is plated on Cm agar plates at high density, clones harvested by washing with LB. Glycerol stocks made of mixed clones; (III) Library screening via HR. An aliquot of the amplified library is grown at 32°C and induced at 42°C. Electrocompetent cells are prepared and transformed with HR cassette (Figure 6(b)). Recombinant clones were identified on kanamycin plates. (b) Recombination cassette. HR cassette consists of a 1.3 Kb kanamycin resistance gene flanked by 50 bp homology arms. kanR cassette is inserted into targeting gene using 50 bp homology arms “H” corresponding to the targeted region. “F” and “Rev” are the flanking screening primers used for screening recombinant clones.
2.10. Library Screening by HR
An aliquot from the amplified library (13.5 × 108 cells) was used to prepare 900 μl of electrocompetent cells induced for recombination as described above. Nine kanR targeting constructs were electroporated 5 times each with 0.1 μg of purified PCR product and incubated in SOB at 32°C for 1.5 hours before plating on kan LB plates.
2.11. Hybridization
For details of hybridization of high density filters see http://bacpac.chori.org/highdensity.htm.
3. Results
3.1. High Efficiency and Selectivity of Homologous Recombination
In order to asses and optimize the efficiency of homologous recombination, we used RP11-622D14 BAC clone (insert size 187 Kb) which contains the complete human β globin locus. The BAC was transferred into DY380 cells by electroporation. A kanR recombination cassette containing 50 bp terminal sequences homologous to nearby locations in the β-globin gene was amplified by PCR as previously described in [20]. The BAC clone was electrotransformed using various amounts of PCR product over a 50-fold range. The number of kanamycin-resistant colonies increased proportionally with the amount of PCR product: 250–500 clones for 10 ng; 5,000–10,000 clones for 100 ng; 20,000–25,000 clones for 500 ng of linear DNA. The 100 ng linear DNA was selected for further transformation experiments as an arbitrary compromise between recombination efficiency and total number of recombinant colonies.
To assess the selectivity of the recombination process, mixtures of the “globin” BAC with unrelated BACs from the “RPCI-11” library (http://bacpac.chori.org/hmale11.htm) were created and propagated in DY380 cells at various levels of complexity. Following the induction of HR at 42°C, electrocompetent cells for the pooled clones were prepared. The results of the transformation of these pools with the recombination cassette are shown in Table 1. The most complex mixture of BACs still resulting in 1-2 kanamycin positive clones had 10,000 unrelated BAC clones with an average 180 Kb insert size. This BAC mixture (1/104) corresponds to a complexity of 1.8 Giga base pairs(Gbp), representing about 60% of the haploid human genome. This 104 efficiency of homologous recombination makes our method compatible with HR screening of full size mammalian genome BAC libraries.
Table 1.
Selectivity of recombination from BAC pools in DY380 cells using 100 ng of linear recombination cassette.
| Complexity of BAC pool | No. of recombinant clones |
|---|---|
| 1 | 4,800–10,000 |
| 1/10 | 450–1,000 |
| 1/100 | 55–100 |
| 1/1,000 | 8–12 |
| 1/10,000 | 1-2 |
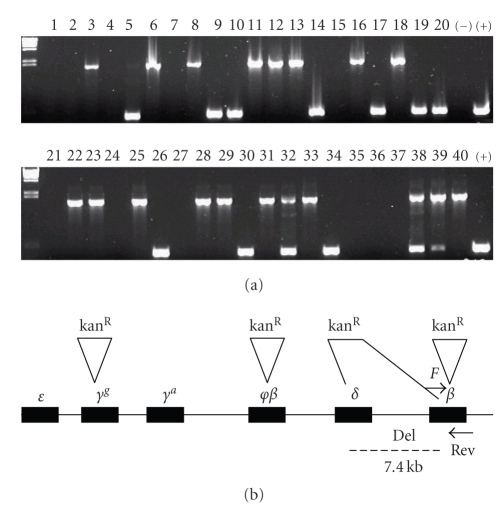
We took 40 random kanamycin positive clones obtained in the recombination selection experiment as a representative sample from different complexities listed in Table 1. All clones tested positive by Southern blot (data not shown) for “globin” gene(s) confirming high selectivity of homologous recombination. Subsequent PCR analysis of these clones revealed various integration sites. Figure 1(a) shows results from PCR screening of various kanamycin resistant clones using primers flanking expected β-globin integration site. Recombinant clones displayed complex recombination events as a result of a partial homology of recombination cassette with duplicated sequences within the globin locus (Figure 1(b)); All clones recovered after recombination selection originated from RP11-622D14 BAC indicating high selectivity for HR.
Figure 1.
PCR colony screening of kanR recombinant clones. (a) The upper band, 1.7 Kb in length (lanes 3, 6, 8, etc.), corresponds to the larger PCR product after insertion of recombination cassette into β-globin gene. 11 clones reveal lower wild-type band, 440 bp in length (lanes 5, 9, 10, etc.) as a result of recombination into other duplicated regions of the globin locus (γg and φβ). No PCR product (11 clones) is due to the δ-β deletion of the F primer binding site. Lanes 32,38 with double bands represent the colonies with heterogeneous BAC population within single colony. (b) Diagram of globin locus illustrating insertion of kanR cassette at four different globin genes.
3.2. Construction of Orangutan Test BAC Library in E. coli DY380: Genomic Representation and Clone Stability
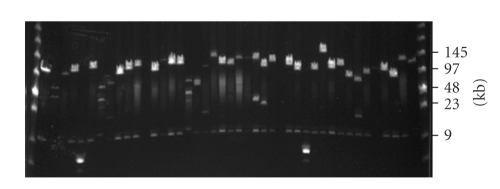
To test the feasibility of HR screening of BAC libraries using multiple probes, we prepared a BAC library from genomic DNA of Sumatran orangutan (Pongo abelii) in the conditional-recombination E. coli strain DY380. We then transferred 16,000 clones with an average size of 130 Kb (2 Gb total or equivalent to 0.7-fold genomic redundancy) (Figure 2) into 384-well plates for size distribution analysis and preparation of high-density filters.
Figure 2.
Insert size determination of orangutan BAC library clones. DNA from 92 random BAC clones was isolated, digested with NotI and separated using pulse field gel electrophoresis (PFGE) on two gels. Low Range PFG marker (NEB) was loaded in the flanking wells. A subset of the analyzed BACs from a single gel is shown. Forty-one recombinant clones and 2 noninsert clones can be seen. The band comigrating in all clones with the 9-Kb marker band is the BAC vector.
To analyze genomic marker representation in the library, the arrayed clones were gridded onto a single nylon membrane. We screened the library using the traditional hybridization method with radioactive probes. Twenty-eight randomly chosen “overgo” genomic probes [21] were designed in correspondence of genomic regions highly conserved between human and mouse. Such probes were radioactively labeled and used to screen the high-density filters made from the orangutan test library. The hybridization results reveal 16 positives each from a different probe, indicating that the library is ~0.6-fold redundant. The consistency of hybridization results with the 0.7-fold estimate based on clone numbers and insert sizes argues for the absence of a major cloning bias in the DY380 strain.
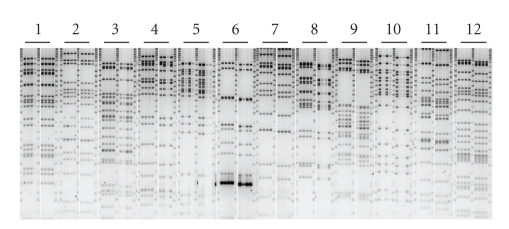
Because the DY380 is a conditional temperature inducible recombination E. coli strain, it is conceivable that some leaky expression of the lambda recombination genes occurs at 32°C and that this may destabilize the BAC clones. To test for genetic instability, 8 isolates for 96 BAC clones have been fingerprinted with EcoRI. Fingerprinting data was used for an automated contig assembly in FPC to bin the clones [22]. We anticipated that for each clone, the 8 isolates fingerprinted would bind together into a contig, so we would expect 96 different contigs (1 per clone) with 8 clones per contig. For 90/96 contigs that was true, but there were a number of exceptions: for 3 clones all 8 isolates contained only a single band, 2 clones were identical, 2 clones failed to grow and in one isolate of clone#2 single band was missing (Figure 3). High-resolution fingerprinting showed that there were no significant differences between the clones in the EcoRI fragments. Therefore, we saw no evidence of major rearrangements of BAC clones during their propagation in DY380 strain in the liquid culture demonstrating genetic stability of the BACs in this strain.
Figure 3.
Fingerprinting of BAC clones 1–12. For each of the 96 BAC clones, 8 isolates were fingerprinted with EcoRI. Due to the gel loading procedure, only 4 clones could run next to each other. Multiple gel photos were assembled into one panel for ease in viewing.
3.3. Library Amplification on Solid Agar
The goal of library amplification is to have enough starting bacteria for preparing competent cells for transformations. For the screening with multiple HR probes (30–300), the nonarrayed library has to be amplified about 104–105-fold without loss of genomic representation. We amplified the library on solid agar making sure that colonies have similar growth conditions, thus maintaining the representation of the nonarrayed BAC library. By plating the library on solid agar (see Section 2 for the details), we achieved 5.6 × 105 amplification rate and collected a total of 9 × 109 cfu. It is noteworthy that with this amount of bacteria one could prepare up to 6 mL of competent cells—enough for ~300 electroporations with 300 HR probes.
The genomic representation of the amplified library was evaluated by PCR analysis of various titers of amplified and nonamplified library for 10 probes. Table 2 illustrates that there was no differential growth during library amplification for any of 10 probes analyzed. Hence, we achieved 5.6 × 105-fold of nonarrayed BAC library amplification without a loss of genomic representation.
Table 2.
PCR analysis of nonarrayed library amplification.
| Gene | Position in human genome | PCR results | |||||
|---|---|---|---|---|---|---|---|
| Before amplification | After amplification | ||||||
| No. of cells per PCR | No. of cells per PCR | ||||||
| 2 × 103 | 2 × 104 | 104 | 2 × 103 | 2 × 104 | 104 | ||
| EMS1 | chr11:69922297–69960337 | − | + | + | + | + | + |
| MGC13034 | chr5:72504796–72506724 | − | + | + | + | + | + |
| SKB1 | chr14:22459578–22468422 | − | + | + | − | + | + |
| GMPPA | chr2:220190181–220197215 | − | + | + | − | + | + |
| FLJ20539 | chr11:60451115–60458665 | − | + | + | − | + | + |
| NFAT5 | chr16:68157387–68288860 | − | + | + | − | + | + |
| GRIA3 | chrX:122043692–122348328 | − | + | + | − | + | + |
| DKFZP564D166 | chr17:58852873–58855920 | − | + | + | − | + | + |
| IDAX | chr4:105750948–105770071 | − | + | + | − | + | + |
| HBB | chr11:5,246,696–5,248,301 | − | + | + | − | + | + |
3.4. Library Screening through HR
To screen the library by homologous integration of a kanR recombination cassette into the genes of interest, nine target cassettes were created by PCR amplification of the kanR gene with 70 nt chimeric oligos. The nine target sequences were a subset of the sixteen conserved regions already confirmed to be present in the library by probe hybridization. Previous reports [23] have demonstrated higher efficiencies of recombinational deletions versus insertions. To make use of this effect while minimizing the size of the deletions, both ends of targeting cassette were separated in the genome by a small distance of 50–200 bp. Hence, homologous integration of the cassette was anticipated to result in a small deletion of genomic target DNA with a concomitant insertion of the 1.3-Kb kanR selection marker.
A stock of amplified nonarrayed library was produced by pooling together 10 μl aliquots from each well of arrayed library (16,000 clones—160 mL, ~5 × 108 cfu/mL) and one of the aliquots was used to produce electrocompetent cells.
For each of the nine HR probes five 20 μl aliquots of electrocompetent cells were transformed with 100 ng of targeting cassettes (45 electroporations total) and plated on selective media. The efficiency of HR screening varied between the probes. We obtained between 1 to 10 kanamycin positive colonies for 7 cassettes. The kanR colonies were confirmed to contain the correct insertion of kanR gene by PCR using screening primers flanking the site of integration (Figures 4(a) and 4(b)). Because all recombineering oligos were of the same length and similar GC content, differences in HR efficiency can be attributed to specific characteristics of the targeting site such as DNA secondary structure or replication asymmetry.
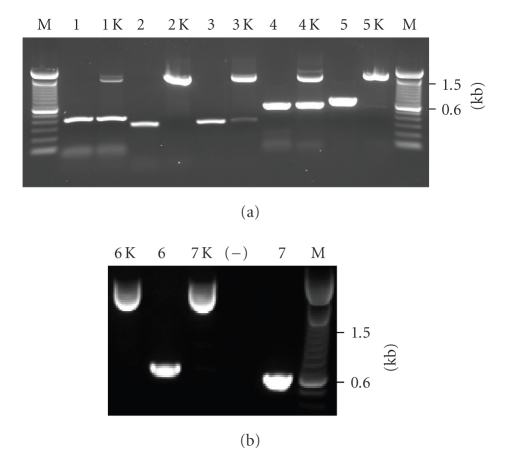
Figure 4.
PCR analysis of 7 BAC clones before and after targeting. Kanamycin-resistant clones (lanes 1–7) were found by targeted recombination screening of the library. The corresponding “original” clones (“1–7 k” lanes) were identified from the arrayed library by radioactive probe screening and served as a control. All clones were screened with gene-specific primers flanking the putative integration site. Lanes 1 through 7 show candidate BACs identified for EMS1, MGC, SKB1, GMPPA, FLJ20539, NFAT5, and GRIA3, respectively. As anticipated, in all seven cases, the size of PCR product was larger than 1.3-Kb compared to the alleged original clone.
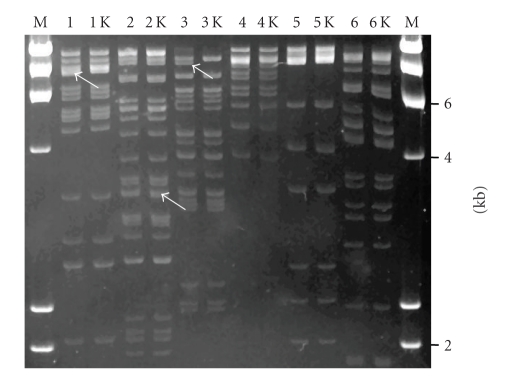
The integrity of targeted BACs after recombination was verified by EcoRI fingerprinting (Figure 5). The fingerprints of the recombinant kanR BAC clones were compared with the original BAC clones after EcoRI digestion. The EcoRI bands containing the 1.3-Kb kanR insert have not been identified for every clone although subtle upward band shifts can be seen in some of the target products (i.e., lanes 1, 2 K, and 3 in Figure 5). Thus, there were no large rearrangements detected confirming the integrity of BAC clones after screening via HR.
Figure 5.
Fingerprinting of recombinant BAC clones. High-resolution fingerpint comparison of six targeted BAC clones with the original clones. EMS1, MGC, GRIA3, GMPPA, SKB1, and FJ20539 retrieved from the orangutan genomic library using screening by HR. Lanes 1–6 contain the original BAC clones and lanes 1 K–6 K show the corresponding targeted BACs.
4. Discussion
Even with advances in new generation sequencing large insert libraries remain indispensable for genome assembly and characterization of structural variations [24]. Furthermore, BACs are a useful tool for cloning and stably maintaining large DNA fragments, including genes and their intact regulatory elements. This is a great advantage, given the growing need for functional evaluation of various genes. Some of the restrictions on manipulation of large insert BACs using conventional cloning techniques have been overcome by HR in E. coli, or “recombineering”. While recombineering simplifies BAC modification and generation of gene targeting constructs, BAC library construction, screening, and maintenance are time consuming and require special facilities. To simplify the process of BAC library operation we took advantage of a method based on homologous recombination.
Although, the HR technique has previously been used for small-insert library screening [14], we developed its application further to screening of large-size genomic BAC libraries. First, we demonstrated the genetic stability of BAC clones during their propagation in DY380 strain. The genomic library created in this strain benefits from the tight regulation of recombination function by a combination of PL promoter with temperature sensitive CI-857 repressor. The library is stably propagated without rearrangements and leaking expression of recombinant proteins. Second, we achieved high efficiency and selectivity of HR compatible with HR screening of BAC libraries. We improved our HR techniques by selecting the clone of interest out of the mixed BACs, created a nonarrayed test library, and obtained seven out of nine positive clones from the library of 16,000 clones through HR screening. Third, the high rate (5.6 × 105) BAC library amplification on solid agar was accomplished without any loss of genomic representation thereby providing enough starting material for screening the library with multiple PCR probes through HR.
The main advantages of isolation of specific clones through HR compared to traditional hybridization screening are its speed and low cost. The process of library construction consists of two main steps: creation of nonarrayed BAC library through a ligation-transformation protocol [18] and then picking, arraying, replicating, and gridding of the library to produce the filters for screening. About 75%–80% of the total cost of the library is associated with the second step. By screening the library through HR we addressed this problem and omitted the second step. After creating the clones, the nonarrayed library is amplified and stored in aliquots ready for screening. A method of library amplification on the agar does not require arrayed clones. Library can be amplified using clones directly from ligation-transformation experiment (Figure 6). To make it more practical and minimize the number of plates used, large (22 × 22 cm) dishes can be utilized. We routinely use such plates to amplify full size genomic libraries without a loss of representation (not published data).
Another advantage of the nonarrayed BAC genomic library method is that BAC clones recovered by screening are ready for further modifications without the need of changing the host or incorporation of additional shuttle vectors to supply recombination function. In addition, HR library screening can be combined with precise targeted BAC modifications to produce reporter constructs for functional studies and creation of animal models.
The 50 bp targeting sequences are relatively easy to choose. Provided that HR oligos are free of repeats, they can be placed within single intron or into different introns flanking critical exon(s) creating insertion or deletion, respectively. The targeting cassette is also simple to modify making it suitable for any specific experiment. For instance, the cassette may include short site-specific recombination sequences such as LoxP, FRT, and Gateway and counter selection markers (pheS, rpsL, tet, sacB) allowing removal of undesired kanR cassette or replacement with mammalian-specific markers and reporters. Cre expressing plasmid transformed into BAC-containing cells will efficiently delete the recombineering cassette even without counter selection.
The low cost of nonarrayed BAC library has the potential of promoting a widespread use of this approach in genomic applications such as identification of genomic regions for comparative analysis, creation of BAC libraries from large number of animals, patients or tumors, cloning of specific haplotypes associated with predisposition to various disorders, and high-throughput engineering of knockout mice using BACs [25]. The drawback of this approach is that it requires large volumes of DY380 electrocompetent cells for library construction. Even though we have seen some large BACs successfully propagated in DY380, home grown cells could never achieve transformation efficiency of commercial cells for BACs over 130 Kb. This limitation could be easily overcome by making the DY380 E. coli cell line commercially available. We pursue alternative approach to construct a BAC vector with inducible λ recombination function on the backbone. Such vector could be transformed into commonly available DH10B E. coli strain.
Supplementary Material
The candidate sequences were chosen from conserved regions in orangutan genome free of repetitive elements. The 50mer portions shown in upper case characters homologous to the sequence of the targeted regions were selected using modified Primer3 software. Using source provided at: http://fokker.wi.mit.edu/primer3/. we increased built-in maximum oligo size up to 50nt.
Acknowledgments
The authors thank N.G. Copeland for DY380 E. coli cell line, L. W. Enquist, E. Eichler, and S. Taylor of Louisville Kentucky Zoo for kindly providing animal DNA sample.
References
- 1.De Gobbi M, Viprakasit V, Hughes JR, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312(5777):1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 2.Lutz CT, Hollifield WC, Seed B, Davie JM, Huang HV. Syrinx 2A: an improved lambda phage vector designed for screening DNA libraries by recombination in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(13):4379–4383. doi: 10.1073/pnas.84.13.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang HV, Little PF, Seed B. Improved suppressor tRNA cloning vectors and plasmid-phage recombination. Biotechnology. 1988;10:269–283. doi: 10.1016/b978-0-409-90042-2.50020-9. [DOI] [PubMed] [Google Scholar]
- 4.Kurnit DM, Seed B. Improved genetic selection for screening bacteriophage libraries by homologous recombination in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(8):3166–3169. doi: 10.1073/pnas.87.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart GD, Hauser MA, Kang H, et al. Plasmids for recombination-based screening. Gene. 1991;106(1):97–101. doi: 10.1016/0378-1119(91)90571-r. [DOI] [PubMed] [Google Scholar]
- 6.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nature Reviews Genetics. 2001;2(10):769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 7.Cox DH, Carvajal JJ, Rigby PWJ. Enhanced efficiency of pSV1-RecA-based BAC recombineering. BioTechniques. 2002;33(6):1206–1208. doi: 10.2144/02336bm04. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Buchholz F, Muyrers JPP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genetics. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 9.Muyrers JPP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Research. 1999;27(6):1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayanan K, Williamson R, Zhang Y, Stewart AF, Ioannou PA. Efficient and precise engineering of a 200 kb β-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Therapy. 1999;6(3):442–447. doi: 10.1038/sj.gt.3300901. [DOI] [PubMed] [Google Scholar]
- 11.Nefedov M, Williamson R, Ioannou PA. Insertion of disease-causing mutations in BACs by homologous recombination in Escherichia coli. Nucleic Acids Research. 2000;28(17, article E79) doi: 10.1093/nar/28.17.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Muyrers JPP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nature Biotechnology. 2000;18(12):1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 13.Lee E-C, Yu D, Martinez De Velasco J, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Li MZ, Elledge SJ. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nature Genetics. 2002;30(1):31–39. doi: 10.1038/ng797. [DOI] [PubMed] [Google Scholar]
- 15.Lee E-C, Yu D, Martinez De Velasco J, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 16.Zeng C, Kouprina N, Zhu B, et al. Large-insert BAC/YAC libraries for selective re-isolation of genomic regions by homologous recombination in yeast. Genomics. 2001;77(1-2):27–34. doi: 10.1006/geno.2001.6616. [DOI] [PubMed] [Google Scholar]
- 17.Ioannou PA, Amemiya CT, Garnes J, et al. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nature Genetics. 1994;6(1):84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 18.Osoegawa K, Woon PY, Zhao B, et al. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52(1):1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- 19.Yu D, Ellis HM, Lee E-C, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamsai D, Orford M, Nefedov M, Fucharoen S, Williamson R, Ioannou PA. Targeted modification of a human β-globin locus BAC clone using GET Recombination and an I-SceI counterselection cassette. Genomics. 2003;82(1):68–77. doi: 10.1016/s0888-7543(03)00100-9. [DOI] [PubMed] [Google Scholar]
- 21.McPherson JD. Genome Analysis: A Laboratory Manual. Vol. 4. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1999. Hybridization probes generated from STS sequence; pp. 207–213. [Google Scholar]
- 22.Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Research. 2000;10(11):1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orford M, Nefedov M, Vadolas J, Zaibak F, Williamson R, Ioannou PA. Engineering EGFP reporter constructs into a 200 kb human beta-globin BAC clone using GET Recombination. Nucleic Acids Research. 2000;28(18, article E84) doi: 10.1093/nar/28.18.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidd JM, Sampas N, Antonacci F, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nature Methods. 2010;7(5):365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valenzuela DM, Murphy AJ, Frendewey D, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nature Biotechnology. 2003;21(6):652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The candidate sequences were chosen from conserved regions in orangutan genome free of repetitive elements. The 50mer portions shown in upper case characters homologous to the sequence of the targeted regions were selected using modified Primer3 software. Using source provided at: http://fokker.wi.mit.edu/primer3/. we increased built-in maximum oligo size up to 50nt.